Genomics Analysis Reveals the Potential Biocontrol Mechanism of Pseudomonas aeruginosa QY43 against Fusarium pseudograminearum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Bacterial Isolation
2.2. Screening and Identification of Antagonistic Strains In Vitro
2.3. Antagonistic Activity of QY43 against Conidia Germination of F. pseudograminearum
2.4. Efficacy of QY43 against FCR in a Glasshouse
2.5. Assessment of Broad-Spectrum Antagonistic Activity of QY43
2.6. Whole-Genome Sequencing of QY43
2.7. Comparative Genomics Analysis to Identify Genes Involved in the Synthesis of Antifungal Compounds Specific to QY43
2.8. Characterization of the Biological Properties and Antifungal Compounds of QY43 In Vitro
2.9. Statistical Analysis
3. Results
3.1. QY43 Was Identified as P. aeruginosa with Biocontrol Ability through Screening
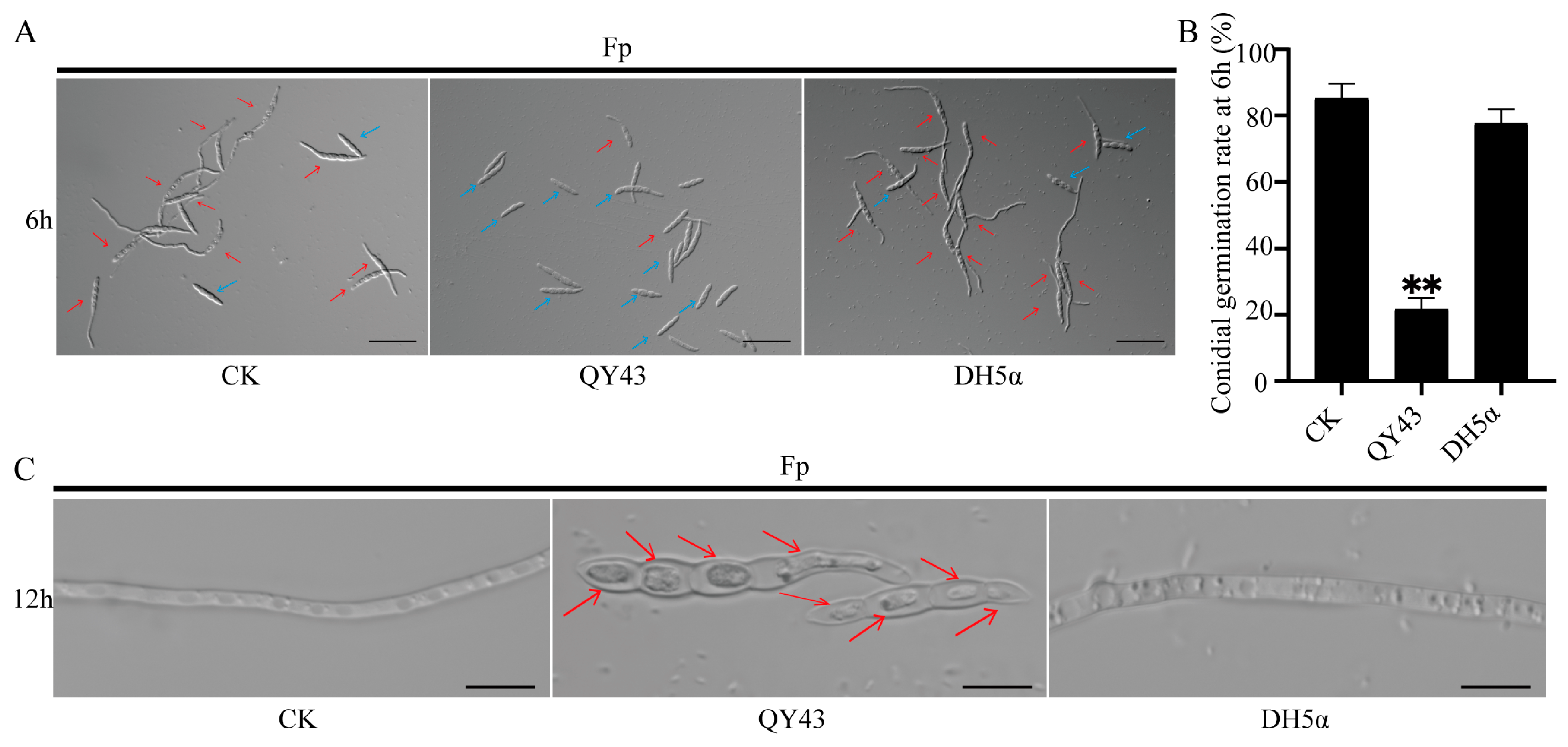
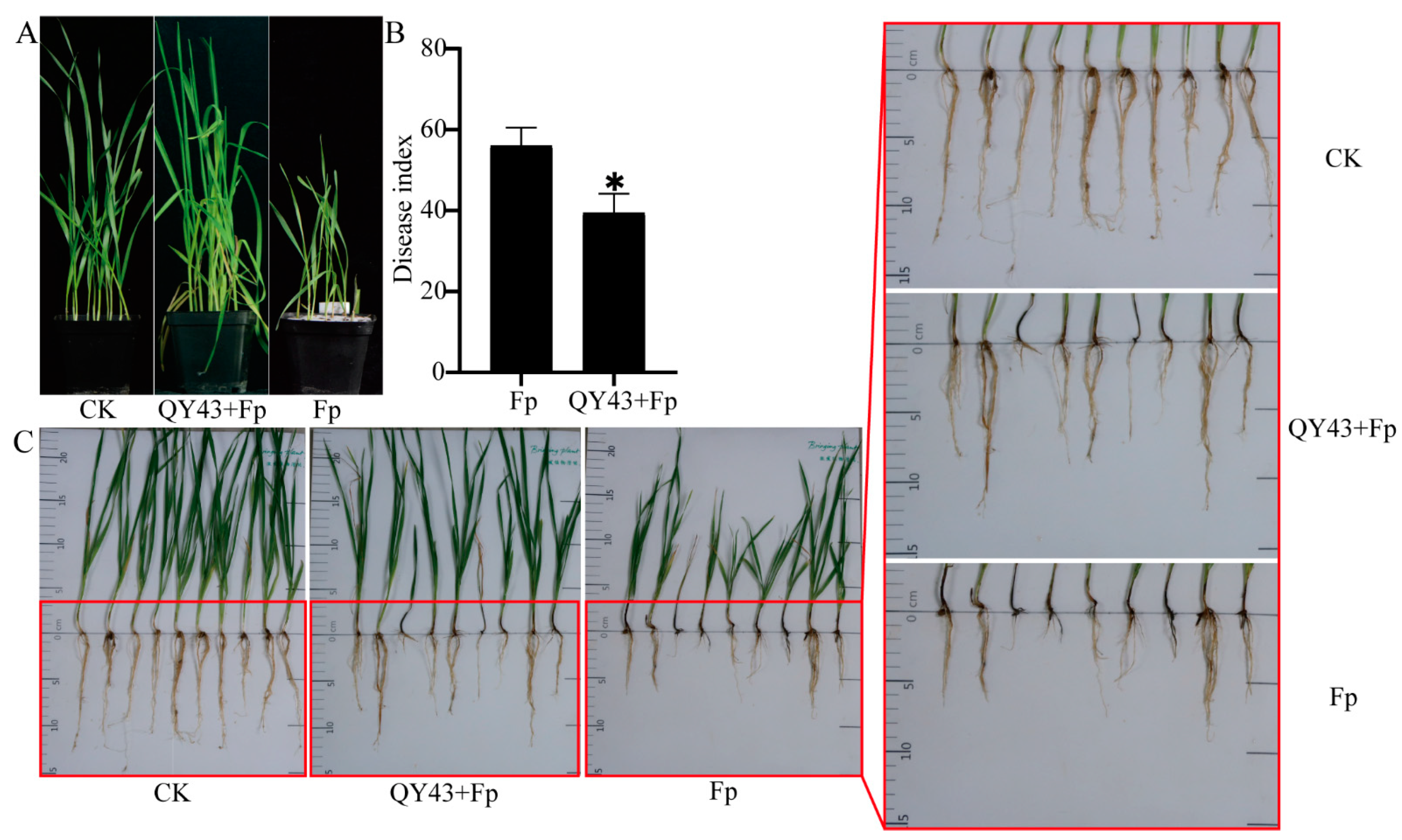
3.2. QY43 Inhibits Conidial Germination, Causes Plasmolysis, and Reduces Pathogenicity of F. pseudograminearum
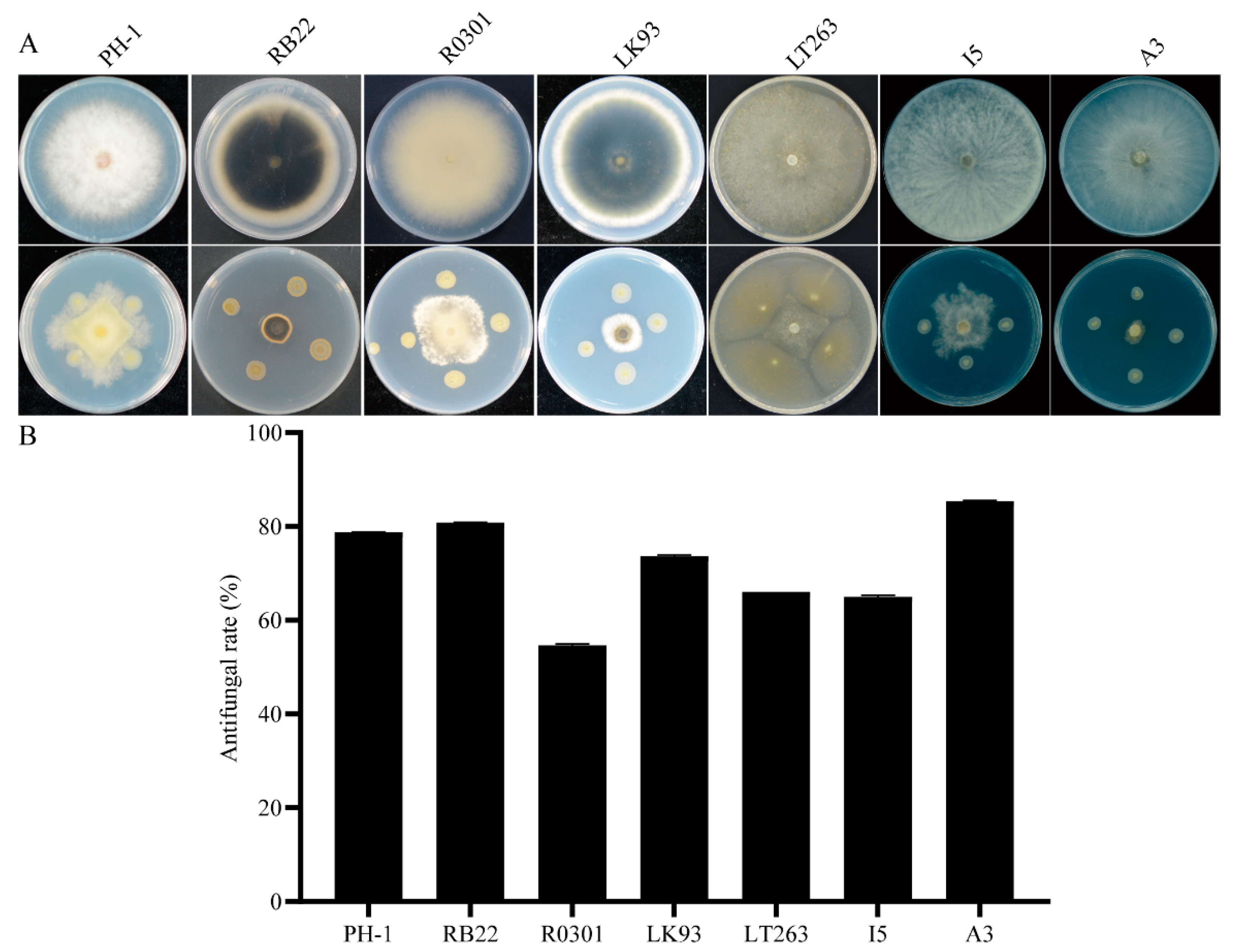
3.3. QY43 Exhibited Broad-Spectrum Antagonistic Activity against Plant Pathogens
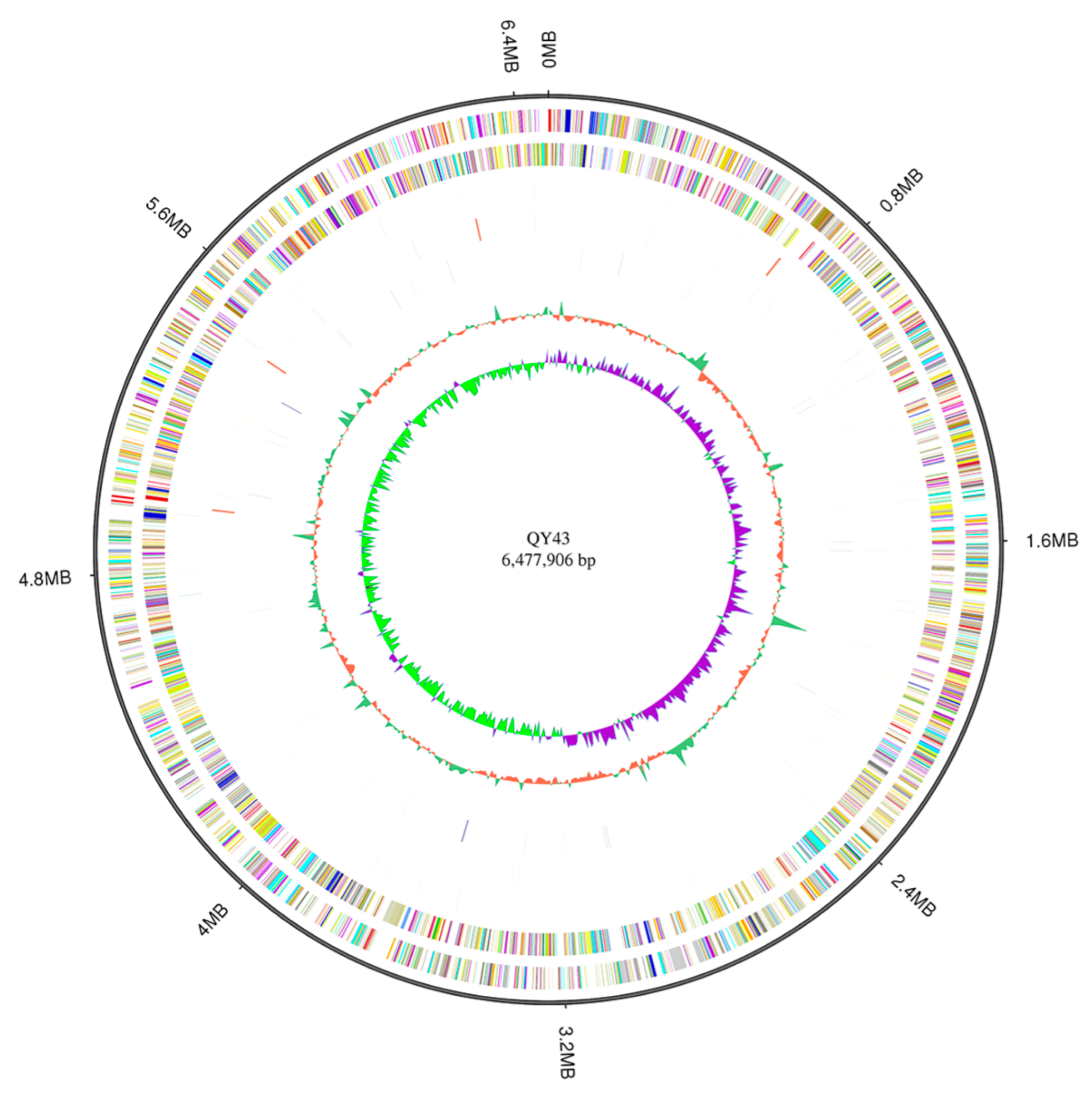
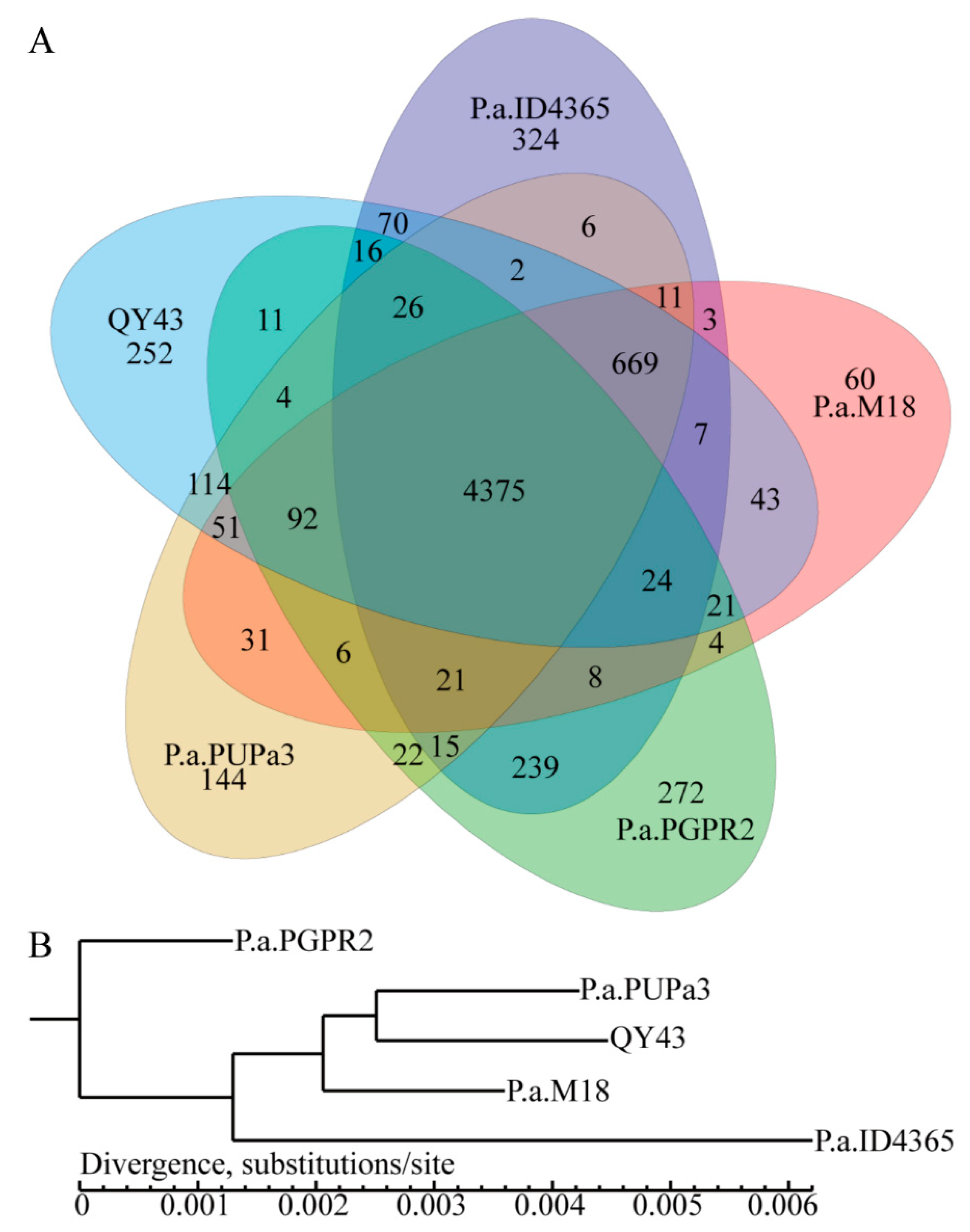
3.4. The Potential Biocontrol Factors of QY43 Were Identified through Genome Analysis
3.5. Comparative Genomics Revealed That QY43 Has Three Unique Genes Related to the Synthesis of Polysaccharide Antifungal Compounds
3.6. Biocontrol Factors of P. aeruginosa QY43 Were Detected by the Experiment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhao, J.; Kang, Z. Fighting Wheat Rusts in China: A Look Back and into the Future. Phytopathol. Res. 2023, 5, 6. [Google Scholar] [CrossRef]
- Zhang, X.; Sun, H.; Shen, C.; Li, W.; Yu, H.; Chen, H. Survey of Fusarium spp. Causing Wheat Crown Rot in Major Winter Wheat Growing Regions of China. Plant Dis. 2015, 99, 1610–1615. [Google Scholar] [CrossRef]
- Xu, F.; Yang, G.; Wang, J.; Song, Y.; Liu, L.; Zhao, K.; Li, Y.; Han, Z. Spatial Distribution of Root and Crown Rot Fungi Associated with Winter Wheat in the North China Plain and Its Relationship with Climate Variables. Front. Microbiol. 2018, 9, 1054. [Google Scholar] [CrossRef] [PubMed]
- Hogg, A.C.; Johnston, R.H.; Dyer, A.T. Applying Real-Time Quantitative Pcr to Fusarium Crown Rot of Wheat. Plant Dis. 2007, 91, 1021–1028. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yuan, H.; Fu, B.; Xing, X.; Sun, B.; Tang, W. First Report of Fusarium pseudograminearum Causing Crown Rot of Wheat in Henan, China. Plant Dis. 2012, 96, 1065. [Google Scholar] [CrossRef]
- Deng, Y.; Li, W.; Zhang, P.; Sun, H.; Zhang, X.; Zhang, A.; Chen, H. Fusarium pseudograminearum as an Emerging Pathogen of Crown Rot of Wheat in Eastern China. Plant Pathol. 2020, 69, 240–248. [Google Scholar] [CrossRef]
- Kazan, K.; Gardiner, D.M. Fusarium Crown Rot Caused by Fusarium pseudograminearum in Cereal Crops: Recent Progress and Future Prospects. Mol. Plant Pathol. 2018, 19, 1547–1562. [Google Scholar] [CrossRef]
- Hou, Y.; Chen, Y.; Wang, L.; Zheng, Y.; Liu, S.; Xu, J. Impact of Pyraclostrobin on the Growth of Fusarium pseudograminearum and Its Control Efficacy against Wheat Crown Rot in Henan Province, China. J. Plant Pathol. 2023, 105, 1499–1510. [Google Scholar] [CrossRef]
- Hou, Y.; Guo, Y.; Wang, L.; He, S.; Zheng, W.; Liu, S.; Xu, J. Impact of Phenamacril on the Growth and Development of Fusarium pseudograminearum and Control of Crown Rot of Wheat. Plant Dis. 2023, 107, 3843–3850. [Google Scholar] [CrossRef]
- Zhang, N.; Xu, Y.; Zhang, Q.; Zhao, L.; Zhu, Y.; Wu, Y.; Li, Z.; Yang, W. Detection of Fungicide Resistance to Fludioxonil and Tebuconazole in Fusarium pseudograminearum, the Causal Agent of Fusarium Crown Rot in Wheat. PeerJ 2023, 11, e14705. [Google Scholar] [CrossRef]
- Malfanova, N.V. Endophytic Bacteria with Plant Growth Promoting and Biocontrol Abilities; Institute Biology of Leiden Faculty of Science Leiden University: Leiden, The Netherlands, 2013; pp. 67–91. [Google Scholar]
- Sarrocco, S. Biological Disease Control by Beneficial (Micro) Organisms: Selected Breakthroughs in the Past 50 Years. Phytopathology 2023, 113, 732–740. [Google Scholar] [CrossRef]
- Kthiri, Z.; Jabeur, M.B.; Machraoui, M.; Gargouri, S.; Hiba, K.; Hamada, W. Coating Seeds with Trichoderma Strains Promotes Plant Growth and Enhance the Systemic Resistance against Fusarium Crown Rot in Durum Wheat. Egypt. J. Biol. Pest Control 2020, 30, 139. [Google Scholar] [CrossRef]
- Dong, Q.; Liu, Q.; Goodwin, P.H.; Deng, X.; Xu, W.; Xia, M.; Zhang, J.; Sun, R.; Wu, C.; Wang, Q.; et al. Isolation and Genome-Based Characterization of Biocontrol Potential of Bacillus Siamensis Yb-1631 against Wheat Crown Rot Caused by Fusarium pseudograminearum. J. Fungi 2023, 9, 547. [Google Scholar] [CrossRef]
- Shoda, M. Bacterial Control of Plant Diseases. J. Biosci. Bioeng. 2000, 89, 515–521. [Google Scholar] [CrossRef]
- Saikia, R.; Srivastava, A.K.; Singh, K.; Arora, D.K.; Lee, M.-W. Effect of Iron Availability on Induction of Systemic Resistance to Fusarium Wilt of Chickpea by Pseudomonas spp. Mycobiology 2005, 33, 35–40. [Google Scholar] [CrossRef]
- Li, B.; Yu, R.; Yu, Q.; Chen, X.; Wu, Z.; Wang, Y.; Xie, G.; Li, H.; Sun, G. Carbon Adaptation Influence the Antagonistic Ability of Pseudomonas aeruginosa against Fusarium oxysporum F. Sp. Melonis. Afr. J. Biotechnol. 2011, 10, 14348–14354. [Google Scholar] [CrossRef]
- Yasmin, S.; Hafeez, F.Y.; Rasul, G. Evaluation of Pseudomonas aeruginosa Z5 for Biocontrol of Cotton Seedling Disease Caused by Fusarium oxysporum. Biocontrol Sci. Technol. 2014, 24, 1227–1242. [Google Scholar] [CrossRef]
- Houshaymi, B.; Awada, R.; Kedees, M.; Soayfane, Z. Pyocyanin, a Metabolite of Pseudomonas aeruginosa, Exhibits Antifungal Drug Activity through Inhibition of a Pleiotropic Drug Resistance Subfamily Fgabc3. Drug Res. 2019, 69, 658–664. [Google Scholar] [CrossRef]
- Dugassa, A.; Alemu, T.; Woldehawariat, Y. In-Vitro Compatibility Assay of Indigenous Trichoderma and Pseudomonas Species and Their Antagonistic Activities against Black Root Rot Disease (Fusarium solani) of Faba Bean (Vicia faba L.). BMC Microbiol. 2021, 21, 115. [Google Scholar] [CrossRef]
- Jatan, R.; Kamboj, R.; Kumar, M.; Kumar, N.; Jain, P.; Lata, C.; Vijayan, J.; Rai, V.; Singh, N.K.; Bisht, D.S. Isolation and Whole Genome Sequencing of Pseudomonas aeruginosa Strain Rk1 and Its Biocontrol Potential against Phytopathogens of Rice. Biologia 2023, 78, 2357–2369. [Google Scholar] [CrossRef]
- Sun, X.; Xu, Y.; Chen, L.; Jin, X.; Ni, H. The Salt-Tolerant Phenazine-1-Carboxamide-Producing Bacterium Pseudomonas aeruginosa Nf011 Isolated from Wheat Rhizosphere Soil in Dry Farmland with Antagonism against Fusarium graminearum. Microbiol. Res. 2021, 245, 126673. [Google Scholar] [CrossRef] [PubMed]
- Rikame, T.; Borde, M. Whole Genome, Functional Annotation and Comparative Genomics of Plant Growth-Promoting Bacteria Pseudomonas aeruginosa (Ng61) with Potential Application in Agro-Industry. Curr. Microbiol. 2022, 79, 169. [Google Scholar] [CrossRef]
- Li, H.; Zhang, Y.; Qin, C.; Wang, Z.; Hao, L.; Zhang, P.; Yuan, Y.; Ding, C.; Wang, M.; Zan, F.; et al. Identification and Characterization of Fprco1 in Regulating Vegetative Growth and Pathogenicity Based on T-DNA Insertion in Fusarium pseudograminearum. J. Integr. Agric. 2024, in press. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhuang, X.; Meng, J.; Zan, F.; Liu, Z.; Qin, C.; Hao, L.; Wang, Z.; Wang, L.; Li, H.; et al. A Putative Zn(Ii)2cys6-Type Transcription Factor Fpume18 Is Required for Development, Conidiation, Cell Wall Integrity, Endocytosis and Full Virulence in Fusarium pseudograminearum. Int. J. Mol. Sci. 2023, 24, 10987. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, S.J.; Al-Musawi, A.T.; Al-Fraji, A.S.; Kareem, H.S. Comparison of Three Culture Media in Assessing the Sensitivity of Antibiotics to Common Foodborne Microorganisms. J. Med. Life 2022, 15, 645–649. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.F.; He, X.L.; Wang, S.; Ma, Q.Z.; Sun, B.J.; Ding, S.L.; Chen, L.L.; Zhang, M.; Li, H.L. Diversity of the Fusarium Pathogens Associated with Crown Rot in the Huanghuai Wheat-Growing Region of China. Environ. Microbiol. 2019, 21, 2740–2754. [Google Scholar] [CrossRef] [PubMed]
- Delcher, A.L.; Bratke, K.A.; Powers, E.C.; Salzberg, S.L. Identifying Bacterial Genes and Endosymbiont DNA with Glimmer. Bioinformatics 2007, 23, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Benson, G. Tandem Repeats Finder: A Program to Analyze DNA Sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. Crisprcasfinder, an Update of Crisrfinder, Includes a Portable Version, Enhanced Performance and Integrates Search for Cas Proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef]
- Lagesen, K.; Hallin, P.; Rodland, E.A.; Stærfeldt, H.H.; Rognes, T.; Ussery, D.W. Rnammer: Consistent and Rapid Annotation of Ribosomal Rna Genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. Trnascan-Se: A Program for Improved Detection of Transfer RNA Genes in Genomic Sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Gardner, P.P.; Daub, J.; Tate, J.G.; Nawrocki, E.P.; Kolbe, D.L.; Lindgreen, S.; Wilkinson, A.C.; Finn, R.D.; Griffiths-Jones, S.; Eddy, S.R.; et al. Rfam: Updates to the Rna Families Database. Nucleic Acids Res. 2009, 37, D136–D140. [Google Scholar] [CrossRef]
- Akhter, S.; Aziz, R.K.; Edwards, R.A. Phispy: A Novel Algorithm for Finding Prophages in Bacterial Genomes That Combines Similarity- and Composition-Based Strategies. Nucleic Acids Res. 2012, 40, e126. [Google Scholar] [CrossRef]
- Chen, L.; Zheng, D.; Liu, B.; Yang, J.; Jin, Q. Vfdb 2016: Hierarchical and Refined Dataset for Big Data Analysis—10 Years on. Nucleic Acids Res. 2016, 44, D694–D697. [Google Scholar] [CrossRef]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and Open Software for Comparing Large Genomes. Genome Biol. 2004, 5, R12. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. Cd-HIT: Accelerated for Clustering the Next-Generation Sequencing Data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Nandi, T.; Ong, C.; Singh, A.P.; Boddey, J.; Atkins, T.; Sarkar-Tyson, M.; Essex-Lopresti, A.E.; Chua, H.H.; Pearson, T.; Kreisberg, J.F. A Genomic Survey of Positive Selection in Burkholderia Pseudomallei Provides Insights into the Evolution of Accidental Virulence. PLoS Pathog. 2010, 6, e1000845. [Google Scholar] [CrossRef]
- Xia, Y.; Yuan, Y.; Li, C.; Sun, Z. Phosphorus-Solubilizing Bacteria Improve the Growth of Nicotiana Benthamiana on Lunar Regolith Simulant by Dissociating Insoluble Inorganic Phosphorus. Commun. Biol. 2023, 6, 1039. [Google Scholar] [CrossRef]
- Li, F.; Xie, Y.J.; Gao, X.; Shan, M.X.; Sun, C.C.; Niu, Y.D.; Shan, A.S. Screening of Cellulose Degradation Bacteria from Min Pigs and Optimization of Its Cellulase Production. Electron. J. Biotechnol. 2020, 48, 29–35. [Google Scholar] [CrossRef]
- Vijayaraghavan, P.; Vincent, S.G.P. A Simple Method for the Detection of Protease Activity on Agar Plates Using Bromocresolgreen Dye. J. Biochem. Technol. 2013, 4, 628–630. [Google Scholar]
- Bellaouchi, R.; Abouloifa, H.; Rokni, Y.; Hasnaoui, A.; Ghabbour, N.; Hakkou, A.; Bechchari, A.; Asehraou, A. Characterization and Optimization of Extracellular Enzymes Production by Aspergillus niger Strains Isolated from Date by-Products. J. Genet. Eng. Biotechnol. 2021, 19, 50. [Google Scholar] [CrossRef]
- Louden, B.C.; Haarmann, D.; Lynne, A.M. Use of Blue Agar Cas Assay for Siderophore Detection. J. Microbiol. Biol. Educ. 2011, 12, 51–53. [Google Scholar] [CrossRef]
- Kamimura, R.; Kanematsu, H.; Ogawa, A.; Kogo, T.; Miura, H.; Kawai, R.; Hirai, N.; Kato, T.; Yoshitake, M.; Barry, D.M. Quantitative Analyses of Biofilm by Using Crystal Violet Staining and Optical Reflection. Materials 2022, 15, 6727. [Google Scholar] [CrossRef]
- Corte, L.; Pierantoni, D.C.; Tascini, C.; Roscini, L.; Cardinali, G. Biofilm Specific Activity: A Measure to Quantify Microbial Biofilm. Microorganisms 2019, 7, 73. [Google Scholar] [CrossRef]
- Wilson, C.; Lukowicz, R.; Merchant, S.; Valquier-Flynn, H.; Caballero, J.; Sandoval, J.; Okuom, M.; Huber, C.; Brooks, T.D.; Wilson, E.; et al. Quantitative and Qualitative Assessment Methods for Biofilm Growth: A Mini-Review. Res. Rev. J. Eng. Technol. 2017, 6, 1–25. [Google Scholar]
- King, E.O.; Ward, M.K.; Raney, D.E. Two Simple Media for the Demonstration of Pyocyanin and Fluorescin. J. Lab. Clin. Med. 1954, 44, 301–307. [Google Scholar]
- Morris, J.D.; Hewitt, J.L.; Wolfe, L.G.; Kamatkar, N.G.; Chapman, S.M.; Diener, J.M.; Courtney, A.J.; Leevy, W.M.; Shrout, J.D. Imaging and Analysis of Pseudomonas aeruginosa Swarming and Rhamnolipid Production. Appl. Environ. Microbiol. 2011, 77, 8310–8317. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Lamont, I.L.; Martin, L.W. Identification and Characterization of Novel Pyoverdine Synthesis Genes in Pseudomonas aeruginosa. Microbiology 2003, 149, 833–842. [Google Scholar] [CrossRef]
- Gross, H.; Loper, J.E. Genomics of Secondary Metabolite Production by Pseudomonas spp. Nat. Prod. Rep. 2009, 26, 1408–1446. [Google Scholar] [CrossRef]
- Kiss, K.; Ng, W.T.; Li, Q. Production of Rhamnolipids-Producing Enzymes of Pseudomonas in E. Coli and Structural Characterization. Front. Chem. Sci. Eng. 2017, 11, 133–138. [Google Scholar] [CrossRef]
- Barreiro, C.; Martínez-Castro, M. Regulation of the phosphate metabolism in Streptomyces genus: Impact on the secondary metabolites. Appl. Microbiol. Biotechnol. 2019, 103, 1643–1658. [Google Scholar] [CrossRef]
- Zeng, Q.W.; Tang, L.S.; Zhang, Y.; Shao, Y.; Wu, W.T.; Wang, J.C.; Ding, X.L.; Han, X.J.; Bilal, M. Isolation and Characterization of Phosphate-Solubilizing Bacteria from Rhizosphere of Poplar on Road Verge and Their Antagonistic Potential against Various Phytopathogens. BMC Microbiol. 2023, 23, 221. [Google Scholar] [CrossRef]
- Breidenstein, E.B.; de la Fuente-Núñez, C.; Hancock, R.E. Pseudomonas aeruginosa: All Roads Lead to Resistance. Trends Microbiol. 2011, 19, 419–426. [Google Scholar] [CrossRef]
- Iswandi, A.; Bossier, P.; Vandenabeele, J.; Verstraete, W. Relation between Soil Microbial Activity and the Effect of Seed Inoculation with the Rhizopseudomonad Strain 7nsk2 on Plant Growth. Biol. Fertil. Soils 1987, 3, 147–151. [Google Scholar] [CrossRef]
- Pierson III, L.S.; Thomashow, L.S. Cloning and Heterologous Expression of the Phenazine Biosynthetic locus from Pseudomonas aureofaciens 30–84. Mol. Plant-Microbe Interact. 1992, 5, 330–339. [Google Scholar] [CrossRef]
- Liu, H.; He, Y.; Jiang, H.; Peng, H.; Huang, X.; Zhang, X.; Thomashow, L.S.; Xu, Y. Characterization of a Phenazine-Producing Strain Pseudomonas chlororaphis Gp72 with Broad-Spectrum Antifungal Activity from Green Pepper Rhizosphere. Curr. Microbiol. 2007, 54, 302–306. [Google Scholar] [CrossRef]
- Wu, D.; Ye, J.; Ou, H.; Wei, X.; Huang, X.; He, Y.; Xu, Y. Genomic Analysis and Temperature-Dependent Transcriptome Profiles of the Rhizosphere Originating Strain Pseudomonas aeruginosa M18. BMC Genom. 2011, 12, 438. [Google Scholar] [CrossRef]
- Stanghellini, M.E.; Miller, R.M. Biosurfactants: Their Identity and Potential Efficacy in the Biological Control of Zoosporic Plant Pathogens. Plant Dis. 1997, 81, 4–12. [Google Scholar] [CrossRef]
- Illakkiam, D.; Ponraj, P.; Shankar, M.; Muthusubramanian, S.; Rajendhran, J.; Gunasekaran, P. Identification and Structure Elucidation of a Novel Antifungal Compound Produced by Pseudomonas aeruginosa Pgpr2 against Macrophomina phaseolina. Appl. Biochem. Biotechnol. 2013, 171, 2176–2185. [Google Scholar] [CrossRef]
- Lee, X.; Azevedo, M.D.; Armstrong, D.J.; Banowetz, G.M.; Reimmann, C. The Pseudomonas aeruginosa antimetabolite L-2-amino-4-methoxy-trans-3-butenoic acid inhibits growth of Erwinia amylovora and acts as a seed germination-arrest factor. Environ. Microbiol. Rep. 2013, 5, 83–89. [Google Scholar] [CrossRef]
- Lam, J.S.; Taylor, V.L.; Islam, S.T.; Hao, Y.; Kocíncová, D. Genetic and Functional Diversity of Pseudomonas aeruginosa Lipopolysaccharide. Front. Microbiol. 2011, 2, 118. [Google Scholar] [CrossRef]
- Shilina, J.; Gushcha, M.; Molozhava, O.; Litvinov, S.; Dmitriev, A. Induction of Arabidopsis Thaliana Resistance to Pathogenic Bacteria by Lipopolysaccharide and Salicylic Acid. Cytol. Genet. 2018, 52, 169–173. [Google Scholar] [CrossRef]
- Islam, S.T.; Huszczynski, S.M.; Nugent, T.; Gold, A.C.; Lam, J.S. Conserved-Residue Mutations in Wzy Affect O-Antigen Polymerization and Wzz-Mediated Chain-Length Regulation in Pseudomonas aeruginosa Pao1. Sci. Rep. 2013, 3, 3441. [Google Scholar] [CrossRef]
- Brencic, A.; Lory, S. Determination of the Regulon and Identification of Novel Mrna Targets of Pseudomonas aeruginosa Rsma. Mol. Microbiol. 2009, 72, 612–632. [Google Scholar] [CrossRef]
- Gebhardt, M.J.; Kambara, T.K.; Ramsey, K.M.; Dove, S.L. Widespread Targeting of Nascent Transcripts by Rsma in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2020, 117, 10520–10529. [Google Scholar] [CrossRef]
- Du, X.; Li, Y.; Zhou, W.; Zhou, Q.; Liu, H.; Xu, Y. Phenazine-1-Carboxylic Acid Production in a Chromosomally Non-Scar Triple-Deleted Mutant Pseudomonas aeruginosa Using Statistical Experimental Designs to Optimize Yield. Appl. Microbiol. Biotechnol. 2013, 97, 7767–7778. [Google Scholar] [CrossRef]


| Gene ID | Gene Annotation |
|---|---|
| QY43GL001843 | Lipopolysccharide (LPS) O-antigen chain length determinant protein, WzzB/FepE family |
| QY43GL001844 | UDP-N-acetyl-D-mannosaminuronate dehydrogenase |
| QY43GL004463 | sRNA-binding carbon storage regulator CsrA |
| Strain | Amylase Productions | Cellulase Production | Protease Production | Inorganic Phosphorus Decomposition | Biofilm Formation | Siderophore Production |
|---|---|---|---|---|---|---|
| QY43 | − | + | + | + | + | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meng, J.; Zan, F.; Liu, Z.; Zhang, Y.; Qin, C.; Hao, L.; Wang, Z.; Wang, L.; Liu, D.; Liang, S.; et al. Genomics Analysis Reveals the Potential Biocontrol Mechanism of Pseudomonas aeruginosa QY43 against Fusarium pseudograminearum. J. Fungi 2024, 10, 298. https://doi.org/10.3390/jof10040298
Meng J, Zan F, Liu Z, Zhang Y, Qin C, Hao L, Wang Z, Wang L, Liu D, Liang S, et al. Genomics Analysis Reveals the Potential Biocontrol Mechanism of Pseudomonas aeruginosa QY43 against Fusarium pseudograminearum. Journal of Fungi. 2024; 10(4):298. https://doi.org/10.3390/jof10040298
Chicago/Turabian StyleMeng, Jiaxing, Feifei Zan, Zheran Liu, Yuan Zhang, Cancan Qin, Lingjun Hao, Zhifang Wang, Limin Wang, Dongmei Liu, Shen Liang, and et al. 2024. "Genomics Analysis Reveals the Potential Biocontrol Mechanism of Pseudomonas aeruginosa QY43 against Fusarium pseudograminearum" Journal of Fungi 10, no. 4: 298. https://doi.org/10.3390/jof10040298





