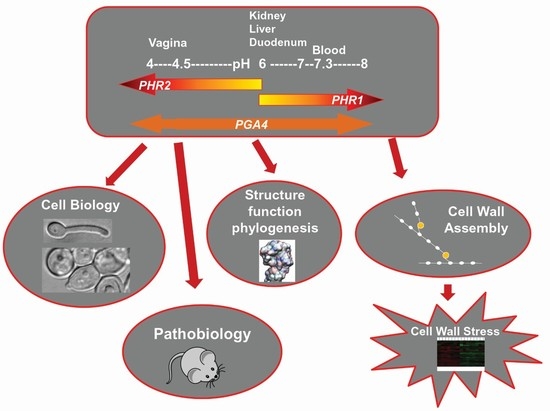
The PHR Family: The Role of Extracellular Transglycosylases in Shaping Candida albicans Cells
Abstract
:1. Introduction
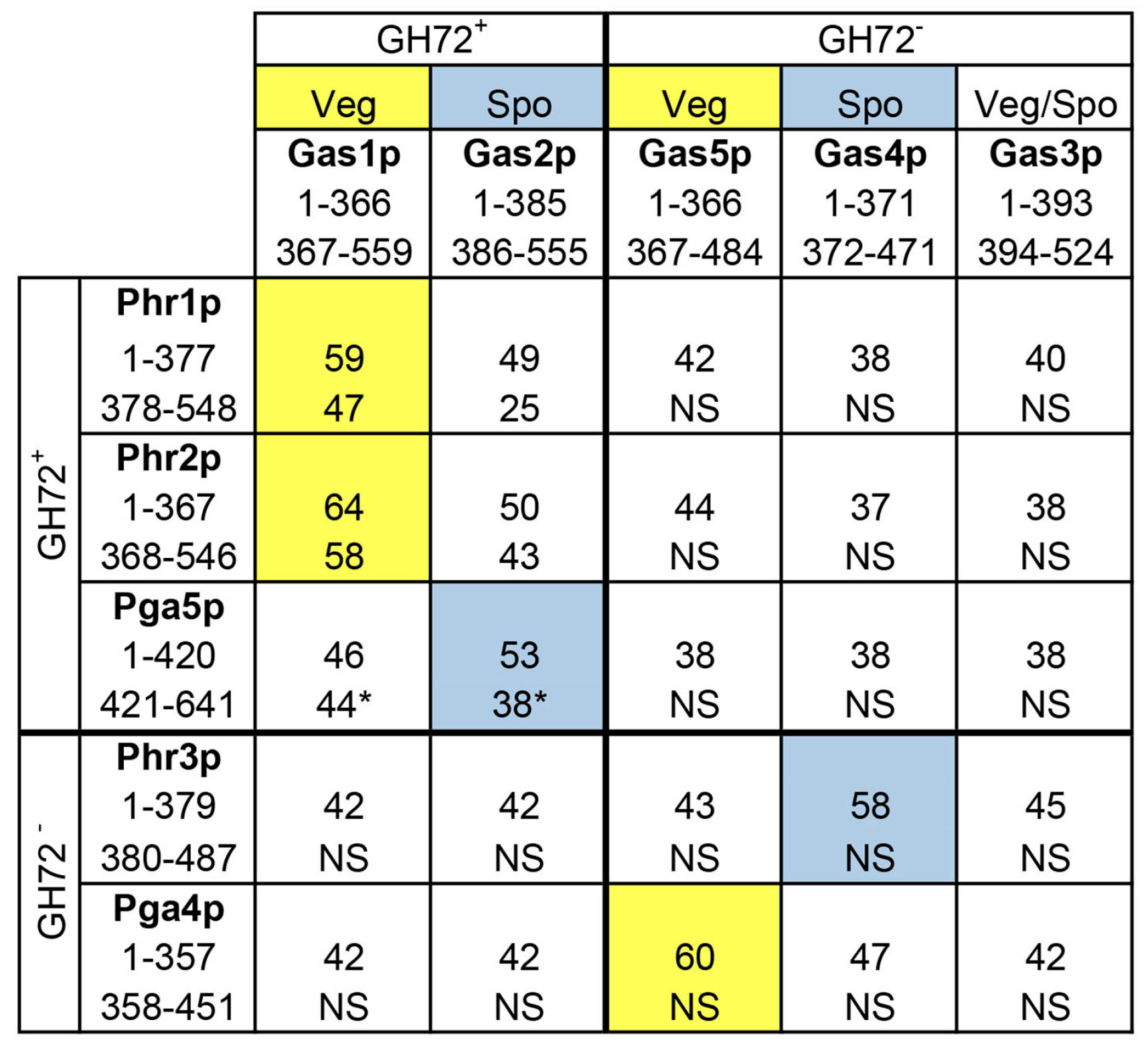
2. Identification of the PHR Multigene Family
Expression Pattern of PHR Multigene Family
3. Molecular Features and Localization of Phr Proteins
3.1. Phr Proteins Are C. albicans Representatives of Family GH72
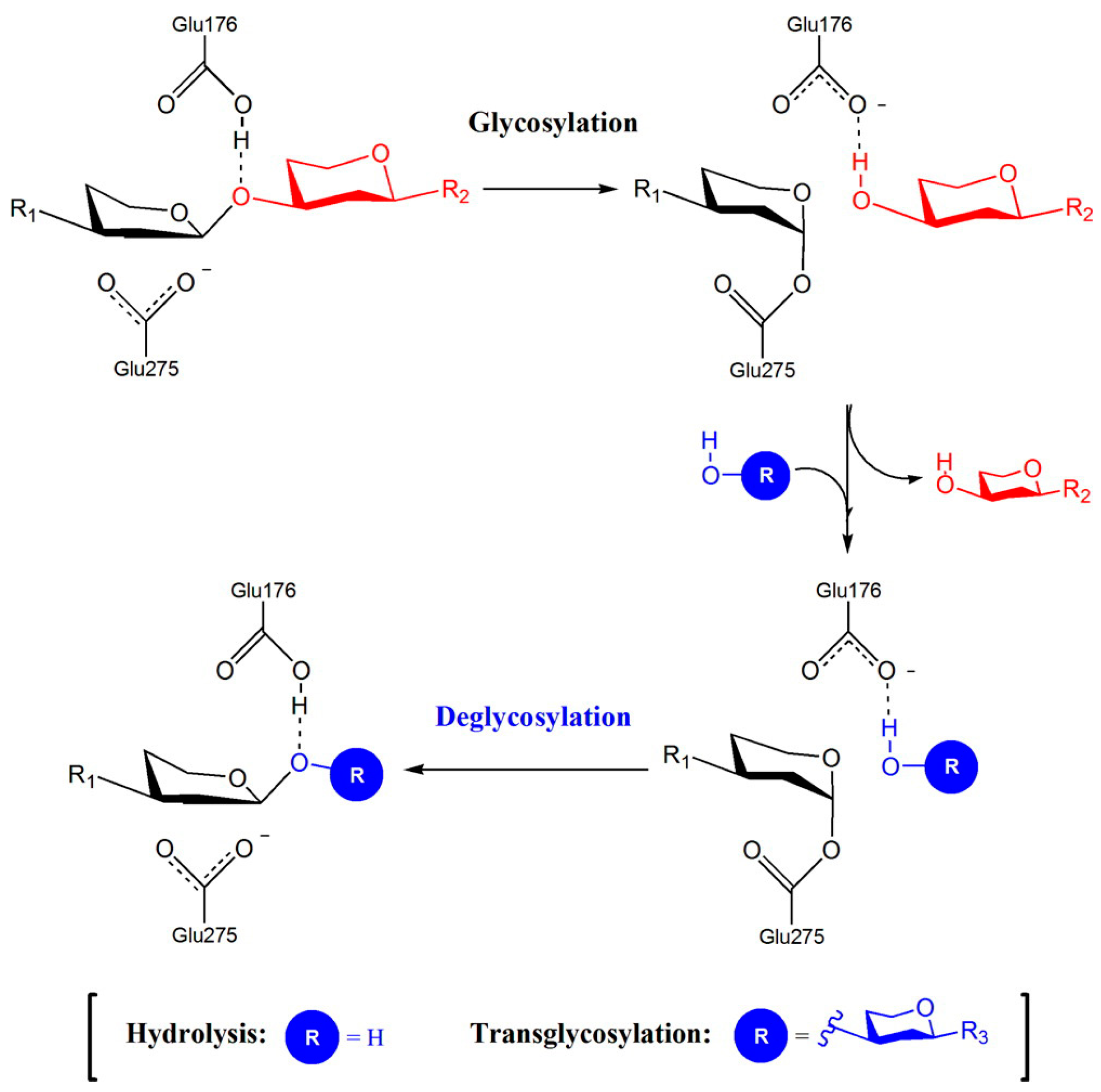
3.1.1. Catalytic Activity of the GH72 Enzymes: AfGel1, Gas1, and Phr1-Phr2 Proteins
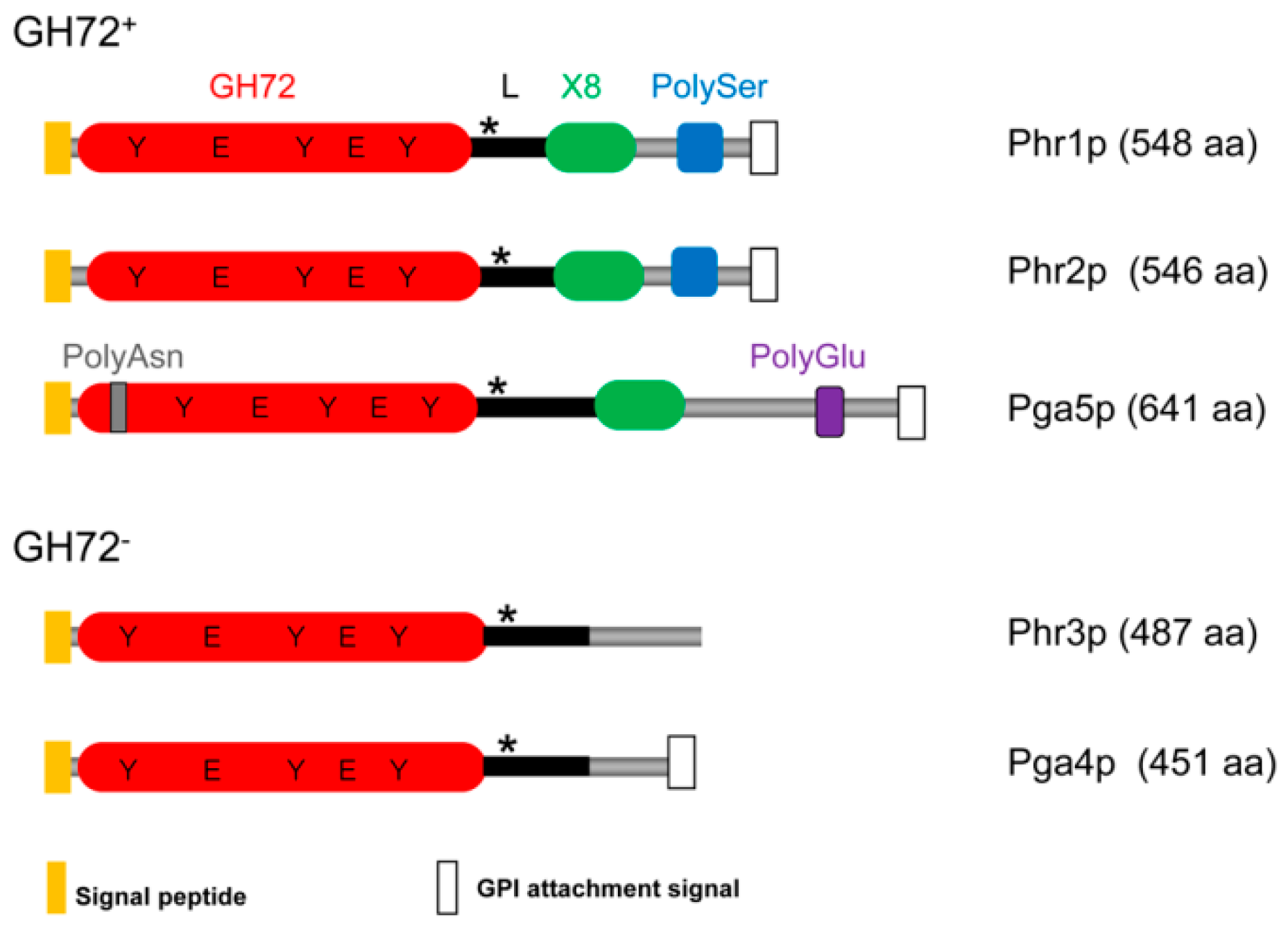
3.1.2. Domain Composition and Structure of the Phr Family of Proteins
3.1.3. Active, Inactive, and Anomalous Members of the Phr Family of Proteins
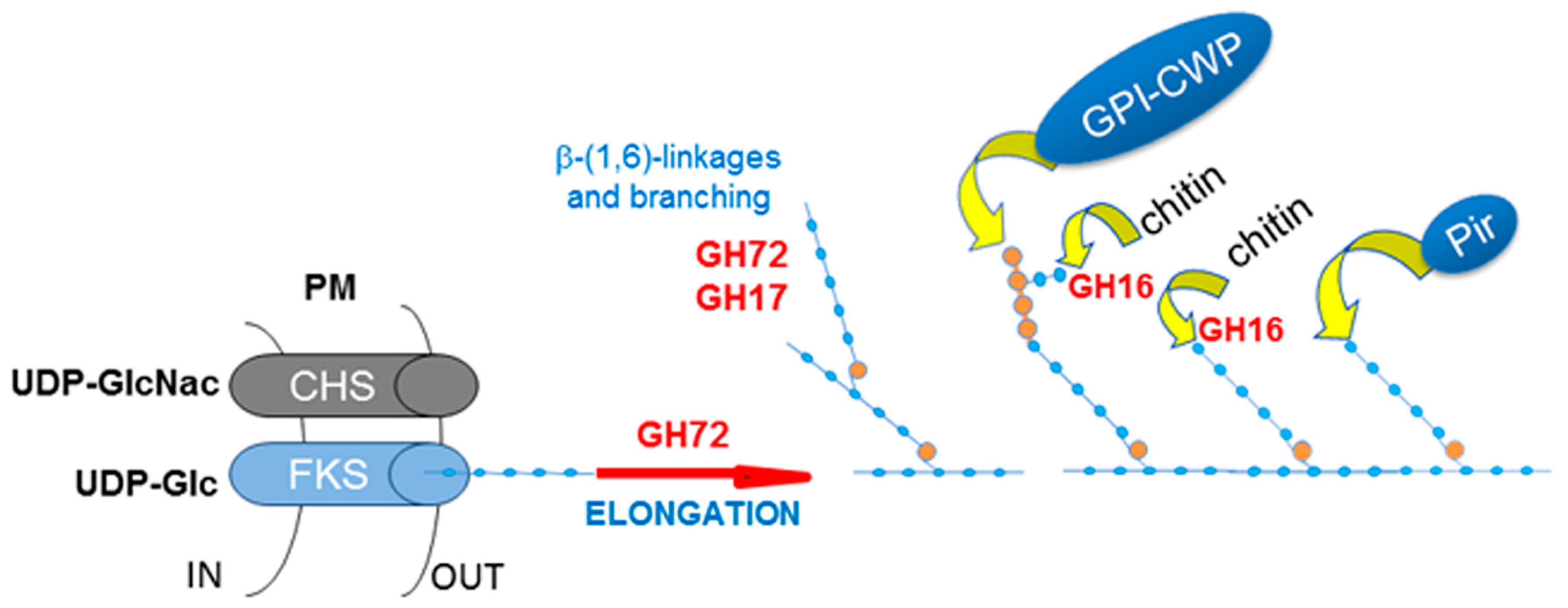
3.1.4. β-(1,3)-glucan Elongation and Branching: Cooperation between Family GH72+ (Gas/Phr/AfGel) and Family GH17 (Bgl/Bgt) Transglycosylases
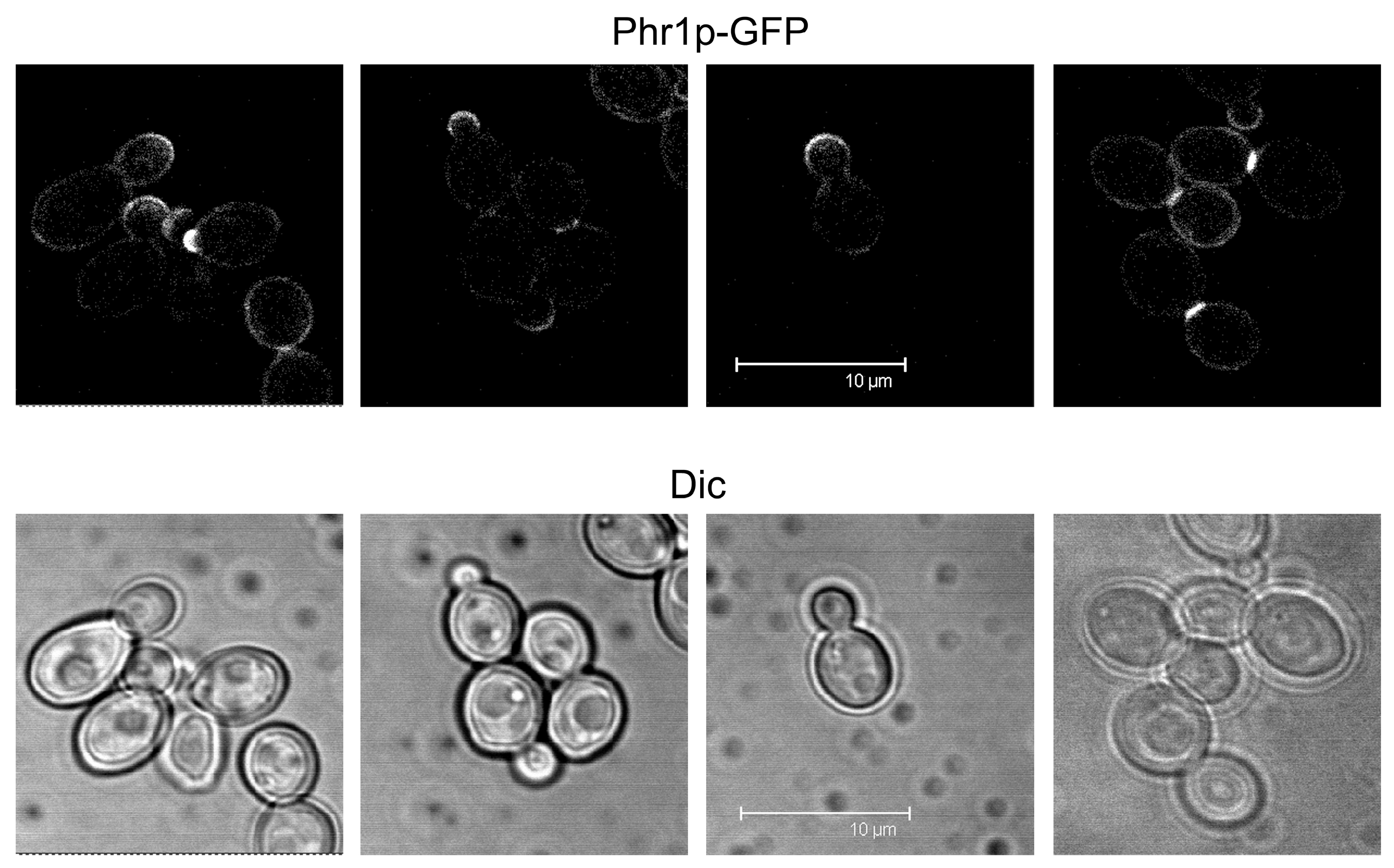
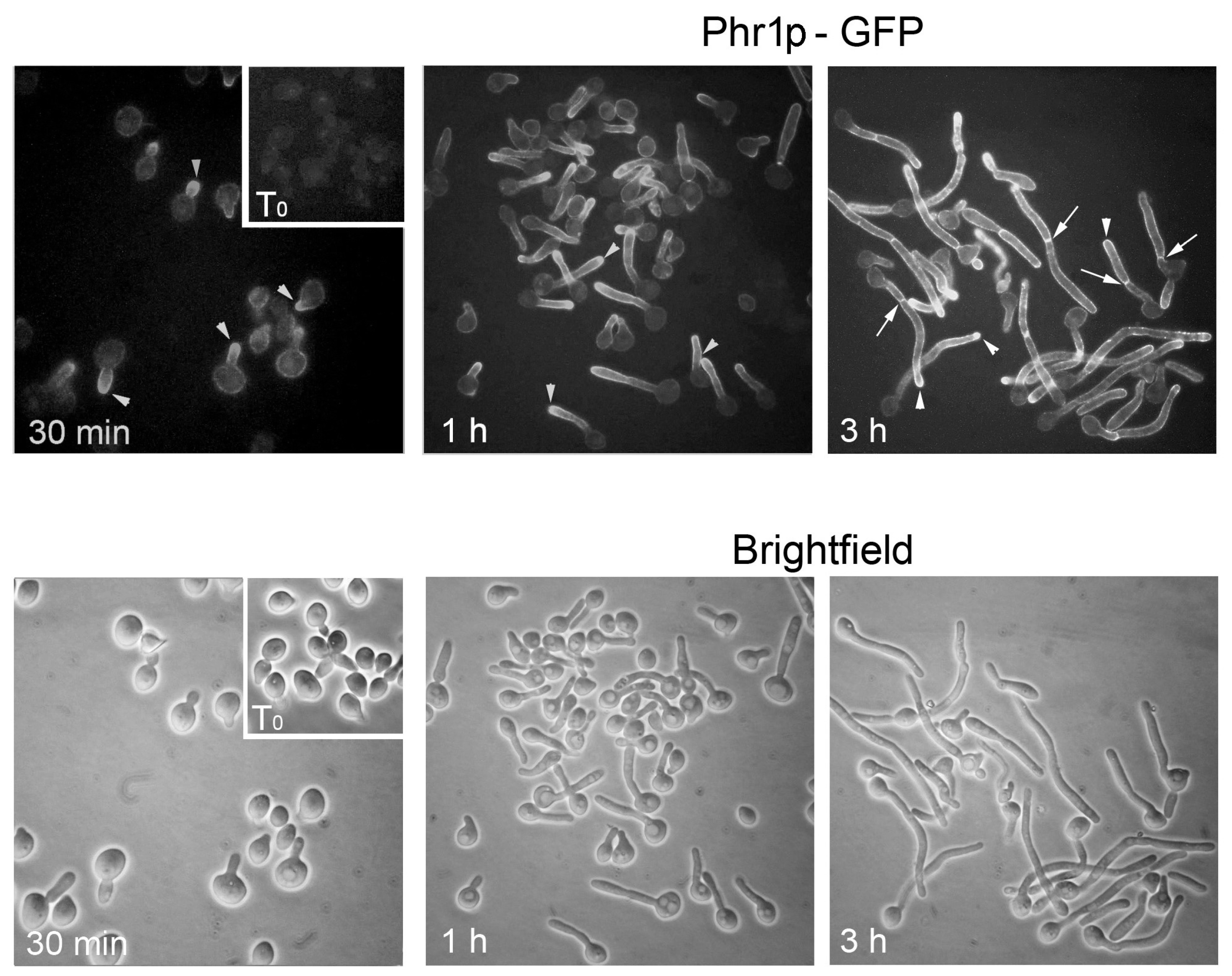
3.2. Where and When Do Phr Proteins Play Their Biological Role in C. albicans Cells?
4. PHR Family in Morphogenesis and Virulence
5. The Adaptive Response to Cell Wall Stress Induced by the Lack of Phr1p
5.1. The Response to Cell Wall Stress
5.2. The Adaptive Response in C. albicans Cells Lacking Phr1p: Protecting Cell Integrity by Preventing Cell Polarization
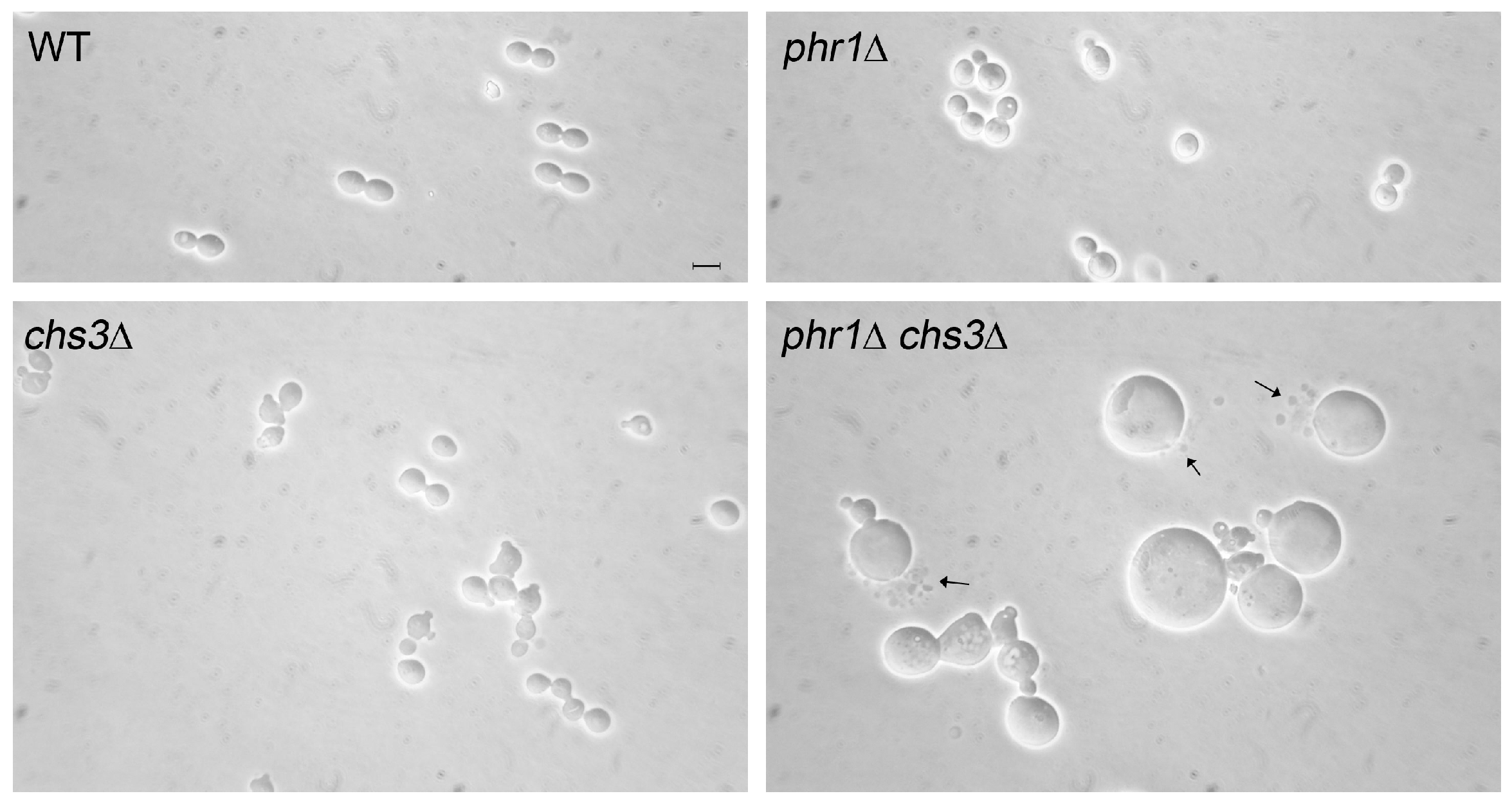
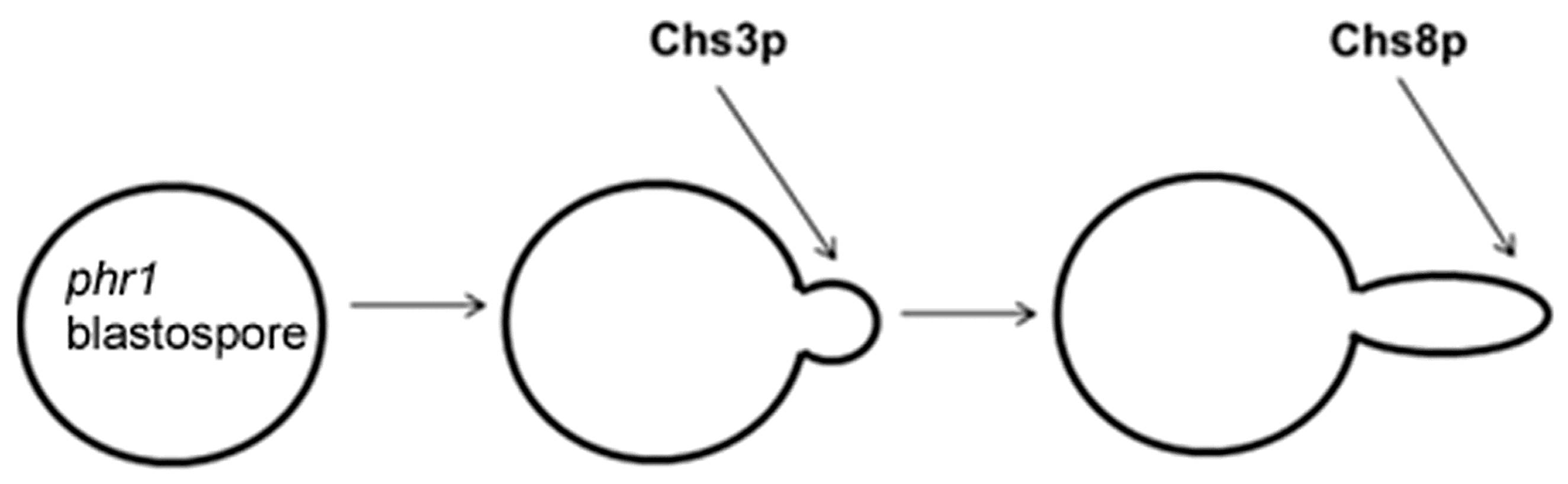
- a set of genes encoding mannoproteins PGA23, orf19.750, RBR1, PGA13, PGA54, RBT4, ECM331 and PGA6, suggesting a qualitative and quantitative change in the pattern of cell wall mannoproteins.
- CHS2 and CHS8 encoding the Class I Chitin synthases, Chs2p and Chs8p, and reported to be subject to Mkc1p and Hog1p-mediated transcriptional increase in the presence of cell wall stress.
- CHS7 encoding the ER-chaperone for Chitin synthase 3 (Chs3p), a post-translational regulated Class IV Chitin synthase.
- CRH11, a homolog of S. cerevisiae CRH1 and a member of C. albicans family GH16 trans-glycosylases together with UTR2/CSF4 and CRH12. GH16 proteins cross-link chitin with glucans by transferring the reducing end of chitin to either the non-reducing end of a β-(1,3)-glucan chain or to a β-(1,3)-glucose side-branch of a β-(1,6)-glucan chain [107] (Figure 5).
6. Conclusions and Future Outlook
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Odds, F.C. Candida and Candidosis: A Review and Bibliography, 2nd ed.; Bailliere Tindal: London, UK, 1988; p. 468. [Google Scholar]
- Pfaller, M.A.; Diekema, D.J. Epidemiology of invasive candidiasis: A persistent public health problem. Clin. Microbiol. Rev. 2007, 20, 133–163. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.D.; Denning, D.W.; Gow, N.A.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012, 4, 165rv13. [Google Scholar] [CrossRef] [PubMed]
- Latge, J.P. The cell wall: A carbohydrate armour for the fungal cell. Mol. Microbiol. 2007, 66, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Latge, J.P.; Beauvais, A. Functional duality of the cell wall. Curr. Opin. Microbiol. 2014, 20, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Calderone, R.A.; Fonzi, W.A. Virulence factors of Candida albicans. Trends Microbiol. 2001, 9, 327–335. [Google Scholar] [CrossRef]
- Douglas, C.M.; D’Ippolito, J.A.; Shei, G.J.; Meinz, M.; Onishi, J.; Marrinan, J.A.; Li, W.; Abruzzo, G.K.; Flattery, A.; Bartizal, K.; et al. Identification of the fks1 gene of Candida albicans as the essential target of 1,3-β-d-glucan synthase inhibitors. Antimicrob. Agents Chemother. 1997, 41, 2471–2479. [Google Scholar] [PubMed]
- Orlean, P. Architecture and biosynthesis of the Saccharomyces cerevisiae cell wall. Genetics 2012, 192, 775–818. [Google Scholar] [CrossRef] [PubMed]
- Popolo, L.; Alberghina, L. Identification of a labile protein involved in the g1-to-s transition in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1984, 81, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Popolo, L.; Vai, M.; Alberghina, L. Identification of a glycoprotein involved in cell cycle progression in yeast. J. Biol. Chem. 1986, 261, 3479–3482. [Google Scholar] [PubMed]
- Vai, M.; Popolo, L.; Grandori, R.; Lacana, E.; Alberghina, L. The cell cycle modulated glycoprotein gp115 is one of the major yeast proteins containing glycosylphosphatidylinositol. Biochim. Biophys. Acta 1990, 1038, 277–285. [Google Scholar] [CrossRef]
- Conzelmann, A.; Riezman, H.; Desponds, C.; Bron, C. A major 125-kd membrane glycoprotein of Saccharomyces cerevisiae is attached to the lipid bilayer through an inositol-containing phospholipid. EMBO J. 1988, 7, 2233–2240. [Google Scholar] [PubMed]
- Birse, C.E.; Irwin, M.Y.; Fonzi, W.A.; Sypherd, P.S. Cloning and characterization of ece1, a gene expressed in association with cell elongation of the dimorphic pathogen Candida albicans. Infect. Immun. 1993, 61, 3648–3655. [Google Scholar] [PubMed]
- Saporito-Irwin, S.M.; Birse, C.E.; Sypherd, P.S.; Fonzi, W.A. Phr1, a pH-regulated gene of Candida albicans, is required for morphogenesis. Mol. Cell. Biol. 1995, 15, 601–613. [Google Scholar] [CrossRef] [PubMed]
- Vai, M.; Orlandi, I.; Cavadini, P.; Alberghina, L.; Popolo, L. Candida albicans homologue of ggp1/gas1 gene is functional in Saccharomyces cerevisiae and contains the determinants for glycosylphosphatidylinositol attachment. Yeast 1996, 12, 361–368. [Google Scholar] [CrossRef]
- Mühlschlegel, F.A.; Fonzi, W.A. Phr2 of Candida albicans encodes a functional homolog of the pH-regulated gene phr1 with an inverted pattern of pH-dependent expression. Mol. Cell. Biol. 1997, 17, 5960–5967. [Google Scholar] [CrossRef] [PubMed]
- Eckert, S.E.; Heinz, W.J.; Zakikhany, K.; Thewes, S.; Haynes, K.; Hube, B.; Muhlschlegel, F.A. Pga4, a gas homologue from Candida albicans, is up-regulated early in infection processes. Fungal. Genet. Biol. 2007, 44, 368–377. [Google Scholar] [CrossRef] [PubMed]
- De Groot, P.W.; Hellingwerf, K.J.; Klis, F.M. Genome-wide identification of fungal GPI proteins. Yeast 2003, 20, 781–796. [Google Scholar] [CrossRef] [PubMed]
- Ramon, A.M.; Porta, A.; Fonzi, W.A. Effect of environmental ph on morphological development of Candida albicans is mediated via the PACC-related transcription factor encoded by prr2. J. Bacteriol. 1999, 181, 7524–7530. [Google Scholar] [PubMed]
- Ramon, A.M.; Fonzi, W.A. Diverged binding specificity of rim101p, the Candida albicans ortholog of pacc. Eukaryot. Cell 2003, 2, 718–728. [Google Scholar] [CrossRef] [PubMed]
- Porta, A.; Wang, Z.; Ramon, A.M.; Mühlschlegel, F.A.; Fonzi, W.A. Spontaneous second site suppressors of the filamentation defect of prr1d mutants define a critical domain of rim101p. Mol. Genet. Genom. 2001, 266, 624–631. [Google Scholar] [CrossRef] [PubMed]
- Davis, D.; Wilson, R.B.; Mitchell, A.P. Rim101-dependent and-independent pathways govern ph responses in Candida albicans. Mol. Cell. Biol. 2000, 20, 971–978. [Google Scholar] [CrossRef] [PubMed]
- Baek, Y.U.; Martin, S.J.; Davis, D.A. Evidence for novel ph-dependent regulation of Candida albicans rim101, a direct transcriptional repressor of the cell wall β-glycosidase phr2. Eukaryot. Cell 2006, 5, 1550–1559. [Google Scholar] [CrossRef] [PubMed]
- Sosinska, G.J.; de Koning, L.J.; de Groot, P.W.; Manders, E.M.; Dekker, H.L.; Hellingwerf, K.J.; de Koster, C.G.; Klis, F.M. Mass spectrometric quantification of the adaptations in the wall proteome of Candida albicans in response to ambient ph. Microbiology 2011, 157, 136–146. [Google Scholar] [CrossRef] [PubMed]
- Fonzi, W.A. Phr1 and phr2 of Candida albicans encode putative glycosidases required for proper cross-linking of β-1,3- and β-1,6-glucans. J. Bacteriol. 1999, 181, 7070–7079. [Google Scholar] [PubMed]
- Samalova, M.; Melida, H.; Vilaplana, F.; Bulone, V.; Soanes, D.M.; Talbot, N.J.; Gurr, S.J. The β-1,3-glucanosyltransferases (gels) affect the structure of the rice blast fungal cell wall during appressorium-mediated plant infection. Cell Microbiol. 2017, 19. [Google Scholar] [CrossRef] [PubMed]
- Sillo, F.; Gissi, C.; Chignoli, D.; Ragni, E.; Popolo, L.; Balestrini, R. Expression and phylogenetic analyses of the gel/gas proteins of tuber melanosporum provide insights into the function and evolution of glucan remodeling enzymes in fungi. Fungal. Genet. Biol. 2013, 53, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Ao, J.; Free, S.J. Genetic and biochemical characterization of the gh72 family of cell wall transglycosylases in neurospora crassa. Fungal. Genet. Biol. 2017, 101, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Rolli, E.; Ragni, E.; de Medina-Redondo, M.; Arroyo, J.; de Aldana, C.R.; Popolo, L. Expression, stability, and replacement of glucan-remodeling enzymes during developmental transitions in Saccharomyces cerevisiae. Mol. Biol. Cell. 2011, 22, 1585–1598. [Google Scholar] [CrossRef] [PubMed]
- De Medina-Redondo, M.; Arnaiz-Pita, Y.; Clavaud, C.; Fontaine, T.; del Rey, F.; Latge, J.P.; Vazquez de Aldana, C.R. β(1,3)-glucanosyl-transferase activity is essential for cell wall integrity and viability of schizosaccharomyces pombe. PLoS ONE 2010, 5, e14046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Medina-Redondo, M.; Arnaiz-Pita, Y.; Fontaine, T.; Del Rey, F.; Latge, J.P.; Vazquez de Aldana, C.R. The β-1,3-glucanosyltransferase GAS4P is essential for ascospore wall maturation and spore viability in schizosaccharomyces pombe. Mol. Microbiol. 2008, 68, 1283–1299. [Google Scholar] [CrossRef] [PubMed]
- Mouyna, I.; Morelle, W.; Vai, M.; Monod, M.; Lechenne, B.; Fontaine, T.; Beauvais, A.; Sarfati, J.; Prevost, M.C.; Henry, C.; et al. Deletion of gel2 encoding for a β(1–3)glucanosyltransferase affects morphogenesis and virulence in Aspergillus fumigatus. Mol. Microbiol. 2005, 56, 1675–1688. [Google Scholar] [CrossRef] [PubMed]
- Mouyna, I.; Fontaine, T.; Vai, M.; Monod, M.; Fonzi, W.A.; Diaquin, M.; Popolo, L.; Hartland, R.P.; Latge, J.P. Glycosylphosphatidylinositol-anchored glucanosyltransferases play an active role in the biosynthesis of the fungal cell wall. J. Biol. Chem. 2000, 275, 14882–14889. [Google Scholar] [CrossRef] [PubMed]
- Lesage, G.; Bussey, H. Cell wall assembly in saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2006, 70, 317–343. [Google Scholar] [CrossRef] [PubMed]
- Mazan, M.; Ragni, E.; Popolo, L.; Farkas, V. Catalytic properties of the gas family β-(1,3)-glucanosyltransferases active in fungal cell-wall biogenesis as determined by a novel fluorescent assay. Biochem. J. 2011, 438, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Kovacova, K.; Degani, G.; Stratilova, E.; Farkas, V.; Popolo, L. Catalytic properties of phr family members of cell wall glucan remodeling enzymes: Implications for the adaptation of Candida albicans to ambient ph. FEMS Yeast Res 2015, 15. [Google Scholar] [CrossRef] [PubMed]
- Hartland, R.P.; Fontaine, T.; Debeaupuis, J.P.; Simenel, C.; Delepierre, M.; Latge, J.P. A novel β-(1–3)-glucanosyltransferase from the cell wall of Aspergillus fumigatus. J. Biol. Chem. 1996, 271, 26843–26849. [Google Scholar] [CrossRef] [PubMed]
- Henriksson, H.; Denman, S.E.; Campuzano, I.D.; Ademark, P.; Master, E.R.; Teeri, T.T.; Brumer, H., 3rd. N-linked glycosylation of native and recombinant cauliflower xyloglucan endotransglycosylase 16a. Biochem. J. 2003, 375, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Raich, L.; Borodkin, V.; Fang, W.; Castro-Lopez, J.; van Aalten, D.M.; Hurtado-Guerrero, R.; Rovira, C. A trapped covalent intermediate of a glycoside hydrolase on the pathway to transglycosylation. Insights from experiments and quantum mechanics/molecular mechanics simulations. J. Am. Chem. Soc. 2016, 138, 3325–3332. [Google Scholar] [CrossRef] [PubMed]
- Ragni, E.; Fontaine, T.; Gissi, C.; Latge, J.P.; Popolo, L. The gas family of proteins of saccharomyces cerevisiae: Characterization and evolutionary analysis. Yeast 2007, 24, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Gatti, E.; Popolo, L.; Vai, M.; Rota, N.; Alberghina, L. O-linked oligosaccharides in yeast glycosyl phosphatidylinositol-anchored protein gp115 are clustered in a serine-rich region not essential for its function. J. Biol. Chem. 1994, 269, 19695–19700. [Google Scholar] [PubMed]
- Popolo, L.; Ragni, E.; Carotti, C.; Palomares, O.; Aardema, R.; Back, J.W.; Dekker, H.L.; de Koning, L.J.; de Jong, L.; de Koster, C.G. Disulfide bond structure and domain organization of yeast β(1,3)-glucanosyltransferases involved in cell wall biogenesis. J. Biol. Chem. 2008, 283, 18553–18565. [Google Scholar] [CrossRef] [PubMed]
- Hurtado-Guerrero, R.; Schuttelkopf, A.W.; Mouyna, I.; Ibrahim, A.F.; Shepherd, S.; Fontaine, T.; Latge, J.P.; van Aalten, D.M. Molecular mechanisms of yeast cell wall glucan remodelling. J. Biol. Chem. 2008, 284, 8461–8469. [Google Scholar] [CrossRef] [PubMed]
- Rolli, E.; Ragni, E.; Rodriguez-Pena, J.M.; Arroyo, J.; Popolo, L. Gas3, a developmentally regulated gene, encodes a highly mannosylated and inactive protein of the gas family of Saccharomyces cerevisiae. Yeast 2010, 27, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Ragni, E.; Coluccio, A.; Rolli, E.; Rodriguez-Pena, J.M.; Colasante, G.; Arroyo, J.; Neiman, A.M.; Popolo, L. Gas2 and gas4, a pair of developmentally regulated genes required for spore wall assembly in Saccharomyces cerevisiae. Eukaryot. Cell 2007, 6, 302–316. [Google Scholar] [CrossRef] [PubMed]
- Sarthy, A.V.; McGonigal, T.; Coen, M.; Frost, D.J.; Meulbroek, J.A.; Goldman, R.C. Phenotype in Candida albicans of a disruption of the Bgl2 gene encoding a 1,3-β-glucosyltransferase. Microbiology 1997, 143, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, R.; Takada, A.; Iwatani, S.; Oka, C.; Kitamoto, T.; Kajiwara, S. The role of bgl2p in the transition to filamentous cells during biofilm formation by Candida albicans. Mycoses 2017, 60, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Mouyna, I.; Hartland, R.P.; Fontaine, T.; Diaquin, M.; Simenel, C.; Delepierre, M.; Henrissat, B.; Latge, J.P. A 1,3-β-glucanosyltransferase isolated from the cell wall of Aspergillus fumigatus is a homologue of the yeast bgl2p. Microbiology 1998, 144, 3171–3180. [Google Scholar] [CrossRef] [PubMed]
- Goldman, R.C.; Sullivan, P.A.; Zakula, D.; Capobianco, J.O. Kinetics of β-1,3 glucan interaction at the donor and acceptor sites of the fungal glucosyltransferase encoded by the bgl2 gene. Eur. J. Biochem. 1995, 227, 372–378. [Google Scholar] [CrossRef] [PubMed]
- Aimanianda, V.; Simenel, C.; Garnaud, C.; Clavaud, C.; Tada, R.; Barbin, L.; Mouyna, I.; Heddergott, C.; Popolo, L.; Ohya, Y.; et al. The dual activity responsible for the elongation and branching of β-(1,3)-glucan in the fungal cell wall. MBIO 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Cabib, E.; Blanco, N.; Arroyo, J. Presence of a large β(1–3)glucan linked to chitin at the Saccharomyces cerevisiae mother-bud neck suggests involvement in localized growth control. Eukaryot. Cell 2012, 11, 388–400. [Google Scholar] [CrossRef] [PubMed]
- Ecker, M.; Deutzmann, R.; Lehle, L.; Mrsa, V.; Tanner, W. Pir proteins of Saccharomyces cerevisiae are attached to β-1,3-glucan by a new protein-carbohydrate linkage. J. Biol. Chem. 2006, 281, 11523–11529. [Google Scholar] [CrossRef] [PubMed]
- Klis, F.M.; Sosinska, G.J.; de Groot, P.W.; Brul, S. Covalently linked cell wall proteins of Candida albicans and their role in fitness and virulence. FEMS Yeast Res. 2009, 9, 1013–1028. [Google Scholar] [CrossRef] [PubMed]
- De Groot, P.W.; de Boer, A.D.; Cunningham, J.; Dekker, H.L.; de Jong, L.; Hellingwerf, K.J.; de Koster, C.; Klis, F.M. Proteomic analysis of Candida albicans cell walls reveals covalently bound carbohydrate-active enzymes and adhesins. Eukaryot. Cell 2004, 3, 955–965. [Google Scholar] [CrossRef] [PubMed]
- Heilmann, C.J.; Sorgo, A.G.; Mohammadi, S.; Sosinska, G.J.; de Koster, C.G.; Brul, S.; de Koning, L.J.; Klis, F.M. Surface stress induces a conserved cell wall stress response in the pathogenic fungus Candida albicans. Eukaryot. Cell 2013, 12, 254–264. [Google Scholar] [CrossRef] [PubMed]
- Marin, E.; Parra-Giraldo, C.M.; Hernandez-Haro, C.; Hernaez, M.L.; Nombela, C.; Monteoliva, L.; Gil, C. Candida albicans shaving to profile human serum proteins on hyphal surface. Front. Microbiol. 2015, 6, 1343. [Google Scholar] [CrossRef] [PubMed]
- Ragni, E.; Calderon, J.; Fascio, U.; Sipiczki, M.; Fonzi, W.A.; Popolo, L. Phr1p, a glycosylphosphatidylinsitol-anchored β(1,3)-glucanosyltransferase critical for hyphal wall formation, localizes to the apical growth sites and septa in Candida albicans. Fungal. Genet. Biol. 2011, 48, 793–805. [Google Scholar] [CrossRef] [PubMed]
- Rolli, E.; Ragni, E.; Calderon, J.; Porello, S.; Fascio, U.; Popolo, L. Immobilization of the glycosylphosphatidylinositol-anchored gas1 protein into the chitin ring and septum is required for proper morphogenesis in yeast. Mol. Biol. Cell 2009, 20, 4856–4870. [Google Scholar] [CrossRef] [PubMed]
- Hopke, A.; Nicke, N.; Hidu, E.E.; Degani, G.; Popolo, L.; Wheeler, R.T. Neutrophil attack triggers extracellular trap-dependent Candida cell wall remodeling and altered immune recognition. PLoS Pathog. 2016, 12, e1005644. [Google Scholar] [CrossRef] [PubMed]
- Popolo, L.; Vai, M.; Gatti, E.; Porello, S.; Bonfante, P.; Balestrini, R.; Alberghina, L. Physiological analysis of mutants indicates involvement of the Saccharomyces cerevisiae gpi-anchored protein gp115 in morphogenesis and cell separation. J. Bacteriol. 1993, 175, 1879–1885. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.M.; Kauffman, S.J.; Hauser, M.; Huang, L.; Lin, M.; Sillaots, S.; Jiang, B.; Xu, D.; Roemer, T. Pathway analysis of Candida albicans survival and virulence determinants in a murine infection model. Proc. Natl. Acad. Sci. USA 2010, 107, 22044–22049. [Google Scholar] [CrossRef] [PubMed]
- Popolo, L.; Vai, M. Defects in assembly of the extracellular matrix are responsible for altered morphogenesis of a Candida albicans phr1 mutant. J. Bacteriol. 1998, 180, 163–166. [Google Scholar] [PubMed]
- Plaine, A.; Walker, L.; Da Costa, G.; Mora-Montes, H.M.; McKinnon, A.; Gow, N.A.; Gaillardin, C.; Munro, C.A.; Richard, M.L. Functional analysis of Candida albicans gpi-anchored proteins: Roles in cell wall integrity and caspofungin sensitivity. Fungal. Genet. Biol. 2008, 45, 1404–1414. [Google Scholar] [CrossRef] [PubMed]
- Ragni, E.; Piberger, H.; Neupert, C.; Garcia-Cantalejo, J.; Popolo, L.; Arroyo, J.; Aebi, M.; Strahl, S. The genetic interaction network of ccw12, a Saccharomyces cerevisiae gene required for cell wall integrity during budding and formation of mating projections. BMC Genom. 2011, 12, 107. [Google Scholar] [CrossRef] [PubMed]
- De Bernardis, F.; Mühlschlegel, F.A.; Cassone, A.; Fonzi, W.A. The ph of the host niche controls gene expression in and virulence of Candida albicans. Infect. Immun. 1998, 66, 3317–3325. [Google Scholar] [PubMed]
- Ghannoum, M.A.; Spellberg, B.; Saporito-Irwin, S.M.; Fonzi, W.A. Reduced virulence of Candida albicans phr1 mutants. Infect. Immun. 1995, 63, 4528–4530. [Google Scholar] [PubMed]
- Zakikhany, K.; Naglik, J.R.; Schmidt-Westhausen, A.; Holland, G.; Schaller, M.; Hube, B. In vivo transcript profiling of Candida albicans identifies a gene essential for interepithelial dissemination. Cell. Microbiol. 2007, 9, 2938–2954. [Google Scholar] [CrossRef] [PubMed]
- Fanning, S.; Xu, W.; Solis, N.; Woolford, C.A.; Filler, S.G.; Mitchell, A.P. Divergent targets of Candida albicans biofilm regulator bcr1 in vitro and in vivo. Eukaryot. Cell 2012, 11, 896–904. [Google Scholar] [CrossRef] [PubMed]
- Thewes, S.; Kretschmar, M.; Park, H.; Schaller, M.; Filler, S.G.; Hube, B. In vivo and ex vivo comparative transcriptional profiling of invasive and non-invasive Candida albicans isolates identifies genes associated with tissue invasion. Mol. Microbiol. 2007, 63, 1606–1628. [Google Scholar] [CrossRef] [PubMed]
- Walker, L.A.; Maccallum, D.M.; Bertram, G.; Gow, N.A.; Odds, F.C.; Brown, A.J. Genome-wide analysis of Candida albicans gene expression patterns during infection of the mammalian kidney. Fungal. Genet. Biol. 2009, 46, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Rosenbach, A.; Dignard, D.; Pierce, J.V.; Whiteway, M.; Kumamoto, C.A. Adaptations of Candida albicans for growth in the mammalian intestinal tract. Eukaryot. Cell 2010, 9, 1075–1086. [Google Scholar] [CrossRef] [PubMed]
- Chao, C.C.; Hsu, P.C.; Jen, C.F.; Chen, I.H.; Wang, C.H.; Chan, H.C.; Tsai, P.W.; Tung, K.C.; Wang, C.H.; Lan, C.Y.; et al. Zebrafish as a model host for Candida albicans infection. Infect. Immun. 2010, 78, 2512–2521. [Google Scholar] [CrossRef] [PubMed]
- Larsen, B.; Markovetz, A.J.; Galask, R.P. The bacterial flora of the female rat genital tract. Proc. Soc. Exp. Biol. Med. 1976, 151, 571–574. [Google Scholar] [CrossRef] [PubMed]
- Calderon, J.; Zavrel, M.; Ragni, E.; Fonzi, W.A.; Rupp, S.; Popolo, L. Phr1, a ph-regulated gene of Candida albicans encoding a glucan remodeling enzyme, is required for adhesion and invasion. Microbiology 2010, 156, 2484–2494. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, R.T.; Fink, G.R. A drug-sensitive genetic network masks fungi from the immune system. PLoS Pathog. 2006, 2. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, R.T.; Kombe, D.; Agarwala, S.D.; Fink, G.R. Dynamic, morphotype-specific Candida albicans β-glucan exposure during infection and drug treatment. PLoS Pathog. 2008, 4. [Google Scholar] [CrossRef] [PubMed]
- Gulati, M.; Nobile, C.J. Candida albicans biofilms: Development, regulation, and molecular mechanisms. Microb. Infect. 2016, 18, 310–321. [Google Scholar] [CrossRef] [PubMed]
- Al-Fattani, M.A.; Douglas, L.J. Biofilm matrix of Candida albicans and Candida tropicalis: Chemical composition and role in drug resistance. J. Med. Microbiol. 2006, 55, 999–1008. [Google Scholar] [CrossRef] [PubMed]
- Murillo, L.A.; Newport, G.; Lan, C.Y.; Habelitz, S.; Dungan, J.; Agabian, N.M. Genome-wide transcription profiling of the early phase of biofilm formation by Candida albicans. Eukaryot. Cell 2005, 4, 1562–1573. [Google Scholar] [CrossRef] [PubMed]
- Nett, J.E.; Lepak, A.J.; Marchillo, K.; Andes, D.R. Time course global gene expression analysis of an in vivo Candida biofilm. J. Infect. Dis. 2009, 200, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Taff, H.T.; Nett, J.E.; Zarnowski, R.; Ross, K.M.; Sanchez, H.; Cain, M.T.; Hamaker, J.; Mitchell, A.P.; Andes, D.R. A Candida biofilm-induced pathway for matrix glucan delivery: Implications for drug resistance. PLoS Pathog. 2012, 8. [Google Scholar] [CrossRef] [PubMed]
- Cabral, V.; Znaidi, S.; Walker, L.A.; Martin-Yken, H.; Dague, E.; Legrand, M.; Lee, K.; Chauvel, M.; Firon, A.; Rossignol, T.; et al. Targeted changes of the cell wall proteome influence Candida albicans ability to form single- and multi-strain biofilms. PLoS Pathog. 2014, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Norice, C.T.; Smith, F.J., Jr.; Solis, N.; Filler, S.G.; Mitchell, A.P. Requirement for Candida albicans sun41 in biofilm formation and virulence. Eukaryot. Cell 2007, 6, 2046–2055. [Google Scholar] [CrossRef] [PubMed]
- Soll, D.R. Why does Candida albicans switch? FEMS Yeast Res. 2009, 9, 973–989. [Google Scholar] [CrossRef] [PubMed]
- Sahni, N.; Yi, S.; Daniels, K.J.; Srikantha, T.; Pujol, C.; Soll, D.R. Genes selectively up-regulated by pheromone in white cells are involved in biofilm formation in Candida albicans. PLoS Pathog. 2009, 5. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Cao, C.; Jia, W.; Tao, L.; Guan, G.; Huang, G. Ph regulates white-opaque switching and sexual mating in Candida albicans. Eukaryot. Cell 2015, 14, 1127–1134. [Google Scholar] [CrossRef] [PubMed]
- Noble, S.M.; French, S.; Kohn, L.A.; Chen, V.; Johnson, A.D. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet. 2010, 42, 590–598. [Google Scholar] [CrossRef] [PubMed]
- Suwunnakorn, S.; Wakabayashi, H.; Rustchenko, E. Chromosome 5 of human pathogen Candida albicans carries multiple genes for negative control of caspofungin and anidulafungin susceptibility. Antimicrob. Agents Chemother. 2016, 60, 7457–7467. [Google Scholar] [PubMed]
- Ene, I.V.; Adya, A.K.; Wehmeier, S.; Brand, A.C.; MacCallum, D.M.; Gow, N.A.; Brown, A.J. Host carbon sources modulate cell wall architecture, drug resistance and virulence in a fungal pathogen. Cell. Microbiol. 2012, 14, 1319–1335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mulhern, S.M.; Logue, M.E.; Butler, G. Candida albicans transcription factor ace2 regulates metabolism and is required for filamentation in hypoxic conditions. Eukaryot. Cell 2006, 5, 2001–2013. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.; Fung, E.; Schlecht, U.; Davis, R.W.; Giaever, G.; St Onge, R.P.; Deutschbauer, A.; Nislow, C. Gene annotation and drug target discovery in Candida albicans with a tagged transposon mutant collection. PLoS Pathog. 2010, 6. [Google Scholar] [CrossRef] [PubMed]
- Walker, L.A.; Gow, N.A.; Munro, C.A. Fungal echinocandin resistance. Fungal. Genet. Biol. 2010, 47, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, K.W. Acidic calcium stores of Saccharomyces cerevisiae. Cell Calcium 2011, 50, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Dichtl, K.; Samantaray, S.; Wagener, J. Cell wall integrity signalling in human pathogenic fungi. Cell Microbiol. 2016, 18, 1228–1238. [Google Scholar] [CrossRef] [PubMed]
- Kumamoto, C.A. A contact-activated kinase signals Candida albicans invasive growth and biofilm development. Proc. Natl. Acad. Sci. USA 2005, 102, 5576–5581. [Google Scholar] [CrossRef] [PubMed]
- De Dios, C.H.; Roman, E.; Monge, R.A.; Pla, J. The role of mapk signal transduction pathways in the response to oxidative stress in the fungal pathogen Candida albicans: Implications in virulence. Curr. Protein Pept. Sci. 2010, 11, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Eisman, B.; Alonso-Monge, R.; Roman, E.; Arana, D.; Nombela, C.; Pla, J. The cek1 and hog1 mitogen-activated protein kinases play complementary roles in cell wall biogenesis and chlamydospore formation in the fungal pathogen Candida albicans. Eukaryot. Cell 2006, 5, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Hou, Y.; Liu, W.; Lu, C.; Wang, W.; Sun, S. Components of the calcium-calcineurin signaling pathway in fungal cells and their potential as antifungal targets. Eukaryot. Cell 2015, 14, 324–334. [Google Scholar] [CrossRef] [PubMed]
- Garcia, R.; Bravo, E.; Diez-Muniz, S.; Nombela, C.; Rodriguez-Pena, J.M.; Arroyo, J. A novel connection between the cell wall integrity and the pka pathways regulates cell wall stress response in yeast. Sci. Rep. 2017, 7, 5703. [Google Scholar] [CrossRef] [PubMed]
- Ha, C.W.; Kim, K.; Chang, Y.J.; Kim, B.; Huh, W.K. The β-1,3-glucanosyltransferase gas1 regulates sir2-mediated rdna stability in Saccharomyces cerevisiae. Nucleic Acids Res. 2014, 42, 8486–8499. [Google Scholar] [CrossRef] [PubMed]
- Ai, W.; Bertram, P.G.; Tsang, C.K.; Chan, T.F.; Zheng, X.F. Regulation of subtelomeric silencing during stress response. Mol. Cell. 2002, 10, 1295–1305. [Google Scholar] [CrossRef]
- Sanz, A.B.; Garcia, R.; Rodriguez-Pena, J.M.; Diez-Muniz, S.; Nombela, C.; Peterson, C.L.; Arroyo, J. Chromatin remodeling by the SWI/SNF complex is essential for transcription mediated by the yeast cell wall integrity mapk pathway. Mol. Biol. Cell. 2012, 23, 2805–2817. [Google Scholar] [CrossRef] [PubMed]
- Koch, M.R.; Pillus, L. The glucanosyltransferase gas1 functions in transcriptional silencing. Proc. Natl. Acad. Sci. USA 2009, 106, 11224–11229. [Google Scholar] [CrossRef] [PubMed]
- Lagorce, A.; Hauser, N.C.; Labourdette, D.; Rodriguez, C.; Martin-Yken, H.; Arroyo, J.; Hoheisel, J.D.; Francois, J. Genome-wide analysis of the response to cell wall mutations in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 2003, 278, 20345–20357. [Google Scholar] [CrossRef] [PubMed]
- Badrane, H.; Nguyen, M.H.; Clancy, C.J. Highly dynamic and specific phosphatidylinositol 4,5-bisphosphate, septin, and cell wall integrity pathway responses correlate with caspofungin activity against Candida albicans. Antimicrob. Agents Chemother. 2016, 60, 3591–3600. [Google Scholar] [CrossRef] [PubMed]
- Degani, G.; Ragni, E.; Botias, P.; Ravasio, D.; Calderon, J.; Pianezzola, E.; Rodriguez-Pena, J.M.; Vanoni, M.A.; Arroyo, J.; Fonzi, W.A.; et al. Genomic and functional analyses unveil the response to hyphal wall stress in Candida albicans cells lacking β(1,3)-glucan remodeling. BMC Genom. 2016, 17, 482. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.; Farkas, V.; Sanz, A.B.; Cabib, E. Strengthening the fungal cell wall through chitin-glucan cross-links: Effects on morphogenesis and cell integrity. Cell Microbiol. 2016, 18, 1239–1250. [Google Scholar] [CrossRef] [PubMed]
- Rigamonti, M.; Groppi, S.; Belotti, F.; Ambrosini, R.; Filippi, G.; Martegani, E.; Tisi, R. Hypotonic stress-induced calcium signaling in Saccharomyces cerevisiae involves trp-like transporters on the endoplasmic reticulum membrane. Cell Calcium 2015, 57, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Gastebois, A.; Fontaine, T.; Latge, J.P.; Mouyna, I. Β(1–3)glucanosyltransferase gel4p is essential for Aspergillus fumigatus. Eukaryot. Cell 2010, 9, 1294–1298. [Google Scholar] [CrossRef] [PubMed]
- Weig, M.; Haynes, K.; Rogers, T.R.; Kurzai, O.; Frosch, M.; Muhlschlegel, F.A. A gas-like gene family in the pathogenic fungus Candida glabrata. Microbiology 2001, 147, 2007–2019. [Google Scholar] [CrossRef] [PubMed]
- Roemer, T.; Jiang, B.; Davison, J.; Ketela, T.; Veillette, K.; Breton, A.; Tandia, F.; Linteau, A.; Sillaots, S.; Marta, C.; et al. Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol. Microbiol. 2003, 50, 167–181. [Google Scholar] [CrossRef] [PubMed]
- Kurzai, O.; Heinz, W.J.; Sullivan, D.J.; Coleman, D.C.; Frosch, M.; Muhlschlegel, F.A. Rapid pcr test for discriminating between Candida albicans and Candida dubliniensis isolates using primers derived from the ph-regulated phr1 and phr2 genes of c. Albicans. J. Clin. Microbiol. 1999, 37, 1587–1590. [Google Scholar] [PubMed]
- Arroyo, J.; Sarfati, J.; Baixench, M.T.; Ragni, E.; Guillen, M.; Rodriguez-Pena, J.M.; Popolo, L.; Latge, J.P. The gpi-anchored gas and crh families are fungal antigens. Yeast 2007, 24, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Bozza, S.; Clavaud, C.; Giovannini, G.; Fontaine, T.; Beauvais, A.; Sarfati, J.; D’Angelo, C.; Perruccio, K.; Bonifazi, P.; Zagarella, S.; et al. Immune sensing of Aspergillus fumigatus proteins, glycolipids, and polysaccharides and the impact on th immunity and vaccination. J. Immunol. 2009, 183, 2407–2414. [Google Scholar] [CrossRef] [PubMed]

| Phr1p | Phr2p | |
|---|---|---|
| Activity in vitro | Endo β-(1,3)-glucanase/transglysosylase | Endo β-(1,3)-glucanase/transglysosylase |
| Temperature optimum | 30 °C | 30 °C |
| pH optimum | 5.8 | 3 |
| Half-time of activity decay at 95 °C | 10 min | 15 min |
| Minimum length of the donor 2 | rG9 | rG10 |
| Preferred acceptor length | ≥L7 | ≥L7 |
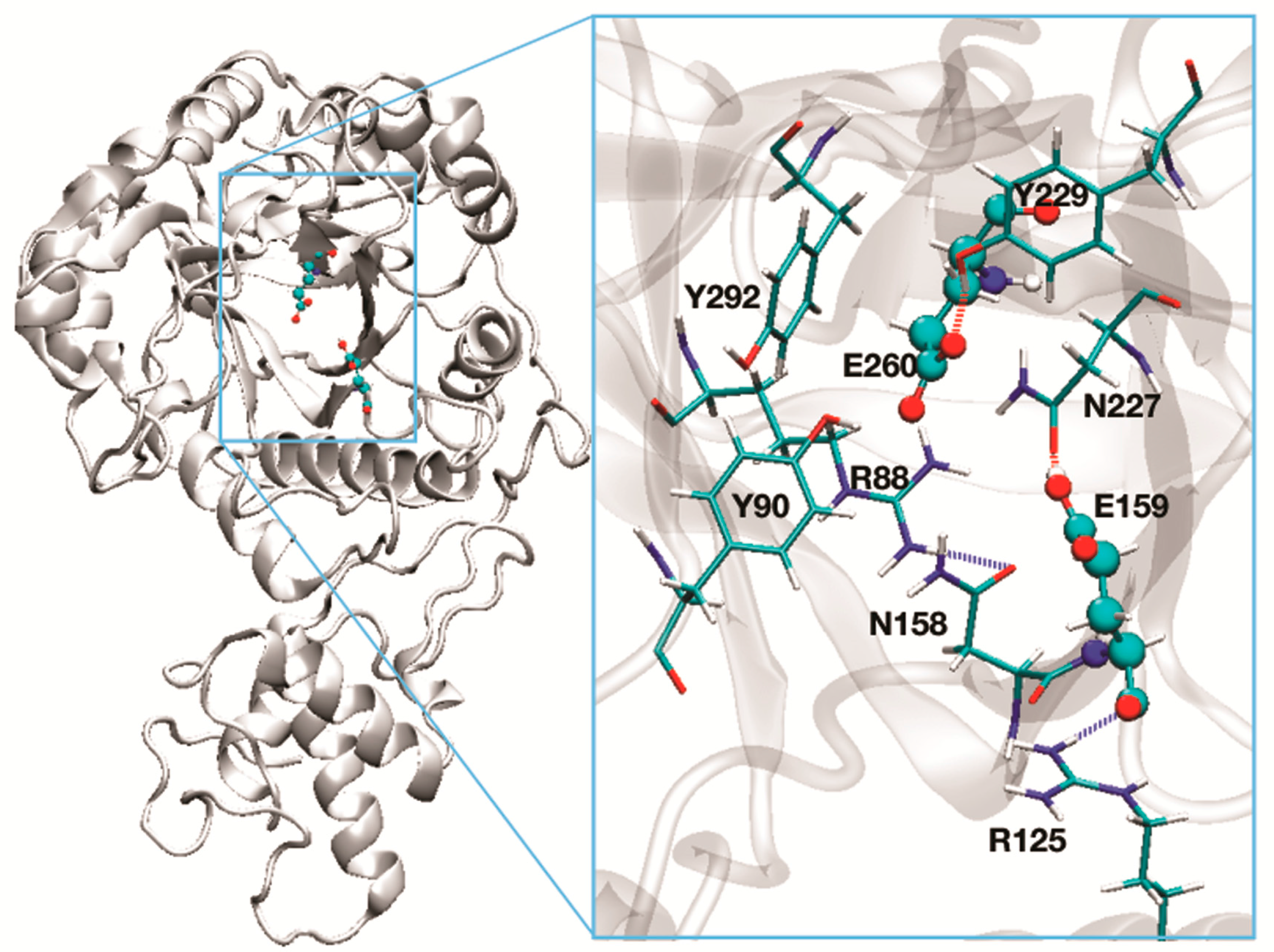
| Residues essential for catalysis 3 | Glu169 and Glu270 | Glu159 and Glu260 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Popolo, L.; Degani, G.; Camilloni, C.; Fonzi, W.A. The PHR Family: The Role of Extracellular Transglycosylases in Shaping Candida albicans Cells. J. Fungi 2017, 3, 59. https://doi.org/10.3390/jof3040059
Popolo L, Degani G, Camilloni C, Fonzi WA. The PHR Family: The Role of Extracellular Transglycosylases in Shaping Candida albicans Cells. Journal of Fungi. 2017; 3(4):59. https://doi.org/10.3390/jof3040059
Chicago/Turabian StylePopolo, Laura, Genny Degani, Carlo Camilloni, and William A. Fonzi. 2017. "The PHR Family: The Role of Extracellular Transglycosylases in Shaping Candida albicans Cells" Journal of Fungi 3, no. 4: 59. https://doi.org/10.3390/jof3040059