Current Insights Regarding the Role of Farm Animals in the Spread of Antimicrobial Resistance from a One Health Perspective
Abstract
:Simple Summary
Abstract
1. Introduction
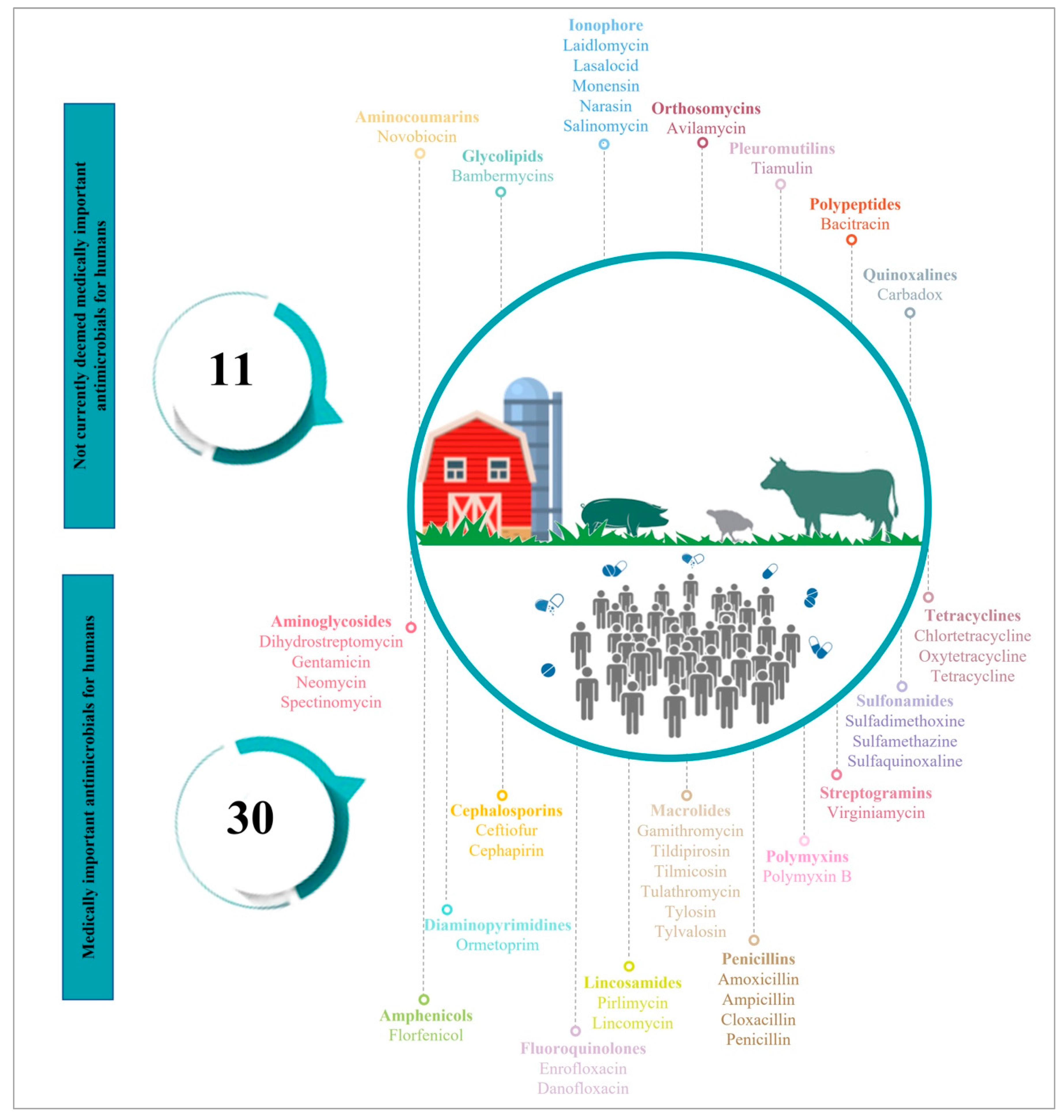
2. Antimicrobials Use (AMU) in Food-Producing Animals
3. The Link between the Use of Antimicrobials in Farm Animals and the Spread of AMR in Both Animals and Humans
3.1. Relationship between AMU and AMR in Farm Animals
3.2. Relationship between AMU/AMR in Farm Animals and AMR in Humans
3.2.1. Direct Transfer of AMR through Zoonotic Bacteria
3.2.2. Indirect Transfer of AMR through Commensal Bacteria
3.2.3. Link between AMU in Farm Animals and AMR in Human Bacteria
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, present and future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef]
- Laxminarayan, R. The overlooked pandemic of antimicrobial resistance. Lancet 2022, 399, 606–607. [Google Scholar] [CrossRef]
- Finlay, B.B.; Conly, J.; Coyte, P.C.; Dillon, J.-A.R.; Douglas, G.; Goddard, E.; Greco, L.; Nicolle, L.E.; Patrick, D.; Prescott, J.F. When Antibiotics Fail: The Expert Panel on the Potential Socio-Economic Impacts of Antimicrobial Resistance in Canada. 2019. Available online: https://cca-reports.ca/reports/the-potential-socio-economic-impacts-of-antimicrobial-resistance-in-canada/ (accessed on 10 August 2022).
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- O’Neill, J. Review on Antimicrobial Resistance. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. 2016. Available online: https://amr-review.org/ (accessed on 10 August 2022).
- Durand, G.A.; Raoult, D.; Dubourg, G. Antibiotic discovery: History, methods and perspectives. Int. J. Antimicrob. Agents 2019, 53, 371–382. [Google Scholar] [CrossRef]
- Moretta, A.; Scieuzo, C.; Petrone, A.M.; Salvia, R.; Manniello, M.D.; Franco, A.; Lucchetti, D.; Vassallo, A.; Vogel, H.; Sgambato, A. Antimicrobial peptides: A new hope in biomedical and pharmaceutical fields. Front. Cell. Infect. Microbiol. 2021, 11, 453. [Google Scholar] [CrossRef]
- Rhouma, M.; Beaudry, F.; Letellier, A. Resistance to colistin: What is the fate for this antibiotic in pig production? Int. J. Antimicrob. Agents 2016, 48, 119–126. [Google Scholar] [CrossRef]
- Butaye, P.; Devriese, L.A.; Haesebrouck, F. Antimicrobial growth promoters used in animal feed: Effects of less well known antibiotics on gram-positive bacteria. Clin. Microbiol. Rev. 2003, 16, 175–188. [Google Scholar] [CrossRef]
- Holmes, A.H.; Moore, L.S.; Sundsfjord, A.; Steinbakk, M.; Regmi, S.; Karkey, A.; Guerin, P.J.; Piddock, L.J. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 2016, 387, 176–187. [Google Scholar] [CrossRef]
- G7 Health Ministers’ Meeting. G7 Health Ministers’ Declaration, Oxford, 4 June 2021. 2021. Available online: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/992268/G7-health_ministers-communique-oxford-4-june-2021_5.pdf (accessed on 4 August 2022).
- Declaration of the G20 Health Ministers. 2021. Available online: https://reliefweb.int/report/world/declaration-g20-health-ministers-rome-5-6-september-2021 (accessed on 2 August 2022).
- Wang, Y.; Xu, C.; Zhang, R.; Chen, Y.; Shen, Y.; Hu, F.; Liu, D.; Lu, J.; Guo, Y.; Xia, X. Changes in colistin resistance and mcr-1 abundance in Escherichia coli of animal and human origins following the ban of colistin-positive additives in China: An epidemiological comparative study. Lancet Infect. Dis. 2020, 20, 1161–1171. [Google Scholar] [CrossRef]
- Xia, X.; Wang, Z.; Fu, Y.; Du, X.-d.; Gao, B.; Zhou, Y.; He, J.; Wang, Y.; Shen, J.; Jiang, H. Association of colistin residues and manure treatment with the abundance of mcr-1 gene in swine feedlots. Environ. Int. 2019, 127, 361–370. [Google Scholar] [CrossRef]
- Havelaar, A.H.; Graveland, H.; van de Kassteele, J.; Zomer, T.P.; Veldman, K.; Bouwknegt, M. A summary index for antimicrobial resistance in food animals in the Netherlands. BMC Vet. Res. 2017, 13, 305. [Google Scholar] [CrossRef] [PubMed]
- Hesp, A.; Veldman, K.; van der Goot, J.; Mevius, D.; van Schaik, G. Monitoring antimicrobial resistance trends in commensal Escherichia coli from livestock, the Netherlands, 1998 to 2016. Eurosurveillance 2019, 24, 1800438. [Google Scholar] [CrossRef]
- Sanders, P.; Mevius, D.; Veldman, K.; van Geijlswijk, I.; Wagenaar, J.A.; Bonten, M.; Heederik, D. Comparison of different antimicrobial use indicators and antimicrobial resistance data in food-producing animals. JAC Antimicrob. Resist. 2021, 3, dlab172. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.L.; Caffrey, N.P.; Nóbrega, D.B.; Cork, S.C.; Ronksley, P.E.; Barkema, H.W.; Polachek, A.J.; Ganshorn, H.; Sharma, N.; Kellner, J.D. Restricting the use of antibiotics in food-producing animals and its associations with antibiotic resistance in food-producing animals and human beings: A systematic review and meta-analysis. Lancet Planet. Health 2017, 1, e316–e327. [Google Scholar] [CrossRef]
- Shi, X.; Li, Y.; Yang, Y.; Shen, Z.; Cai, C.; Wang, Y.; Walsh, T.R.; Shen, J.; Wu, Y.; Wang, S. High prevalence and persistence of carbapenem and colistin resistance in livestock farm environments in China. J. Hazard. Mater. 2021, 406, 124298. [Google Scholar] [CrossRef]
- Rhouma, M.; Tessier, M.; Aenishaenslin, C.; Sanders, P.; Carabin, H. Should the increased awareness of the One Health approach brought by the COVID-19 pandemic be used to further tackle the challenge of antimicrobial resistance? Antibiotics 2021, 10, 464. [Google Scholar] [CrossRef]
- World Health Organization. Tripartite and UNEP Support OHHLEP’s Definition of “One Health”. 2021. Available online: https://www.who.int/news/item/01-12-2021-tripartite-and-unep-support-ohhlep-s-definition-of-one-health (accessed on 11 July 2022).
- Laxminarayan, R.; Van Boeckel, T.; Frost, I.; Kariuki, S.; Khan, E.A.; Limmathurotsakul, D.; Larsson, D.J.; Levy-Hara, G.; Mendelson, M.; Outterson, K. The Lancet Infectious Diseases Commission on antimicrobial resistance: 6 years later. Lancet Infect. Dis. 2020, 20, e51–e60. [Google Scholar] [CrossRef]
- Sharma, C.; Rokana, N.; Chandra, M.; Singh, B.P.; Gulhane, R.D.; Gill, J.P.S.; Ray, P.; Puniya, A.K.; Panwar, H. Antimicrobial resistance: Its surveillance, impact, and alternative management strategies in dairy animals. Front. Vet. Sci. 2018, 4, 237. [Google Scholar] [CrossRef]
- Rhouma, M.; Fairbrother, J.M.; Beaudry, F.; Letellier, A. Post weaning diarrhea in pigs: Risk factors and non-colistin-based control strategies. Acta Vet. Scand. 2017, 59, 31. [Google Scholar] [CrossRef]
- More, S.J. European perspectives on efforts to reduce antimicrobial usage in food animal production. Ir. Vet. J. 2020, 73, 2. [Google Scholar] [CrossRef] [Green Version]
- Chambers, J.A.; Crumlish, M.; Comerford, D.A.; O’Carroll, R.E. Antimicrobial resistance in humans and animals: Rapid review of psychological and behavioral determinants. Antibiotics 2020, 9, 285. [Google Scholar] [CrossRef]
- Vidovic, N.; Vidovic, S. Antimicrobial resistance and food animals: Influence of livestock environment on the emergence and dissemination of antimicrobial resistance. Antibiotics 2020, 9, 52. [Google Scholar] [CrossRef]
- Haenni, M.; Dagot, C.; Chesneau, O.; Bibbal, D.; Labanowski, J.; Vialette, M.; Bouchard, D.; Martin-Laurent, F.; Calsat, L.; Nazaret, S. Environmental contamination in a high-income country (France) by antibiotics, antibiotic-resistant bacteria, and antibiotic resistance genes: Status and possible causes. Environ. Int. 2022, 159, 107047. [Google Scholar] [CrossRef]
- Wang, F.; Fu, Y.-H.; Sheng, H.-J.; Topp, E.; Jiang, X.; Zhu, Y.-G.; Tiedje, J.M. Antibiotic resistance in the soil ecosystem: A One Health perspective. Curr. Opin. Environ. Sci. 2021, 20, 100230. [Google Scholar] [CrossRef]
- Collignon, P.J.; McEwen, S.A. One health—Its importance in helping to better control antimicrobial resistance. Trop. Med. Infect. Dis. 2019, 4, 22. [Google Scholar] [CrossRef]
- Olaitan, A.O.; Dandachi, I.; Baron, S.A.; Daoud, Z.; Morand, S.; Rolain, J.-M. Banning colistin in feed additives: A small step in the right direction. Lancet Infect. Dis. 2021, 21, 29–30. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. Summary Report on Antimicrobials Sold or Distributed for Use in Food-Producing Animals. 2020. Available online: https://www.fda.gov/media/154820/download (accessed on 3 August 2022).
- Aidara-Kane, A.; Angulo, F.J.; Conly, J.M.; Minato, Y.; Silbergeld, E.K.; McEwen, S.A.; Collignon, P.J. World Health Organization (WHO) guidelines on use of medically important antimicrobials in food-producing animals. Antimicrob. Resist. Infect. Control 2018, 7, 7. [Google Scholar] [CrossRef]
- World Organisation for Animal Health (OIE). OIE List of Antimicrobial Agents of Veterinary Importance. 2018. Available online: https://www.oie.int/app/uploads/2021/03/a-oie-list-antimicrobials-may2018.pdf (accessed on 10 April 2022).
- Pinto Ferreira, J.; Gochez, D.; Jeannin, M.; Magongo, M.W.; Loi, C.; Bucher, K.; Moulin, G.; Erlacher-Vindel, E. From OIE standards to responsible and prudent use of antimicrobials: Supporting stewardship for the use of antimicrobial agents in animals. JAC Antimicrob. Resist. 2022, 4, dlac017. [Google Scholar] [CrossRef]
- WHO. Critically Important Antimicrobials for Human Medicine. 2018. Available online: https://apps.who.int/iris/bitstream/handle/10665/312266/9789241515528-eng.pdf (accessed on 15 July 2022).
- McEwen, S.A.; Collignon, P.J. Antimicrobial Resistance: A One Health Perspective. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Tiseo, K.; Huber, L.; Gilbert, M.; Robinson, T.P.; Van Boeckel, T.P. Global trends in antimicrobial use in food animals from 2017 to 2030. Antibiotics 2020, 9, 918. [Google Scholar] [CrossRef]
- Van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef] [PubMed]
- Góchez, D.; Raicek, M.; Pinto Ferreira, J.; Jeannin, M.; Moulin, G.; Erlacher-Vindel, E. OIE Annual Report on Antimicrobial Agents Intended for Use in Animals: Methods Used. Front. Vet. Sci. 2019, 6, 317. [Google Scholar] [CrossRef]
- Kempf, I.; Jouy, E.; Chauvin, C. Colistin use and colistin resistance in bacteria from animals. Int. J. Antimicrob. Agents 2016, 48, 598–606. [Google Scholar] [CrossRef]
- OIE. OIE Annual Report on the Use of Antimicrobial Agents in Animals (Fifth Report). 2021. Available online: https://www.woah.org/app/uploads/2021/05/a-fifth-annual-report-amr.pdf (accessed on 27 July 2022).
- Rhouma, M.; Beaudry, F.; Theriault, W.; Letellier, A. Colistin in pig production: Chemistry, mechanism of antibacterial action, microbial resistance emergence, and One Health perspectives. Front. Microbiol. 2016, 7, 1789. [Google Scholar] [CrossRef]
- Ma, F.; Xu, S.; Tang, Z.; Li, Z.; Zhang, L. Use of antimicrobials in food animals and impact of transmission of antimicrobial resistance on humans. Biosaf. Health 2021, 3, 32–38. [Google Scholar] [CrossRef]
- Public Health Agency of Canada. Canadian Antimicrobial Resistance Surveillance System—Update. 2020. Available online: https://www.canada.ca/content/dam/hc-sc/documents/services/drugs-health-products/canadian-antimicrobial-resistance-surveillance-system-2020-report/CARSS-2020-report-2020-eng.pdf (accessed on 20 July 2022).
- McCubbin, K.D.; Anholt, R.M.; De Jong, E.; Ida, J.A.; Nóbrega, D.B.; Kastelic, J.P.; Conly, J.M.; Götte, M.; McAllister, T.A.; Orsel, K. Knowledge Gaps in the Understanding of Antimicrobial Resistance in Canada. Front. Public Health 2021, 9, 1523. [Google Scholar] [CrossRef]
- ECDC; EFSA; EMA. Antimicrobial Consumption and Resistance in Bacteria from Humans and Animals. Third Joint Inter-Agency Report on Integrated Analysis of Antimicrobial Agent Consumption and Occurrence of Antimicrobial Resistance in Bacteria from Humans and Food-Producing Animals in the EU/EEA. 2021. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/JIACRA-III-Antimicrobial-Consumption-and-Resistance-in-Bacteria-from-Humans-and-Animals.pdf (accessed on 20 July 2022).
- ECDC; EFSA; EMA. ECDC/EFSA/EMA Second Joint Report on the Integrated Analysis of the Consumption of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Bacteria from Humans and Food-Producing Animals: Joint Interagency Antimicrobial Consumption and Resistance Analysis (JIACRA) Report. EFSA J. 2017, 15, e04872. [Google Scholar]
- Van Boeckel, T.P.; Glennon, E.E.; Chen, D.; Gilbert, M.; Robinson, T.P.; Grenfell, B.T.; Levin, S.A.; Bonhoeffer, S.; Laxminarayan, R. Reducing antimicrobial use in food animals. Science 2017, 357, 1350–1352. [Google Scholar] [CrossRef] [PubMed]
- Werner, N.; McEwen, S.; Kreienbrock, L. Monitoring antimicrobial drug usage in animals: Methods and applications. In Antimicrobial Resistance in Bacteria from Livestock and Companion Animals; Møller Aarestrup, F., Schwarz, S., Shen, J., Cavaco, L.M., Eds.; ASM Press: Washington, DC, USA, 2018; pp. 569–594. [Google Scholar]
- Sanders, P.; Vanderhaeghen, W.; Fertner, M.; Fuchs, K.; Obritzhauser, W.; Agunos, A.; Carson, C.; Borck Høg, B.; Dalhoff Andersen, V.; Chauvin, C. Monitoring of farm-level antimicrobial use to guide stewardship: Overview of existing systems and analysis of key components and processes. Front. Vet. Sci. 2020, 7, 540. [Google Scholar] [CrossRef]
- Bulut, E.; Ivanek, R. Comparison of different biomass methodologies to adjust sales data on veterinary antimicrobials in the USA. J. Antimicrob. Chemother. 2022, 77, 827–842. [Google Scholar] [CrossRef]
- Lardé, H.; Francoz, D.; Roy, J.P.; Massé, J.; Archambault, M.; Paradis, M.; Dufour, S. Comparison of quantification methods to estimate farm-level usage of antimicrobials other than in medicated feed in dairy farms from Québec, Canada. Microorganisms 2021, 9, 1106. [Google Scholar] [CrossRef] [PubMed]
- Walsh, T.R.; Wu, Y. China bans colistin as a feed additive for animals. Lancet Infect. Dis. 2016, 16, 1102–1103. [Google Scholar] [CrossRef]
- Shen, Y.; Zhou, H.; Xu, J.; Wang, Y.; Zhang, Q.; Walsh, T.R.; Shao, B.; Wu, C.; Hu, Y.; Yang, L. Anthropogenic and environmental factors associated with high incidence of mcr-1 carriage in humans across China. Nat. Microbiol. 2018, 3, 1054–1062. [Google Scholar] [CrossRef] [PubMed]
- European Medicines Agency. Sales of Veterinary Antimicrobial Agents in 31 European Countries in 2019 and 2020. 2021. Available online: https://www.ema.europa.eu/en/documents/report/sales-veterinary-antimicrobial-agents-31-european-countries-2019-2020-trends-2010-2020-eleventh_en.pdf (accessed on 19 July 2022).
- Wallinga, D.; Smit, L.A.; Davis, M.F.; Casey, J.A.; Nachman, K.E. A review of the effectiveness of current US policies on antimicrobial use in meat and poultry production. Curr. Environ. Health Rep. 2022, 9, 339–354. [Google Scholar] [CrossRef] [PubMed]
- Roy, J.-P.; Archambault, M.; Desrochers, A.; Dubuc, J.; Dufour, S.; Francoz, D.; Paradis, M.-È.; Rousseau, M. New Quebec regulation on the use of antimicrobials of very high importance in food animals: Implementation and impacts in dairy cattle practice. Can. Vet. J. 2020, 61, 193. [Google Scholar]
- Millar, N.; Aenishaenslin, C.; Lardé, H.; Roy, J.P.; Fourichon, C.; Francoz, D.; Paradis, M.È.; Dufour, S. Evidence of a decrease in sales of antimicrobials of very high importance for humans in dairy herds after a new regulation restricting their use in Quebec, Canada. Zoonoses Public Health 2022, 69, 370–381. [Google Scholar] [CrossRef]
- Firth, C.L.; Käsbohrer, A.; Pless, P.; Koeberl-Jelovcan, S.; Obritzhauser, W. Analysis of Antimicrobial Use and the Presence of Antimicrobial-Resistant Bacteria on Austrian Dairy Farms—A Pilot Study. Antibiotics 2022, 11, 124. [Google Scholar] [CrossRef]
- Viel, A.; Henri, J.; Perrin-Guyomard, A.; Laroche, J.; Couet, W.; Gregoire, N.; Laurentie, M. Lack of experimental evidence to support mcr-1-positive Escherichia coli strain selection during oral administration of colistin at recommended and higher dose given by gavage in weaned piglets. Int. J. Antimicrob. Agents 2017, 51, 128–131. [Google Scholar] [CrossRef]
- Fleury, M.; Jouy, E.; Eono, F.; Cariolet, R.; Couet, W.; Gobin, P.; Le Goff, O.; Blanquet-Diot, S.; Alric, M.; Kempf, I. Impact of two different colistin dosing strategies on healthy piglet fecal microbiota. Res. Vet. Sci. 2016, 107, 152–160. [Google Scholar] [CrossRef]
- Rhouma, M.; Beaudry, F.; Theriault, W.; Bergeron, N.; Beauchamp, G.; Laurent-Lewandowski, S.; Fairbrother, J.M.; Letellier, A. In vivo therapeutic efficacy and pharmacokinetics of colistin sulfate in an experimental model of enterotoxigenic Escherichia coli infection in weaned pigs. Vet. Res. 2016, 47, 58. [Google Scholar] [CrossRef]
- Santman-Berends, I.; Gonggrijp, M.A.; Hage, J.J.; Heuvelink, A.E.; Velthuis, A.; Lam, T.; van Schaik, G. Prevalence and risk factors for extended-spectrum β-lactamase or AmpC-producing Escherichia coli in organic dairy herds in the Netherlands. J. Dairy Sci. 2017, 100, 562–571. [Google Scholar] [CrossRef]
- Gonggrijp, M.A.; Santman-Berends, I.; Heuvelink, A.E.; Buter, G.J.; van Schaik, G.; Hage, J.J.; Lam, T. Prevalence and risk factors for extended-spectrum β-lactamase- and AmpC-producing Escherichia coli in dairy farms. J. Dairy Sci. 2016, 99, 9001–9013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiki, M.; Kawanishi, M.; Abo, H.; Kojima, A.; Koike, R.; Hamamoto, S.; Asai, T. Decreased resistance to broad-spectrum cephalosporin in Escherichia coli from healthy broilers at farms in Japan after voluntary withdrawal of ceftiofur. Foodborne Pathog. Dis. 2015, 12, 639–643. [Google Scholar] [CrossRef] [PubMed]
- De Lagarde, M.; Fairbrother, J.M.; Archambault, M.; Dufour, S.; Francoz, D.; Massé, J.; Lardé, H.; Aenishaenslin, C.; Paradis, M.-È.; Roy, J.-P. Impact of a regulation restricting critical antimicrobial usage on prevalence of antimicrobial resistance in Escherichia coli isolates from fecal and manure pit samples on dairy farms in Québec, Canada. Front. Vet. Sci. 2022, 9, 838498. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Zhong, L.-L.; Yang, Y.; Doi, Y.; Paterson, D.L.; Stoesser, N.; Ma, F.; El-Sayed, M.A.E.-G.; Feng, S.; Huang, S. Dynamics of mcr-1 prevalence and mcr-1-positive Escherichia coli after the cessation of colistin use as a feed additive for animals in China: A prospective cross-sectional and whole genome sequencing-based molecular epidemiological study. Lancet Microbe 2020, 1, e34–e43. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, T.; Wang, C.; Liang, G.; Lu, Q.; Wen, G.; Guo, Y.; Cheng, Y.; Wang, Z.; Shao, H. Prevalence of colistin resistance gene mcr-1 in Escherichia coli isolated from chickens in central China, 2014 to 2019. J. Glob. Antimicrob. Resist. 2022, 29, 241–246. [Google Scholar] [CrossRef]
- Aarestrup, F.M.; Seyfarth, A.M.; Emborg, H.-D.; Pedersen, K.; Hendriksen, R.S.; Bager, F. Effect of abolishment of the use of antimicrobial agents for growth promotion on occurrence of antimicrobial resistance in fecal enterococci from food animals in Denmark. Antimicrob. Agents Chemother. 2001, 45, 2054–2059. [Google Scholar] [CrossRef] [PubMed]
- Aarestrup, F.M.; Bager, F.; Andersen, J.S. Association between the use of avilamycin for growth promotion and the occurrence of resistance among Enterococcus faecium from broilers: Epidemiological study and changes over time. Microb. Drug Resist. 2000, 6, 71–75. [Google Scholar] [CrossRef]
- Boerlin, P.; Wissing, A.; Aarestrup, F.M.; Frey, J.; Nicolet, J. Antimicrobial growth promoter ban and resistance to macrolides and vancomycin in enterococci from pigs. J. Clin. Microbiol. 2001, 39, 4193–4195. [Google Scholar] [CrossRef]
- Chauvin, C.; Gicquel-Bruneau, M.; Perrin-Guyomard, A.; Humbert, F.; Salvat, G.; Guillemot, D.; Sanders, P. Use of avilamycin for growth promotion and avilamycin-resistance among Enterococcus faecium from broilers in a matched case-control study in France. Prev. Vet. Med. 2005, 70, 155–163. [Google Scholar] [CrossRef]
- Tang, K.L.; Caffrey, N.P.; Nóbrega, D.B.; Cork, S.C.; Ronksley, P.E.; Barkema, H.W.; Polachek, A.J.; Ganshorn, H.; Sharma, N.; Kellner, J.D.; et al. Comparison of different approaches to antibiotic restriction in food-producing animals: Stratified results from a systematic review and meta-analysis. BMJ Glob. Health 2019, 4, e001710. [Google Scholar] [CrossRef]
- Butaye, P.; Devriese, L.A.; Haesebrouck, F. Effect of avilamycin fed to chickens on E. faecium counts and on the selection of avilamycin-resistant E. faecium populations. Microb. Drug Resist. 2005, 11, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Nobrega, D.B.; Tang, K.L.; Caffrey, N.P.; De Buck, J.; Cork, S.C.; Ronksley, P.E.; Polachek, A.J.; Ganshorn, H.; Sharma, N.; Kastelic, J.P. Prevalence of antimicrobial resistance genes and its association with restricted antimicrobial use in food-producing animals: A systematic review and meta-analysis. J. Antimicrob. Chemother. 2021, 76, 561–575. [Google Scholar] [CrossRef] [PubMed]
- Österberg, J.; Wingstrand, A.; Nygaard Jensen, A.; Kerouanton, A.; Cibin, V.; Barco, L.; Denis, M.; Aabo, S.; Bengtsson, B. Antibiotic resistance in Escherichia coli from pigs in organic and conventional farming in four European countries. PLoS ONE 2016, 11, e0157049. [Google Scholar] [CrossRef] [PubMed]
- De Lucia, A.; Card, R.; Duggett, N.; Smith, R.; Davies, R.; Cawthraw, S.; Anjum, M.; Rambaldi, M.; Ostanello, F.; Martelli, F. Reduction in antimicrobial resistance prevalence in Escherichia coli from a pig farm following withdrawal of group antimicrobial treatment. Vet. Microbiol. 2021, 258, 109125. [Google Scholar] [CrossRef]
- Carson, C.; Li, X.-Z.; Agunos, A.; Loest, D.; Chapman, B.; Finley, R.; Mehrotra, M.; Sherk, L.M.; Gaumond, R.; Irwin, R. Ceftiofur-resistant Salmonella enterica serovar Heidelberg of poultry origin–a risk profile using the Codex framework. Epidemiol. Infect. 2019, 147, E296. [Google Scholar] [CrossRef]
- Trujillo-Soto, T.; Machuca, J.; Arca-Suárez, J.; Rodríguez-Iglesias, M.; Galán-Sánchez, F. Co-Occurrence of mcr-1 and qnrS1 on an IncHI2 plasmid in clinical isolates of Salmonella typhimurium in Spain. Vector Borne Zoonotic Dis. 2019, 19, 662–665. [Google Scholar] [CrossRef]
- Xavier, B.; Lammens, C.; Butaye, P.; Goossens, H.; Malhotra-Kumar, S. Complete sequence of an IncFII plasmid harbouring the colistin resistance gene mcr-1 isolated from Belgian pig farms. J. Antimicrob. Chemother. 2016, 71, 2342–2344. [Google Scholar] [CrossRef]
- Green, A.G.; Yoon, C.H.; Chen, M.L.; Ektefaie, Y.; Fina, M.; Freschi, L.; Gröschel, M.I.; Kohane, I.; Beam, A.; Farhat, M. A convolutional neural network highlights mutations relevant to antimicrobial resistance in Mycobacterium tuberculosis. Nat. Commun. 2022, 13, 3817. [Google Scholar] [CrossRef]
- Li, X.; Zhang, Z.; Xu, D.; Wu, C.; Li, J.; Zheng, Y. A prediction method for animal-derived drug resistance trend using a grey-bp neural network combination model. Antibiotics 2021, 10, 692. [Google Scholar] [CrossRef]
- Rhouma, M.; Letellier, A. Extended-spectrum beta-lactamases, carbapenemases and the mcr-1 gene: Is there a historical link? Int. J. Antimicrob. Agents 2017, 49, 269–271. [Google Scholar] [CrossRef]
- Oliveira, K.; Viegas, C.; Ribeiro, E. MRSA Colonization in Workers from Different Occupational Environments—A One Health Approach Perspective. Atmosphere 2022, 13, 658. [Google Scholar] [CrossRef]
- Crespo-Piazuelo, D.; Lawlor, P.G. Livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) prevalence in humans in close contact with animals and measures to reduce on-farm colonisation. Ir. Vet. J. 2021, 74, 21. [Google Scholar] [CrossRef] [PubMed]
- Fetsch, A.; Etter, D.; Johler, S. Livestock-associated meticillin-resistant Staphylococcus aureus—Current situation and impact from a One Health perspective. Curr. Clin. Microbiol. Rep. 2021, 8, 103–113. [Google Scholar] [CrossRef]
- Price, L.B.; Stegger, M.; Hasman, H.; Aziz, M.; Larsen, J.; Andersen, P.S.; Pearson, T.; Waters, A.E.; Foster, J.T.; Schupp, J.; et al. Staphylococcus aureus CC398: Host adaptation and emergence of methicillin resistance in livestock. mBio 2012, 3, e00305-11. [Google Scholar] [CrossRef] [PubMed]
- Dorado-García, A.; Dohmen, W.; Bos, M.E.; Verstappen, K.M.; Houben, M.; Wagenaar, J.A.; Heederik, D.J. Dose-response relationship between antimicrobial drugs and livestock-associated MRSA in pig farming. Emerg. Infect. Dis. 2015, 21, 950. [Google Scholar] [CrossRef] [PubMed]
- Mather, A.; Reid, S.; Maskell, D.; Parkhill, J.; Fookes, M.; Harris, S.; Brown, D.; Coia, J.; Mulvey, M.; Gilmour, M. Distinguishable epidemics of multidrug-resistant Salmonella Typhimurium DT104 in different hosts. Science 2013, 341, 1514–1517. [Google Scholar] [CrossRef]
- Lester, C.H.; Frimodt-Møller, N.; Sørensen, T.L.; Monnet, D.L.; Hammerum, A.M. In vivo transfer of the vanA resistance gene from an Enterococcus faecium isolate of animal origin to an E. faecium isolate of human origin in the intestines of human volunteers. Antimicrob. Agents Chemother. 2006, 50, 596–599. [Google Scholar] [CrossRef]
- Smet, A.; Rasschaert, G.; Martel, A.; Persoons, D.; Dewulf, J.; Butaye, P.; Catry, B.; Haesebrouck, F.; Herman, L.; Heyndrickx, M. In situ ESBL conjugation from avian to human Escherichia coli during cefotaxime administration. J. Appl. Microbiol. 2011, 110, 541–549. [Google Scholar] [CrossRef]
- Dorado-García, A.; Smid, J.H.; van Pelt, W.; Bonten, M.J.M.; Fluit, A.C.; van den Bunt, G.; Wagenaar, J.A.; Hordijk, J.; Dierikx, C.M.; Veldman, K.T.; et al. Molecular relatedness of ESBL/AmpC-producing Escherichia coli from humans, animals, food and the environment: A pooled analysis. J. Antimicrob. Chemother. 2018, 73, 339–347. [Google Scholar] [CrossRef]
- Mughini-Gras, L.; Dorado-García, A.; van Duijkeren, E.; van den Bunt, G.; Dierikx, C.M.; Bonten, M.J.; Bootsma, M.C.; Schmitt, H.; Hald, T.; Evers, E.G. Attributable sources of community-acquired carriage of Escherichia coli containing β-lactam antibiotic resistance genes: A population-based modelling study. Lancet Planet Health 2019, 3, e357–e369. [Google Scholar] [CrossRef]
- De Freitas Costa, E.; Hagenaars, T.J.; Dame-Korevaar, A.; Brouwer, M.S.; de Vos, C.J. Multidirectional dynamic model for the spread of extended-spectrum-β-lactamase-producing Escherichia coli in the Netherlands. Microb. Risk Anal. 2022, 100230. [Google Scholar] [CrossRef]
- Day, M.J.; Hopkins, K.L.; Wareham, D.W.; Toleman, M.A.; Elviss, N.; Randall, L.; Teale, C.; Cleary, P.; Wiuff, C.; Doumith, M. Extended-spectrum β-lactamase-producing Escherichia coli in human-derived and foodchain-derived samples from England, Wales, and Scotland: An epidemiological surveillance and typing study. Lancet Infect. Dis. 2019, 19, 1325–1335. [Google Scholar] [CrossRef]
- Lazarus, B.; Paterson, D.L.; Mollinger, J.L.; Rogers, B.A. Do human extraintestinal Escherichia coli infections resistant to expanded-spectrum cephalosporins originate from food-producing animals? A systematic review. Clin. Infect. Dis. 2015, 60, 439–452. [Google Scholar] [CrossRef] [PubMed]
- Ludden, C.; Raven, K.E.; Jamrozy, D.; Gouliouris, T.; Blane, B.; Coll, F.; de Goffau, M.; Naydenova, P.; Horner, C.; Hernandez-Garcia, J. One health genomic surveillance of Escherichia coli demonstrates distinct lineages and mobile genetic elements in isolates from humans versus livestock. mBio 2019, 10, e02693-18. [Google Scholar] [CrossRef]
- Miltgen, G.; Martak, D.; Valot, B.; Kamus, L.; Garrigos, T.; Verchere, G.; Gbaguidi-Haore, H.; Ben Cimon, C.; Ramiandrisoa, M.; Picot, S. One Health compartmental analysis of ESBL-producing Escherichia coli on Reunion Island reveals partitioning between humans and livestock. J. Antimicrob. Chemother. 2022, 77, 1254–1262. [Google Scholar] [CrossRef]
- Nguyen, M.N.; Hoang, H.T.T.; Xavier, B.B.; Lammens, C.; Le, H.T.; Hoang, N.T.B.; Nguyen, S.T.; Pham, N.T.; Goossens, H.; Dang, A.D.; et al. Prospective One Health genetic surveillance in Vietnam identifies distinct bla(CTX-M)-harbouring Escherichia coli in food-chain and human-derived samples. Clin. Microbiol. Infect. 2021, 27, 1515.e1–1515.e8. [Google Scholar] [CrossRef]
- Nguyen, V.T.; Jamrozy, D.; Matamoros, S.; Carrique-Mas, J.J.; Ho, H.M.; Thai, Q.H.; Nguyen, T.N.M.; Wagenaar, J.A.; Thwaites, G.; Parkhill, J.; et al. Limited contribution of non-intensive chicken farming to ESBL-producing Escherichia coli colonization in humans in Vietnam: An epidemiological and genomic analysis. J. Antimicrob. Chemother. 2019, 74, 561–570. [Google Scholar] [CrossRef]
- Leverstein-van Hall, M.; Dierikx, C.; Stuart, J.C.; Voets, G.; Van Den Munckhof, M.; van Essen-Zandbergen, A.; Platteel, T.; Fluit, A.; Van De Sande-Bruinsma, N.; Scharinga, J. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin. Microbiol. Infect. 2011, 17, 873–880. [Google Scholar] [CrossRef]
- Büdel, T.; Kuenzli, E.; Campos-Madueno, E.I.; Mohammed, A.H.; Hassan, N.K.; Zinsstag, J.; Hatz, C.; Endimiani, A. On the island of Zanzibar people in the community are frequently colonized with the same MDR Enterobacterales found in poultry and retailed chicken meat. J. Antimicrob. Chemother. 2020, 75, 2432–2441. [Google Scholar] [CrossRef]
- Olaitan, A.O.; Thongmalayvong, B.; Akkhavong, K.; Somphavong, S.; Paboriboune, P.; Khounsy, S.; Morand, S.; Rolain, J.M. Clonal transmission of a colistin-resistant Escherichia coli from a domesticated pig to a human in Laos. J. Antimicrob. Chemother. 2015, 70, 3402–3404. [Google Scholar] [CrossRef]
- Zhang, X.-F. Possible transmission of mcr-1–harboring Escherichia coli between companion animals and human. Emerg. Infect. Dis. 2016, 22, 1679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, Z.; Maciel-Guerra, A.; Baker, M.; Zhang, X.; Hu, Y.; Wang, W.; Rong, J.; Zhang, J.; Xue, N.; Barrow, P. Whole-genome sequencing and gene sharing network analysis powered by machine learning identifies antibiotic resistance sharing between animals, humans and environment in livestock farming. PLoS Comput. Biol. 2022, 18, e1010018. [Google Scholar] [CrossRef] [PubMed]
- Klare, I.; Badstübner, D.; Konstabel, C.; Böhme, G.; Claus, H.; Witte, W. Decreased incidence of VanA-type vancomycin-resistant enterococci isolated from poultry meat and from fecal samples of humans in the community after discontinuation of avoparcin usage in animal husbandry. Microb. Drug Resist. 1999, 5, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Bruinsma, N.; Willems, R.J.; van den Bogaard, A.E.; van Santen-Verheuvel, M.; London, N.; Driessen, C.; Stobberingh, E.E. Different levels of genetic homogeneity in vancomycin-resistant and -susceptible Enterococcus faecium isolates from different human and animal sources analyzed by amplified-fragment length polymorphism. Antimicrob. Agents Chemother. 2002, 46, 2779–2783. [Google Scholar] [CrossRef]
- Willems, R.J.; Homan, W.; Top, J.; van Santen-Verheuvel, M.; Tribe, D.; Manzioros, X.; Gaillard, C.; Vandenbroucke-Grauls, C.M.; Mascini, E.M.; van Kregten, E.; et al. Variant esp gene as a marker of a distinct genetic lineage of vancomycin-resistant Enterococcus faecium spreading in hospitals. Lancet 2001, 357, 853–855. [Google Scholar] [CrossRef]
- Leavis, H.L.; Bonten, M.J.; Willems, R.J. Identification of high-risk enterococcal clonal complexes: Global dispersion and antibiotic resistance. Curr. Opin. Microbiol. 2006, 9, 454–460. [Google Scholar] [CrossRef] [PubMed]
- Dutil, L.; Irwin, R.; Finley, R.; Ng, L.K.; Avery, B.; Boerlin, P.; Bourgault, A.-M.; Cole, L.; Daignault, D.; Desruisseau, A. Ceftiofur resistance in Salmonella enterica serovar Heidelberg from chicken meat and humans, Canada. Emerg. Infect. Dis. 2010, 16, 48. [Google Scholar] [CrossRef]
- NethMap. Consumption of Antimicrobial Agents and Antimicrobial Resistance among Medically Important Bacteria in the Netherlands. 2021. Available online: https://swab.nl/nl/exec/file/download/197 (accessed on 9 August 2022).

| Category | Antimicrobial Classes |
|---|---|
| Veterinary Critically Important Antimicrobial Agents (VCIA) | Aminoglycosides Amphenicols Cephalosporins (third and fourth generation) Diaminopyrimidines MacrolidesPenicillins Fluoroquinolones (Quinolones second generation) Sulfonamides Tetracyclines |
| Veterinary Highly Important Antimicrobial Agents (VHIA) | Ansamycin–Rifamycins Cephalosporins first and second generations Ionophores Lincosamides Phosphonic acid Pleuromutilins Polypeptides (e.g., colistin) Quinolones first generation |
| Veterinary Important Antimicrobial Agents (VIA) | Aminocoumarin Arsenical Bicyclomycin Fusidic Acid Orthosomycins Quinoxalines Streptogramins Thiostrepton |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rhouma, M.; Soufi, L.; Cenatus, S.; Archambault, M.; Butaye, P. Current Insights Regarding the Role of Farm Animals in the Spread of Antimicrobial Resistance from a One Health Perspective. Vet. Sci. 2022, 9, 480. https://doi.org/10.3390/vetsci9090480
Rhouma M, Soufi L, Cenatus S, Archambault M, Butaye P. Current Insights Regarding the Role of Farm Animals in the Spread of Antimicrobial Resistance from a One Health Perspective. Veterinary Sciences. 2022; 9(9):480. https://doi.org/10.3390/vetsci9090480
Chicago/Turabian StyleRhouma, Mohamed, Leila Soufi, Schlasiva Cenatus, Marie Archambault, and Patrick Butaye. 2022. "Current Insights Regarding the Role of Farm Animals in the Spread of Antimicrobial Resistance from a One Health Perspective" Veterinary Sciences 9, no. 9: 480. https://doi.org/10.3390/vetsci9090480





