Gaining Insights from Candida Biofilm Heterogeneity: One Size Does Not Fit All
Abstract
:1. What Is Biofilm Heterogeneity?
2. How Do We Investigate Biofilm Formation?
3. Is Heterogeneity Clinically Important?
4. How Does Heterogeneity Impact Antifungal Treatment?
5. Do Non-Albicans Species Play a Role?
6. Interkingdom Interactions Support Biofilm Defects
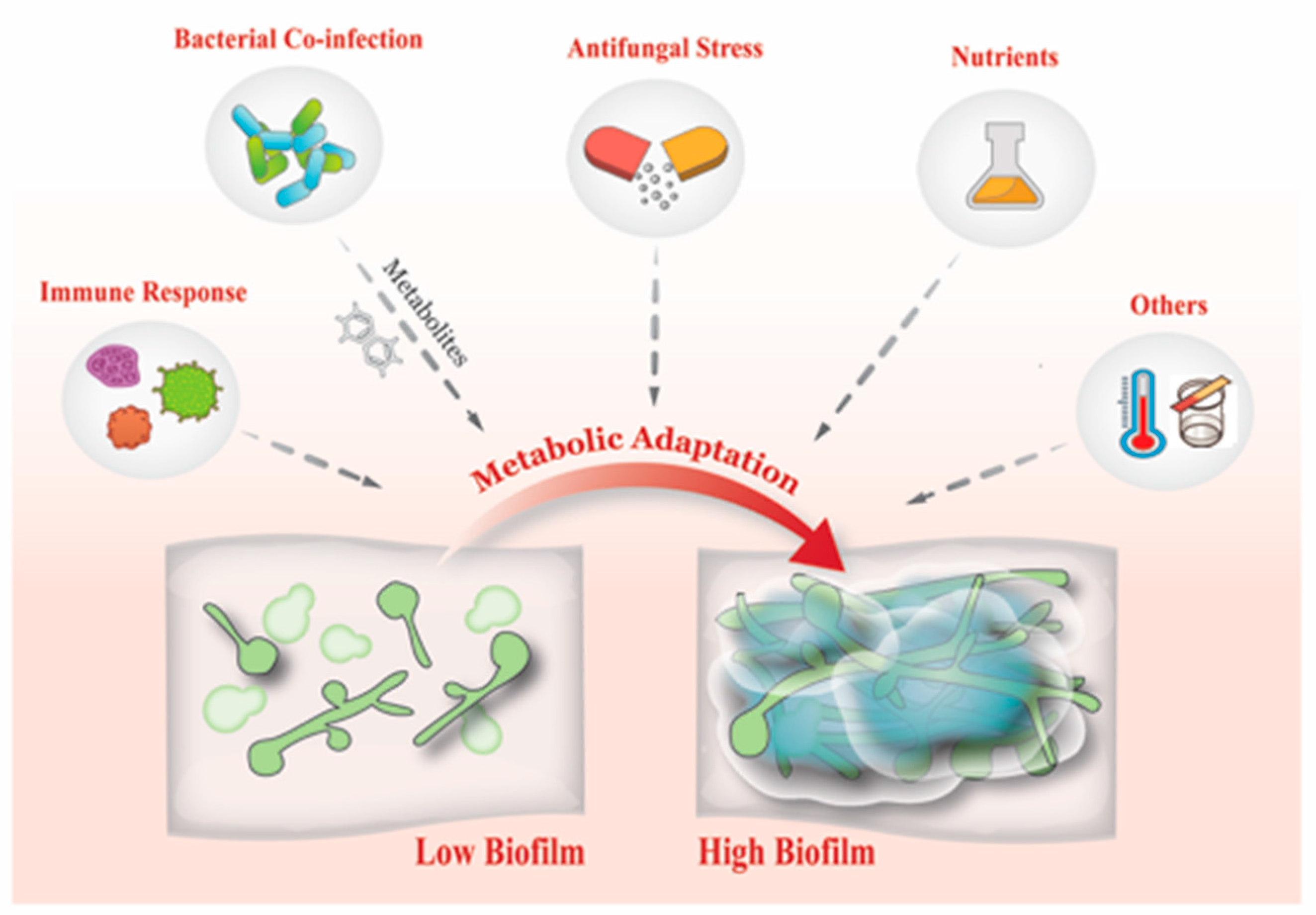
7. What Drives Biofilm Heterogeneity?
8. Conclusions and Future Outlook
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Fux, C.A.; Shirtliff, M.; Stoodley, P.; Costerton, J.W. Can laboratory reference strains mirror “real-world” pathogenesis? Trends Microbiol. 2005, 13, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Azeredo, J.; Azevedo, N.F.; Briandet, R.; Cerca, N.; Coenye, T.; Costa, A.R.; Desvaux, M.; Di Bonaventura, G.; Hebraud, M.; Jaglic, Z.; et al. Critical review on biofilm methods. Crit. Rev. Microbiol. 2017, 43, 313–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lohse, M.B.; Gulati, M.; Valle Arevalo, A.; Fishburn, A.; Johnson, A.D.; Nobile, C.J. Assessment and Optimizations of Candida albicans In Vitro Biofilm Assays. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Montelongo-Jauregui, D.; Srinivasan, A.; Ramasubramanian, A.K.; Lopez-Ribot, J.L. An In Vitro Model for Oral Mixed Biofilms of Candida albicans and Streptococcus gordonii in Synthetic Saliva. Front. Microbiol. 2016, 7, 686. [Google Scholar] [CrossRef] [PubMed]
- Uppuluri, P.; Dinakaran, H.; Thomas, D.P.; Chaturvedi, A.K.; Lopez-Ribot, J.L. Characteristics of Candida albicans biofilms grown in a synthetic urine medium. J. Clin. Microbiol. 2009, 47, 4078–4083. [Google Scholar] [CrossRef] [PubMed]
- Kean, R.; Rajendran, R.; Haggarty, J.; Townsend, E.M.; Short, B.; Burgess, K.E.; Lang, S.; Millington, O.; Mackay, W.G.; Williams, C.; et al. Candida albicans Mycofilms Support Staphylococcus aureus Colonization and Enhances Miconazole Resistance in Dual-Species Interactions. Front. Microbiol. 2017, 8, 258. [Google Scholar] [CrossRef] [PubMed]
- Hawser, S. Comparisons of the susceptibilities of planktonic and adherent Candida albicans to antifungal agents: A modified XTT tetrazolium assay using synchronised C. albicans cells. J. Med. Vet. Mycol. 1996, 34, 149–152. [Google Scholar] [CrossRef] [PubMed]
- Hawser, S.P.; Norris, H.; Jessup, C.J.; Ghannoum, M.A. Comparison of a 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino)carbonyl]-2H-tetrazolium hydroxide (XTT) colorimetric method with the standardized National Committee for Clinical Laboratory Standards method of testing clinical yeast isolates for susceptibility to antifungal agents. J. Clin. Microbiol. 1998, 36, 1450–1452. [Google Scholar] [PubMed]
- Ramage, G.; Vande Walle, K.; Wickes, B.L.; Lopez-Ribot, J.L. Biofilm formation by Candida dubliniensis. J. Clin. Microbiol. 2001, 39, 3234–3240. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, D.M.; Balkis, M.; Chandra, J.; Mukherjee, P.K.; Ghannoum, M.A. Uses and limitations of the XTT assay in studies of Candida growth and metabolism. J. Clin. Microbiol. 2003, 41, 506–508. [Google Scholar] [CrossRef] [PubMed]
- Taff, H.T.; Nett, J.E.; Andes, D.R. Comparative analysis of Candida biofilm quantitation assays. Med. Mycol. 2012, 50, 214–218. [Google Scholar] [CrossRef] [PubMed]
- Nett, J.; Lincoln, L.; Marchillo, K.; Massey, R.; Holoyda, K.; Hoff, B.; VanHandel, M.; Andes, D. Putative role of β-1,3 glucans in Candida albicans biofilm resistance. Antimicrob. Agents Chemother. 2007, 51, 510–520. [Google Scholar] [CrossRef] [PubMed]
- Pongracz, J.; Benedek, K.; Juhasz, E.; Ivan, M.; Kristof, K. In vitro biofilm production of Candida bloodstream isolates: Any association with clinical characteristics? J. Med. Microbiol. 2016, 65, 272–277. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, R.; Sherry, L.; Nile, C.J.; Sherriff, A.; Johnson, E.M.; Hanson, M.F.; Williams, C.; Munro, C.A.; Jones, B.J.; Ramage, G. Biofilm formation is a risk factor for mortality in patients with Candida albicans bloodstream infection-Scotland, 2012–2013. Clin. Microbiol. Infect. 2016, 22, 87–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soldini, S.; Posteraro, B.; Vella, A.; De Carolis, E.; Borghi, E.; Falleni, M.; Losito, A.R.; Maiuro, G.; Trecarichi, E.M.; Sanguinetti, M.; et al. Microbiological and clinical characteristics of biofilm-forming Candida parapsilosis isolates associated with fungaemia and their impact on mortality. Clin. Microbiol. Infect. 2017. [Google Scholar] [CrossRef] [PubMed]
- Tumbarello, M.; Fiori, B.; Trecarichi, E.M.; Posteraro, P.; Losito, A.R.; De Luca, A.; Sanguinetti, M.; Fadda, G.; Cauda, R.; Posteraro, B. Risk factors and outcomes of candidemia caused by biofilm-forming isolates in a tertiary care hospital. PLoS ONE 2012, 7, e33705. [Google Scholar] [CrossRef] [PubMed]
- Tumbarello, M.; Posteraro, B.; Trecarichi, E.M.; Fiori, B.; Rossi, M.; Porta, R.; de Gaetano Donati, K.; La Sorda, M.; Spanu, T.; Fadda, G.; et al. Biofilm production by Candida species and inadequate antifungal therapy as predictors of mortality for patients with candidemia. J. Clin. Microbiol. 2007, 45, 1843–1850. [Google Scholar] [CrossRef] [PubMed]
- Kumar, C.P.; Menon, T. Biofilm production by clinical isolates of Candida species. Med. Mycol. 2006, 44, 99–101. [Google Scholar] [CrossRef] [PubMed]
- Sherry, L.; Rajendran, R.; Lappin, D.F.; Borghi, E.; Perdoni, F.; Falleni, M.; Tosi, D.; Smith, K.; Williams, C.; Jones, B.; et al. Biofilms formed by Candida albicans bloodstream isolates display phenotypic and transcriptional heterogeneity that are associated with resistance and pathogenicity. BMC Microbiol. 2014, 14, 182. [Google Scholar] [CrossRef] [PubMed]
- Kojic, E.M.; Darouiche, R.O. Candida infections of medical devices. Clin. Microbiol. Rev. 2004, 17, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, S.; Mitchell, A.P. Mucosal biofilms of Candida albicans. Curr. Opin. Microbiol. 2011, 14, 380–385. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, L.E.; Alalwan, H.K.; Kean, R.; Calvert, G.; Nile, C.J.; Lappin, D.F.; Robertson, D.; Williams, C.; Ramage, G.; Sherry, L. Candida albicans biofilm heterogeneity does not influence denture stomatitis but strongly influences denture cleansing capacity. J. Med. Microbiol. 2017, 66, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Sherry, L.; Kean, R.; McKloud, E.; O’Donnell, L.E.; Metcalfe, R.; Jones, B.L.; Ramage, G. Biofilms Formed by Isolates from Recurrent Vulvovaginal Candidiasis Patients Are Heterogeneous and Insensitive to Fluconazole. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Jain, N.; Kohli, R.; Cook, E.; Gialanella, P.; Chang, T.; Fries, B.C. Biofilm formation by and antifungal susceptibility of Candida isolates from urine. Appl. Environ. Microbiol. 2007, 73, 1697–1703. [Google Scholar] [CrossRef] [PubMed]
- Bitar, I.; Khalaf, R.A.; Harastani, H.; Tokajian, S. Identification, typing, antifungal resistance profile, and biofilm formation of Candida albicans isolates from Lebanese hospital patients. Biomed. Res. Int. 2014, 2014, 931372. [Google Scholar] [CrossRef] [PubMed]
- Alnuaimi, A.D.; O’Brien-Simpson, N.M.; Reynolds, E.C.; McCullough, M.J. Clinical isolates and laboratory reference Candida species and strains have varying abilities to form biofilms. FEMS Yeast Res. 2013, 13, 689–699. [Google Scholar] [CrossRef] [PubMed]
- De Carvalho, F.G.; Silva, D.S.; Hebling, J.; Spolidorio, L.C.; Spolidorio, D.M. Presence of mutans streptococci and Candida spp. in dental plaque/dentine of carious teeth and early childhood caries. Arch. Oral Biol. 2006, 51, 1024–1028. [Google Scholar] [CrossRef] [PubMed]
- Jenkinson, H.F.; Lala, H.C.; Shepherd, M.G. Coaggregation of Streptococcus sanguis and other streptococci with Candida albicans. Infect. Immun. 1990, 58, 1429–1436. [Google Scholar] [PubMed]
- Reynaud, A.H.; Nygaard-Ostby, B.; Boygard, G.K.; Eribe, E.R.; Olsen, I.; Gjermo, P. Yeasts in periodontal pockets. J. Clin. Periodontol. 2001, 28, 860–864. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Redding, S.; Dongari-Bagtzoglou, A. Candida glabrata: An emerging oral opportunistic pathogen. J. Dent. Res. 2007, 86, 204–215. [Google Scholar] [CrossRef] [PubMed]
- McCormack, M.G.; Smith, A.J.; Akram, A.N.; Jackson, M.; Robertson, D.; Edwards, G. Staphylococcus aureus and the oral cavity: An overlooked source of carriage and infection? Am. J. Infect. Control 2015, 43, 35–37. [Google Scholar] [CrossRef] [PubMed]
- Bauernfeind, A.; Bertele, R.M.; Harms, K.; Horl, G.; Jungwirth, R.; Petermuller, C.; Przyklenk, B.; Weisslein-Pfister, C. Qualitative and quantitative microbiological analysis of sputa of 102 patients with cystic fibrosis. Infection 1987, 15, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Nseir, S.; Jozefowicz, E.; Cavestri, B.; Sendid, B.; Di Pompeo, C.; Dewavrin, F.; Favory, R.; Roussel-Delvallez, M.; Durocher, A. Impact of antifungal treatment on Candida-Pseudomonas interaction: A preliminary retrospective case-control study. Intensive Care Med. 2007, 33, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Garsin, D.A.; Lorenz, M.C. Candida albicans and Enterococcus faecalis in the gut: Synergy in commensalism? Gut Microbes 2013, 4, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Raponi, G.; Visconti, V.; Brunetti, G.; Ghezzi, M.C. Clostridium difficile infection and Candida colonization of the gut: Is there a correlation? Clin. Infect. Dis. 2014, 59, 1648–1649. [Google Scholar] [CrossRef] [PubMed]
- Falagas, M.E.; Betsi, G.I.; Athanasiou, S. Probiotics for prevention of recurrent vulvovaginal candidiasis: A review. J. Antimicrob. Chemother. 2006, 58, 266–272. [Google Scholar] [CrossRef] [PubMed]
- Borman, A.M.; Szekely, A.; Johnson, E.M. Comparative Pathogenicity of United Kingdom Isolates of the Emerging Pathogen Candida auris and Other Key Pathogenic Candida Species. mSphere 2016, 1. [Google Scholar] [CrossRef] [PubMed]
- Malani, P.N.; McNeil, S.A.; Bradley, S.F.; Kauffman, C.A. Candida albicans sternal wound infections: A chronic and recurrent complication of median sternotomy. Clin. Infect. Dis. 2002, 35, 1316–1320. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, L.E.; Robertson, D.; Nile, C.J.; Cross, L.J.; Riggio, M.; Sherriff, A.; Bradshaw, D.; Lambert, M.; Malcolm, J.; Buijs, M.J.; et al. The Oral Microbiome of Denture Wearers Is Influenced by Levels of Natural Dentition. PLoS ONE 2015, 10, e0137717. [Google Scholar] [CrossRef] [PubMed]
- Coco, B.J.; Bagg, J.; Cross, L.J.; Jose, A.; Cross, J.; Ramage, G. Mixed Candida albicans and Candida glabrata populations associated with the pathogenesis of denture stomatitis. Oral Microbiol. Immunol. 2008, 23, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Elving, G.J.; van der Mei, H.C.; Busscher, H.J.; van Weissenbruch, R.; Albers, F.W. Comparison of the microbial composition of voice prosthesis biofilms from patients requiring frequent versus infrequent replacement. Ann. Otol. Rhinol. Laryngol. 2002, 111, 200–203. [Google Scholar] [CrossRef] [PubMed]
- Bauters, T.G.; Moerman, M.; Vermeersch, H.; Nelis, H.J. Colonization of voice prostheses by albicans and non-albicans Candida species. Laryngoscope 2002, 112, 708–712. [Google Scholar] [CrossRef] [PubMed]
- Mylonakis, E.; Calderwood, S.B. Infective endocarditis in adults. N. Engl. J. Med. 2001, 345, 1318–1330. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Burner, K.D.; Fealey, M.E.; Edwards, W.D.; Tazelaar, H.D.; Orszulak, T.A.; Wright, A.J.; Baddour, L.M. Prosthetic valve endocarditis: Clinicopathological correlates in 122 surgical specimens from 116 patients (1985–2004). Cardiovasc. Pathol. 2011, 20, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Schelenz, S.; Hagen, F.; Rhodes, J.L.; Abdolrasouli, A.; Chowdhary, A.; Hall, A.; Ryan, L.; Shackleton, J.; Trimlett, R.; Meis, J.F.; et al. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob. Resist. Infect. Control 2016, 5, 35. [Google Scholar] [CrossRef] [PubMed]
- Tchekmedyian, N.S.; Newman, K.; Moody, M.R.; Costerton, J.W.; Aisner, J.; Schimpff, S.C.; Reed, W.P. Special studies of the Hickman catheter of a patient with recurrent bacteremia and candidemia. Am. J. Med. Sci. 1986, 291, 419–424. [Google Scholar] [CrossRef] [PubMed]
- Adler, A.; Yaniv, I.; Steinberg, R.; Solter, E.; Samra, Z.; Stein, J.; Levy, I. Infectious complications of implantable ports and Hickman catheters in paediatric haematology-oncology patients. J. Hosp. Infect. 2006, 62, 358–365. [Google Scholar] [CrossRef] [PubMed]
- Vallabhaneni, S.; Kallen, A.; Tsay, S.; Chow, N.; Welsh, R.; Kerins, J.; Kemble, S.K.; Pacilli, M.; Black, S.R.; Landon, E.; et al. Investigation of the First Seven Reported Cases of Candida auris, a Globally Emerging Invasive, Multidrug-Resistant Fungus—United States, May 2013–August 2016. MMWR Morb. Mortal Wkly. Rep. 2016, 65, 1234–1237. [Google Scholar] [CrossRef] [PubMed]
- Weiner, L.M.; Webb, A.K.; Limbago, B.; Dudeck, M.A.; Patel, J.; Kallen, A.J.; Edwards, J.R.; Sievert, D.M. Antimicrobial-Resistant Pathogens Associated With Healthcare-Associated Infections: Summary of Data Reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect. Control Hosp. Epidemiol. 2016, 37, 1288–1301. [Google Scholar] [CrossRef] [PubMed]
- Uppuluri, P.; Chaturvedi, A.K.; Srinivasan, A.; Banerjee, M.; Ramasubramaniam, A.K.; Kohler, J.R.; Kadosh, D.; Lopez-Ribot, J.L. Dispersion as an important step in the Candida albicans biofilm developmental cycle. PLoS Pathog. 2010, 6, e1000828. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, R.; Sherry, L.; Deshpande, A.; Johnson, E.M.; Hanson, M.F.; Williams, C.; Munro, C.A.; Jones, B.L.; Ramage, G. A Prospective Surveillance Study of Candidaemia: Epidemiology, Risk Factors, Antifungal Treatment and Outcome in Hospitalized Patients. Front. Microbiol. 2016, 7, 915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tascini, C.; Sozio, E.; Corte, L.; Sbrana, F.; Scarparo, C.; Ripoli, A.; Bertolino, G.; Merelli, M.; Tagliaferri, E.; Corcione, A.; et al. The role of biofilm forming on mortality in patients with candidemia: A study derived from real world data. Infect. Dis. 2017. [Google Scholar] [CrossRef] [PubMed]
- Cornely, O.A.; Bassetti, M.; Calandra, T.; Garbino, J.; Kullberg, B.J.; Lortholary, O.; Meersseman, W.; Akova, M.; Arendrup, M.C.; Arikan-Akdagli, S.; et al. ESCMID* guideline for the diagnosis and management of Candida diseases 2012: Non-neutropenic adult patients. Clin. Microbiol. Infect. 2012, 18 (Suppl. S7), 19–37. [Google Scholar] [CrossRef] [PubMed]
- Koehler, P.; Tacke, D.; Cornely, O.A. Our 2014 approach to candidaemia. Mycoses 2014, 57, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Mermel, L.A.; Allon, M.; Bouza, E.; Craven, D.E.; Flynn, P.; O’Grady, N.P.; Raad, I.I.; Rijnders, B.J.; Sherertz, R.J.; Warren, D.K. Clinical practice guidelines for the diagnosis and management of intravascular catheter-related infection: 2009 Update by the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 49, 1–45. [Google Scholar] [CrossRef] [PubMed]
- Andes, D.R.; Safdar, N.; Baddley, J.W.; Playford, G.; Reboli, A.C.; Rex, J.H.; Sobel, J.D.; Pappas, P.G.; Kullberg, B.J.; Mycoses Study Group. Impact of treatment strategy on outcomes in patients with candidemia and other forms of invasive candidiasis: A patient-level quantitative review of randomized trials. Clin. Infect. Dis. 2012, 54, 1110–1122. [Google Scholar] [CrossRef] [PubMed]
- Nucci, M.; Anaissie, E.; Betts, R.F.; Dupont, B.F.; Wu, C.; Buell, D.N.; Kovanda, L.; Lortholary, O. Early removal of central venous catheter in patients with candidemia does not improve outcome: Analysis of 842 patients from 2 randomized clinical trials. Clin. Infect. Dis. 2010, 51, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Naglik, J.R.; Moyes, D.; Makwana, J.; Kanzaria, P.; Tsichlaki, E.; Weindl, G.; Tappuni, A.R.; Rodgers, C.A.; Woodman, A.J.; Challacombe, S.J.; et al. Quantitative expression of the Candida albicans secreted aspartyl proteinase gene family in human oral and vaginal candidiasis. Microbiology 2008, 154 Pt 11, 3266–3280. [Google Scholar] [CrossRef] [PubMed]
- Joo, M.Y.; Shin, J.H.; Jang, H.C.; Song, E.S.; Kee, S.J.; Shin, M.G.; Suh, S.P.; Ryang, D.W. Expression of SAP5 and SAP9 in Candida albicans biofilms: Comparison of bloodstream isolates with isolates from other sources. Med. Mycol. 2013, 51, 892–896. [Google Scholar] [CrossRef] [PubMed]
- Ramage, G.; Coco, B.; Sherry, L.; Bagg, J.; Lappin, D.F. In vitro Candida albicans biofilm induced proteinase activity and SAP8 expression correlates with in vivo denture stomatitis severity. Mycopathologia 2012, 174, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Naglik, J.R.; Rodgers, C.A.; Shirlaw, P.J.; Dobbie, J.L.; Fernandes-Naglik, L.L.; Greenspan, D.; Agabian, N.; Challacombe, S.J. Differential expression of Candida albicans secreted aspartyl proteinase and phospholipase B genes in humans correlates with active oral and vaginal infections. J. Infect. Dis. 2003, 188, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Winter, M.B.; Salcedo, E.C.; Lohse, M.B.; Hartooni, N.; Gulati, M.; Sanchez, H.; Takagi, J.; Hube, B.; Andes, D.R.; Johnson, A.D.; et al. Global Identification of Biofilm-Specific Proteolysis in Candida albicans. mBio 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Moyes, D.L.; Wilson, D.; Richardson, J.P.; Mogavero, S.; Tang, S.X.; Wernecke, J.; Hofs, S.; Gratacap, R.L.; Robbins, J.; Runglall, M.; et al. Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature 2016, 532, 64–68. [Google Scholar] [CrossRef] [PubMed]
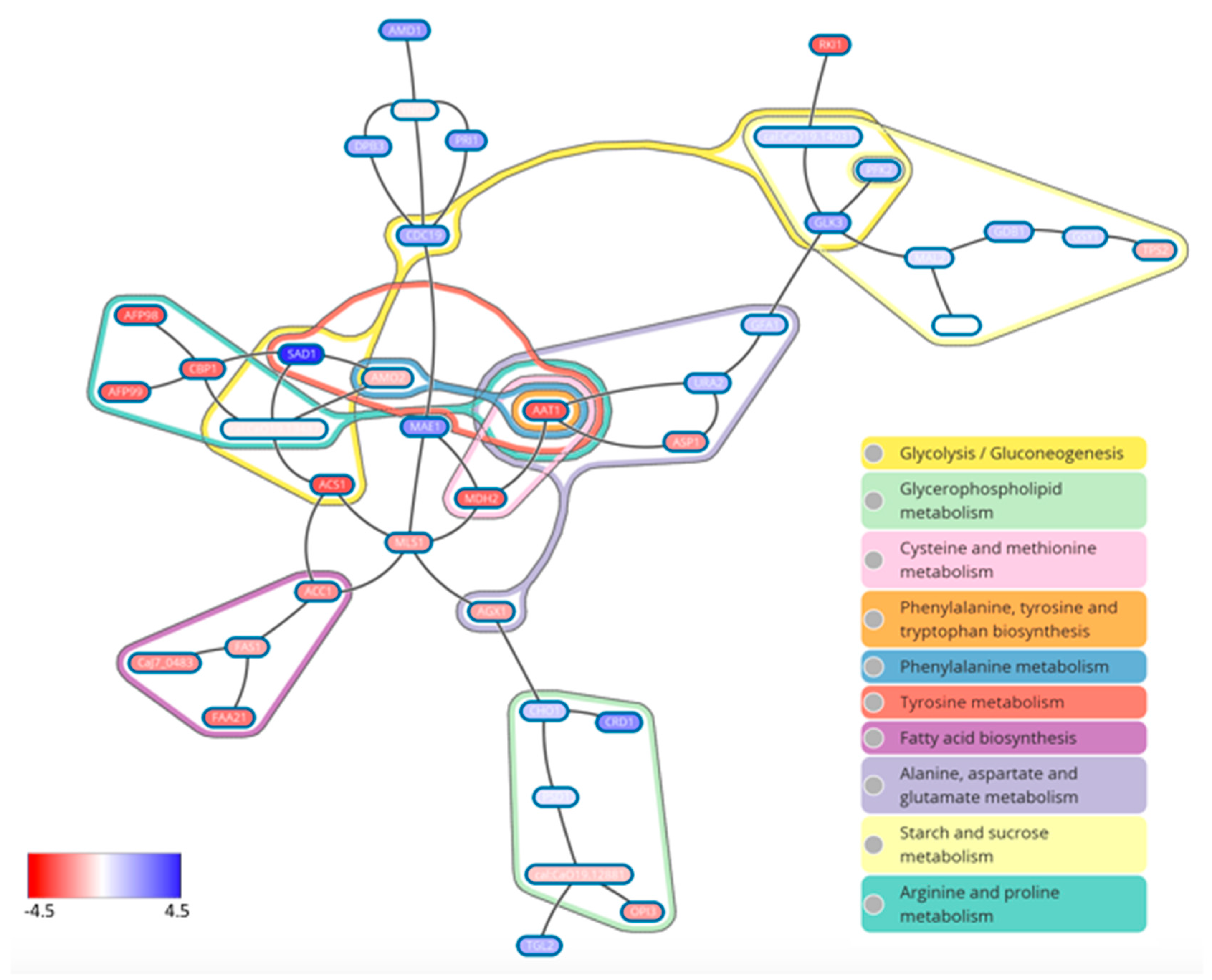
- Rajendran, R.; May, A.; Sherry, L.; Kean, R.; Williams, C.; Jones, B.L.; Burgess, K.V.; Heringa, J.; Abeln, S.; Brandt, B.W.; et al. Integrating Candida albicans metabolism with biofilm heterogeneity by transcriptome mapping. Sci. Rep. 2016, 6, 35436. [Google Scholar] [CrossRef] [PubMed]
- Sobel, J.D. Vulvovaginal candidosis. Lancet 2007, 369, 1961–1971. [Google Scholar] [CrossRef]
- Sobel, J.D. Recurrent vulvovaginal candidiasis. Am. J. Obstet. Gynecol. 2016, 214, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Harriott, M.M.; Lilly, E.A.; Rodriguez, T.E.; Fidel, P.L.; Noverr, M.C. Candida albicans forms biofilms on the vaginal mucosa. Microbiology 2010, 156 Pt 12, 3635–3644. [Google Scholar] [CrossRef] [PubMed]
- Muzny, C.A.; Schwebke, J.R. Biofilms: An Underappreciated Mechanism of Treatment Failure and Recurrence in Vaginal Infections. Clin. Infect. Dis. 2015, 61, 601–606. [Google Scholar] [CrossRef] [PubMed]
- Sobel, J.D. Editorial Commentary: Vaginal Biofilm: Much Ado About Nothing, or a New Therapeutic Challenge? Clin. Infect. Dis. 2015, 61, 607–608. [Google Scholar] [CrossRef] [PubMed]
- Chassot, F.; Negri, M.F.; Svidzinski, A.E.; Donatti, L.; Peralta, R.M.; Svidzinski, T.I.; Consolaro, M.E. Can intrauterine contraceptive devices be a Candida albicans reservoir? Contraception 2008, 77, 355–359. [Google Scholar] [CrossRef] [PubMed]
- Ramage, G.; Vande Walle, K.; Wickes, B.L.; Lopez-Ribot, J.L. Standardized method for in vitro antifungal susceptibility testing of Candida albicans biofilms. Antimicrob. Agents Chemother. 2001, 45, 2475–2479. [Google Scholar] [CrossRef] [PubMed]
- Kucharikova, S.; Tournu, H.; Holtappels, M.; Van Dijck, P.; Lagrou, K. In vivo efficacy of anidulafungin against mature Candida albicans biofilms in a novel rat model of catheter-associated Candidiasis. Antimicrob. Agents Chemother. 2010, 54, 4474–4475. [Google Scholar] [CrossRef] [PubMed]
- Marcos-Zambrano, L.J.; Escribano, P.; Bouza, E.; Guinea, J. Susceptibility of Candida albicans biofilms to caspofungin and anidulafungin is not affected by metabolic activity or biomass production. Med. Mycol. 2016, 54, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Muadcheingka, T.; Tantivitayakul, P. Distribution of Candida albicans and non-albicans Candida species in oral candidiasis patients: Correlation between cell surface hydrophobicity and biofilm forming activities. Arch. Oral Biol. 2015, 60, 894–901. [Google Scholar] [CrossRef] [PubMed]
- Hasan, F.; Xess, I.; Wang, X.; Jain, N.; Fries, B.C. Biofilm formation in clinical Candida isolates and its association with virulence. Microbes Infect. 2009, 11, 753–761. [Google Scholar] [CrossRef] [PubMed]
- Perlin, D.S.; Shor, E.; Zhao, Y. Update on Antifungal Drug Resistance. Curr. Clin. Microbiol. Rep. 2015, 2, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Thompson, G.R., 3rd; Wiederhold, N.P.; Vallor, A.C.; Villareal, N.C.; Lewis, J.S., 2nd; Patterson, T.F. Development of caspofungin resistance following prolonged therapy for invasive candidiasis secondary to Candida glabrata infection. Antimicrob. Agents Chemother. 2008, 52, 3783–3785. [Google Scholar] [CrossRef] [PubMed]
- Ben-Ami, R.; Zimmerman, O.; Finn, T.; Amit, S.; Novikov, A.; Wertheimer, N.; Lurie-Weinberger, M.; Berman, J. Heteroresistance to Fluconazole Is a Continuously Distributed Phenotype among Candida glabrata Clinical Strains Associated with In Vivo Persistence. mBio 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- El-Halfawy, O.M.; Valvano, M.A. Antimicrobial heteroresistance: An emerging field in need of clarity. Clin. Microbiol. Rev. 2015, 28, 191–207. [Google Scholar] [CrossRef] [PubMed]
- Mondon, P.; Petter, R.; Amalfitano, G.; Luzzati, R.; Concia, E.; Polacheck, I.; Kwon-Chung, K.J. Heteroresistance to fluconazole and voriconazole in Cryptococcus neoformans. Antimicrob. Agents Chemother. 1999, 43, 1856–1861. [Google Scholar] [PubMed]
- Marr, K.A.; Lyons, C.N.; Ha, K.; Rustad, T.R.; White, T.C. Inducible azole resistance associated with a heterogeneous phenotype in Candida albicans. Antimicrob. Agents Chemother. 2001, 45, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Ramage, G.; Bachmann, S.; Patterson, T.F.; Wickes, B.L.; Lopez-Ribot, J.L. Investigation of multidrug efflux pumps in relation to fluconazole resistance in Candida albicans biofilms. J. Antimicrob. Chemother. 2002, 49, 973–980. [Google Scholar] [CrossRef] [PubMed]
- Song, J.W.; Shin, J.H.; Kee, S.J.; Kim, S.H.; Shin, M.G.; Suh, S.P.; Ryang, D.W. Expression of CgCDR1, CgCDR2, and CgERG11 in Candida glabrata biofilms formed by bloodstream isolates. Med. Mycol. 2009, 47, 545–548. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. Persister cells. Annu. Rev. Microbiol. 2010, 64, 357–372. [Google Scholar] [CrossRef] [PubMed]
- Balaban, N.Q.; Merrin, J.; Chait, R.; Kowalik, L.; Leibler, S. Bacterial persistence as a phenotypic switch. Science 2004, 305, 1622–1625. [Google Scholar] [CrossRef] [PubMed]
- Truong, T.; Zeng, G.; Qingsong, L.; Kwang, L.T.; Tong, C.; Chan, F.Y.; Wang, Y.; Seneviratne, C.J. Comparative Ploidy Proteomics of Candida albicans Biofilms Unraveled the Role of the AHP1 Gene in the Biofilm Persistence Against Amphotericin B. Mol. Cell. Proteom. 2016, 15, 3488–3500. [Google Scholar] [CrossRef] [PubMed]
- LaFleur, M.D.; Kumamoto, C.A.; Lewis, K. Candida albicans biofilms produce antifungal-tolerant persister cells. Antimicrob. Agents Chemother. 2006, 50, 3839–3846. [Google Scholar] [CrossRef] [PubMed]
- Bojsen, R.; Regenberg, B.; Gresham, D.; Folkesson, A. A common mechanism involving the TORC1 pathway can lead to amphotericin B-persistence in biofilm and planktonic Saccharomyces cerevisiae populations. Sci. Rep. 2016, 6, 21874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, P.; Seneviratne, C.J.; Alpi, E.; Vizcaino, J.A.; Jin, L. Delicate Metabolic Control and Coordinated Stress Response Critically Determine Antifungal Tolerance of Candida albicans Biofilm Persisters. Antimicrob. Agents Chemother. 2015, 59, 6101–6112. [Google Scholar] [CrossRef] [PubMed]
- Robbins, N.; Uppuluri, P.; Nett, J.; Rajendran, R.; Ramage, G.; Lopez-Ribot, J.L.; Andes, D.; Cowen, L.E. Hsp90 governs dispersion and drug resistance of fungal biofilms. PLoS Pathog. 2011, 7, e1002257. [Google Scholar] [CrossRef] [PubMed]
- Al-Dhaheri, R.S.; Douglas, L.J. Absence of amphotericin B-tolerant persister cells in biofilms of some Candida species. Antimicrob. Agents Chemother. 2008, 52, 1884–1887. [Google Scholar] [CrossRef] [PubMed]
- Bink, A.; Vandenbosch, D.; Coenye, T.; Nelis, H.; Cammue, B.P.; Thevissen, K. Superoxide dismutases are involved in Candida albicans biofilm persistence against miconazole. Antimicrob. Agents Chemother. 2011, 55, 4033–4037. [Google Scholar] [CrossRef] [PubMed]
- Fortun, J.; Martin-Davila, P.; de la Gomez-Garcia Pedrosa, E.; Pintado, V.; Cobo, J.; Fresco, G.; Meije, Y.; Ros, L.; Alvarez, M.E.; Luengo, J.; et al. Emerging trends in candidemia: A higher incidence but a similar outcome. J. Infect. 2012, 65, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.; Henriques, M.; Martins, A.; Oliveira, R.; Williams, D.; Azeredo, J. Biofilms of non-Candida albicans Candida species: Quantification, structure and matrix composition. Med. Mycol. 2009, 47, 681–689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vipulanandan, G.; Herrera, M.; Wiederhold, N.P.; Li, X.; Mintz, J.; Wickes, B.L.; Kadosh, D. Dynamics of Mixed- Candida Species Biofilms in Response to Antifungals. J. Dent. Res. 2018, 97, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Tati, S.; Davidow, P.; McCall, A.; Hwang-Wong, E.; Rojas, I.G.; Cormack, B.; Edgerton, M. Candida glabrata Binding to Candida albicans Hyphae Enables Its Development in Oropharyngeal Candidiasis. PLoS Pathog. 2016, 12, e1005522. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.; Henriques, M.; Hayes, A.; Oliveira, R.; Azeredo, J.; Williams, D.W. Candida glabrata and Candida albicans co-infection of an in vitro oral epithelium. J. Oral Pathol. Med. 2011, 40, 421–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeffery-Smith, A.; Taori, S.K.; Schelenz, S.; Jeffery, K.; Johnson, E.M.; Borman, A.; Candida auris Incident Management, T.; Manuel, R.; Brown, C.S. Candida auris: A Review of the Literature. Clin. Microbiol. Rev. 2017, 31. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, S.R.; Etienne, K.A.; Vallabhaneni, S.; Farooqi, J.; Chowdhary, A.; Govender, N.P.; Colombo, A.L.; Calvo, B.; Cuomo, C.A.; Desjardins, C.A.; et al. Simultaneous Emergence of Multidrug-Resistant Candida auris on 3 Continents Confirmed by Whole-Genome Sequencing and Epidemiological Analyses. Clin. Infect. Dis. 2017, 64, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Sarma, S.; Upadhyay, S. Current perspective on emergence, diagnosis and drug resistance in Candida auris. Infect. Drug Resist. 2017, 10, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Ben-Ami, R.; Berman, J.; Novikov, A.; Bash, E.; Shachor-Meyouhas, Y.; Zakin, S.; Maor, Y.; Tarabia, J.; Schechner, V.; Adler, A.; et al. Multidrug-Resistant Candida haemulonii and C. auris, Tel Aviv, Israel. Emerg. Infect. Dis 2017, 23, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Oh, B.J.; Shin, J.H.; Kim, M.N.; Sung, H.; Lee, K.; Joo, M.Y.; Shin, M.G.; Suh, S.P.; Ryang, D.W. Biofilm formation and genotyping of Candida haemulonii, Candida pseudohaemulonii, and a proposed new species (Candida auris) isolates from Korea. Med. Mycol. 2011, 49, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Sherry, L.; Ramage, G.; Kean, R.; Borman, A.; Johnson, E.M.; Richardson, M.D.; Rautemaa-Richardson, R. Biofilm-Forming Capability of Highly Virulent, Multidrug-Resistant Candida auris. Emerg. Infect. Dis. 2017, 23, 328–331. [Google Scholar] [CrossRef] [PubMed]
- Larkin, E.; Hager, C.; Chandra, J.; Mukherjee, P.K.; Retuerto, M.; Salem, I.; Long, L.; Isham, N.; Kovanda, L.; Borroto-Esoda, K.; et al. The Emerging Pathogen Candida auris: Growth Phenotype, Virulence Factors, Activity of Antifungals, and Effect of SCY-078, a Novel Glucan Synthesis Inhibitor, on Growth Morphology and Biofilm Formation. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Piedrahita, C.T.; Cadnum, J.L.; Jencson, A.L.; Shaikh, A.A.; Ghannoum, M.A.; Donskey, C.J. Environmental Surfaces in Healthcare Facilities are a Potential Source for Transmission of Candida auris and Other Candida Species. Infect. Control Hosp. Epidemiol. 2017, 38, 1107–1109. [Google Scholar] [CrossRef] [PubMed]
- Welsh, R.M.; Bentz, M.L.; Shams, A.; Houston, H.; Lyons, A.; Rose, L.J.; Litvintseva, A.P. Survival, Persistence, and Isolation of the Emerging Multidrug-Resistant Pathogenic Yeast Candida auris on a Plastic Health Care Surface. J. Clin. Microbiol. 2017, 55, 2996–3005. [Google Scholar] [CrossRef] [PubMed]
- Cadnum, J.L.; Shaikh, A.A.; Piedrahita, C.T.; Sankar, T.; Jencson, A.L.; Larkin, E.L.; Ghannoum, M.A.; Donskey, C.J. Effectiveness of Disinfectants Against Candida auris and Other Candida Species. Infect. Control Hosp. Epidemiol. 2017, 38, 1240–1243. [Google Scholar] [CrossRef] [PubMed]
- Kean, R.; Sherry, L.; Townsend, E.; McKloud, E.; Short, B.; Akinbobola, A.; Mackay, W.G.; Williams, C.; Jones, B.L.; Ramage, G. Surface disinfection challenges for Candida auris: An in vitro study. J. Hosp. Infect. 2017. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, L.E.; Millhouse, E.; Sherry, L.; Kean, R.; Malcolm, J.; Nile, C.J.; Ramage, G. Polymicrobial Candida biofilms: Friends and foe in the oral cavity. FEMS Yeast Res. 2015, 15. [Google Scholar] [CrossRef]
- Orsi, C.F.; Sabia, C.; Ardizzoni, A.; Colombari, B.; Neglia, R.G.; Peppoloni, S.; Morace, G.; Blasi, E. Inhibitory effects of different lactobacilli on Candida albicans hyphal formation and biofilm development. J. Biol. Regul. Homeost. Agents 2014, 28, 743–752. [Google Scholar] [PubMed]
- Dubsky, M.; Jirkovska, A.; Bem, R.; Fejfarova, V.; Skibova, J.; Schaper, N.C.; Lipsky, B.A. Risk factors for recurrence of diabetic foot ulcers: Prospective follow-up analysis in the Eurodiale subgroup. Int. Wound J. 2013, 10, 555–561. [Google Scholar] [CrossRef] [PubMed]
- James, G.A.; Swogger, E.; Wolcott, R.; Pulcini, E.; Secor, P.; Sestrich, J.; Costerton, J.W.; Stewart, P.S. Biofilms in chronic wounds. Wound Repair Regen. 2008, 16, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Neut, D.; Tijdens-Creusen, E.J.; Bulstra, S.K.; van der Mei, H.C.; Busscher, H.J. Biofilms in chronic diabetic foot ulcers—A study of 2 cases. Acta Orthop. 2011, 82, 383–385. [Google Scholar] [CrossRef] [PubMed]
- Townsend, E.M.; Sherry, L.; Rajendran, R.; Hansom, D.; Butcher, J.; Mackay, W.G.; Williams, C.; Ramage, G. Development and characterisation of a novel three-dimensional inter-kingdom wound biofilm model. Biofouling 2016, 32, 1259–1270. [Google Scholar] [CrossRef] [PubMed]
- Chellan, G.; Shivaprakash, S.; Karimassery Ramaiyar, S.; Varma, A.K.; Varma, N.; Thekkeparambil Sukumaran, M.; Rohinivilasam Vasukutty, J.; Bal, A.; Kumar, H. Spectrum and prevalence of fungi infecting deep tissues of lower-limb wounds in patients with type 2 diabetes. J. Clin. Microbiol. 2010, 48, 2097–2102. [Google Scholar] [CrossRef] [PubMed]
- Townsend, E.M.; Sherry, L.; Kean, R.; Hansom, D.; Mackay, W.G.; Williams, C.; Butcher, J.; Ramage, G. Implications of Antimicrobial Combinations in Complex Wound Biofilms Containing Fungi. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Kart, D.; Tavernier, S.; Van Acker, H.; Nelis, H.J.; Coenye, T. Activity of disinfectants against multispecies biofilms formed by Staphylococcus aureus, Candida albicans and Pseudomonas aeruginosa. Biofouling 2014, 30, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Ovchinnikova, E.S.; Krom, B.P.; Busscher, H.J.; van der Mei, H.C. Evaluation of adhesion forces of Staphylococcus aureus along the length of Candida albicans hyphae. BMC Microbiol. 2012, 12, 281. [Google Scholar] [CrossRef] [PubMed]
- Peters, B.M.; Ovchinnikova, E.S.; Krom, B.P.; Schlecht, L.M.; Zhou, H.; Hoyer, L.L.; Busscher, H.J.; van der Mei, H.C.; Jabra-Rizk, M.A.; Shirtliff, M.E. Staphylococcus aureus adherence to Candida albicans hyphae is mediated by the hyphal adhesin Als3p. Microbiology 2012, 158 Pt 12, 2975–2986. [Google Scholar] [CrossRef] [PubMed]
- Harriott, M.M.; Noverr, M.C. Candida albicans and Staphylococcus aureus form polymicrobial biofilms: Effects on antimicrobial resistance. Antimicrob. Agents Chemother. 2009, 53, 3914–3922. [Google Scholar] [CrossRef] [PubMed]
- Kong, E.F.; Tsui, C.; Kucharikova, S.; Andes, D.; Van Dijck, P.; Jabra-Rizk, M.A. Commensal Protection of Staphylococcus aureus against Antimicrobials by Candida albicans Biofilm Matrix. mBio 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- De Brucker, K.; Tan, Y.; Vints, K.; De Cremer, K.; Braem, A.; Verstraeten, N.; Michiels, J.; Vleugels, J.; Cammue, B.P.; Thevissen, K. Fungal β-1,3-glucan increases ofloxacin tolerance of Escherichia coli in a polymicrobial E. coli/Candida albicans biofilm. Antimicrob. Agents Chemother. 2015, 59, 3052–3058. [Google Scholar] [CrossRef] [PubMed]
- Ramage, G.; Saville, S.P.; Wickes, B.L.; Lopez-Ribot, J.L. Inhibition of Candida albicans biofilm formation by farnesol, a quorum-sensing molecule. Appl. Environ. Microbiol. 2002, 68, 5459–5463. [Google Scholar] [CrossRef] [PubMed]
- Jabra-Rizk, M.A.; Meiller, T.F.; James, C.E.; Shirtliff, M.E. Effect of farnesol on Staphylococcus aureus biofilm formation and antimicrobial susceptibility. Antimicrob. Agents Chemother. 2006, 50, 1463–1469. [Google Scholar] [CrossRef] [PubMed]
- Unnanuntana, A.; Bonsignore, L.; Shirtliff, M.E.; Greenfield, E.M. The effects of farnesol on Staphylococcus aureus biofilms and osteoblasts. An in vitro study. J. Bone Joint Surg. Am. 2009, 91, 2683–2692. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Sengupta, A.; Niepa, T.H.; Lee, B.H.; Weljie, A.; Freitas-Blanco, V.S.; Murata, R.M.; Stebe, K.J.; Lee, D.; Koo, H. Candida albicans stimulates Streptococcus mutans microcolony development via cross-kingdom biofilm-derived metabolites. Sci. Rep. 2017, 7, 41332. [Google Scholar] [CrossRef] [PubMed]
- Van Leeuwen, P.T.; van der Peet, J.M.; Bikker, F.J.; Hoogenkamp, M.A.; Oliveira Paiva, A.M.; Kostidis, S.; Mayboroda, O.A.; Smits, W.K.; Krom, B.P. Interspecies Interactions between Clostridium difficile and Candida albicans. mSphere 2016, 1. [Google Scholar] [CrossRef] [PubMed]
- Fox, E.P.; Cowley, E.S.; Nobile, C.J.; Hartooni, N.; Newman, D.K.; Johnson, A.D. Anaerobic bacteria grow within Candida albicans biofilms and induce biofilm formation in suspension cultures. Curr. Biol. 2014, 24, 2411–2416. [Google Scholar] [CrossRef] [PubMed]
- Hoarau, G.; Mukherjee, P.K.; Gower-Rousseau, C.; Hager, C.; Chandra, J.; Retuerto, M.A.; Neut, C.; Vermeire, S.; Clemente, J.; Colombel, J.F.; et al. Bacteriome and Mycobiome Interactions Underscore Microbial Dysbiosis in Familial Crohn’s Disease. mBio 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Zarnowski, R.; Westler, W.M.; Lacmbouh, G.A.; Marita, J.M.; Bothe, J.R.; Bernhardt, J.; Lounes-Hadj Sahraoui, A.; Fontaine, J.; Sanchez, H.; Hatfield, R.D.; et al. Novel entries in a fungal biofilm matrix encyclopedia. mBio 2014, 5, e01333-14. [Google Scholar] [CrossRef] [PubMed]
- Nett, J.E.; Crawford, K.; Marchillo, K.; Andes, D.R. Role of Fks1p and matrix glucan in Candida albicans biofilm resistance to an echinocandin, pyrimidine, and polyene. Antimicrob. Agents Chemother. 2010, 54, 3505–3508. [Google Scholar] [CrossRef] [PubMed]
- Nett, J.E.; Sanchez, H.; Cain, M.T.; Andes, D.R. Genetic basis of Candida biofilm resistance due to drug-sequestering matrix glucan. J. Infect. Dis. 2010, 202, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, K.F.; Zarnowski, R.; Andes, D.R. Fungal Super Glue: The Biofilm Matrix and Its Composition, Assembly, and Functions. PLoS Pathog. 2016, 12, e1005828. [Google Scholar] [CrossRef] [PubMed]
- Martins, M.; Uppuluri, P.; Thomas, D.P.; Cleary, I.A.; Henriques, M.; Lopez-Ribot, J.L.; Oliveira, R. Presence of extracellular DNA in the Candida albicans biofilm matrix and its contribution to biofilms. Mycopathologia 2010, 169, 323–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martins, M.; Henriques, M.; Lopez-Ribot, J.L.; Oliveira, R. Addition of DNase improves the in vitro activity of antifungal drugs against Candida albicans biofilms. Mycoses 2012, 55, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, R.; Sherry, L.; Lappin, D.F.; Nile, C.J.; Smith, K.; Williams, C.; Munro, C.A.; Ramage, G. Extracellular DNA release confers heterogeneity in Candida albicans biofilm formation. BMC Microbiol. 2014, 14, 303. [Google Scholar] [CrossRef] [PubMed]
- Nobile, C.J.; Fox, E.P.; Nett, J.E.; Sorrells, T.R.; Mitrovich, Q.M.; Hernday, A.D.; Tuch, B.B.; Andes, D.R.; Johnson, A.D. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 2012, 148, 126–138. [Google Scholar] [CrossRef] [PubMed]
- Fox, E.P.; Bui, C.K.; Nett, J.E.; Hartooni, N.; Mui, M.C.; Andes, D.R.; Nobile, C.J.; Johnson, A.D. An expanded regulatory network temporally controls Candida albicans biofilm formation. Mol. Microbiol. 2015, 96, 1226–1239. [Google Scholar] [CrossRef] [PubMed]
- Holland, L.M.; Schroder, M.S.; Turner, S.A.; Taff, H.; Andes, D.; Grozer, Z.; Gacser, A.; Ames, L.; Haynes, K.; Higgins, D.G.; et al. Comparative phenotypic analysis of the major fungal pathogens Candida parapsilosis and Candida albicans. PLoS Pathog. 2014, 10, e1004365. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Wang, H.; Shang, Q.; Jiang, Y.; Cao, Y.; Chai, Y. Time course analysis of Candida albicans metabolites during biofilm development. J. Proteome Res. 2013, 12, 2375–2385. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.J.; Brown, G.D.; Netea, M.G.; Gow, N.A. Metabolism impacts upon Candida immunogenicity and pathogenicity at multiple levels. Trends Microbiol. 2014, 22, 614–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nett, J.E. The Host’s Reply to Candida Biofilm. Pathogens 2016, 5, 33. [Google Scholar] [CrossRef] [PubMed]
- Chandra, J.; McCormick, T.S.; Imamura, Y.; Mukherjee, P.K.; Ghannoum, M.A. Interaction of Candida albicans with adherent human peripheral blood mononuclear cells increases C. albicans biofilm formation and results in differential expression of pro- and anti-inflammatory cytokines. Infect. Immun. 2007, 75, 2612–2620. [Google Scholar] [CrossRef] [PubMed]
- Nett, J.E.; Zarnowski, R.; Cabezas-Olcoz, J.; Brooks, E.G.; Bernhardt, J.; Marchillo, K.; Mosher, D.F.; Andes, D.R. Host contributions to construction of three device-associated Candida albicans biofilms. Infect. Immun. 2015, 83, 4630–4638. [Google Scholar] [CrossRef] [PubMed]
- Urban, C.F.; Reichard, U.; Brinkmann, V.; Zychlinsky, A. Neutrophil extracellular traps capture and kill Candida albicans yeast and hyphal forms. Cell. Microbiol. 2006, 8, 668–676. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.J.; Cabezas-Olcoz, J.; Kernien, J.F.; Wang, S.X.; Beebe, D.J.; Huttenlocher, A.; Ansari, H.; Nett, J.E. The Extracellular Matrix of Candida albicans Biofilms Impairs Formation of Neutrophil Extracellular Traps. PLoS Pathog. 2016, 12, e1005884. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.J.; Kernien, J.F.; Hoyer, A.R.; Nett, J.E. Mechanisms involved in the triggering of neutrophil extracellular traps (NETs) by Candida glabrata during planktonic and biofilm growth. Sci. Rep. 2017, 7, 13065. [Google Scholar] [CrossRef] [PubMed]
- Katragkou, A.; Kruhlak, M.J.; Simitsopoulou, M.; Chatzimoschou, A.; Taparkou, A.; Cotten, C.J.; Paliogianni, F.; Diza-Mataftsi, E.; Tsantali, C.; Walsh, T.J.; et al. Interactions between human phagocytes and Candida albicans biofilms alone and in combination with antifungal agents. J. Infect. Dis. 2010, 201, 1941–1949. [Google Scholar] [CrossRef] [PubMed]
- Katragkou, A.; Chatzimoschou, A.; Simitsopoulou, M.; Georgiadou, E.; Roilides, E. Additive antifungal activity of anidulafungin and human neutrophils against Candida parapsilosis biofilms. J. Antimicrob. Chemother. 2011, 66, 588–591. [Google Scholar] [CrossRef] [PubMed]
- Rocha, F.A.C.; Alves, A.; Rocha, M.F.G.; Cordeiro, R.A.; Brilhante, R.S.N.; Pinto, A.; Nunes, R.M.; Girao, V.C.C.; Sidrim, J.J.C. Tumor necrosis factor prevents Candida albicans biofilm formation. Sci. Rep. 2017, 7, 1206. [Google Scholar] [CrossRef] [PubMed]
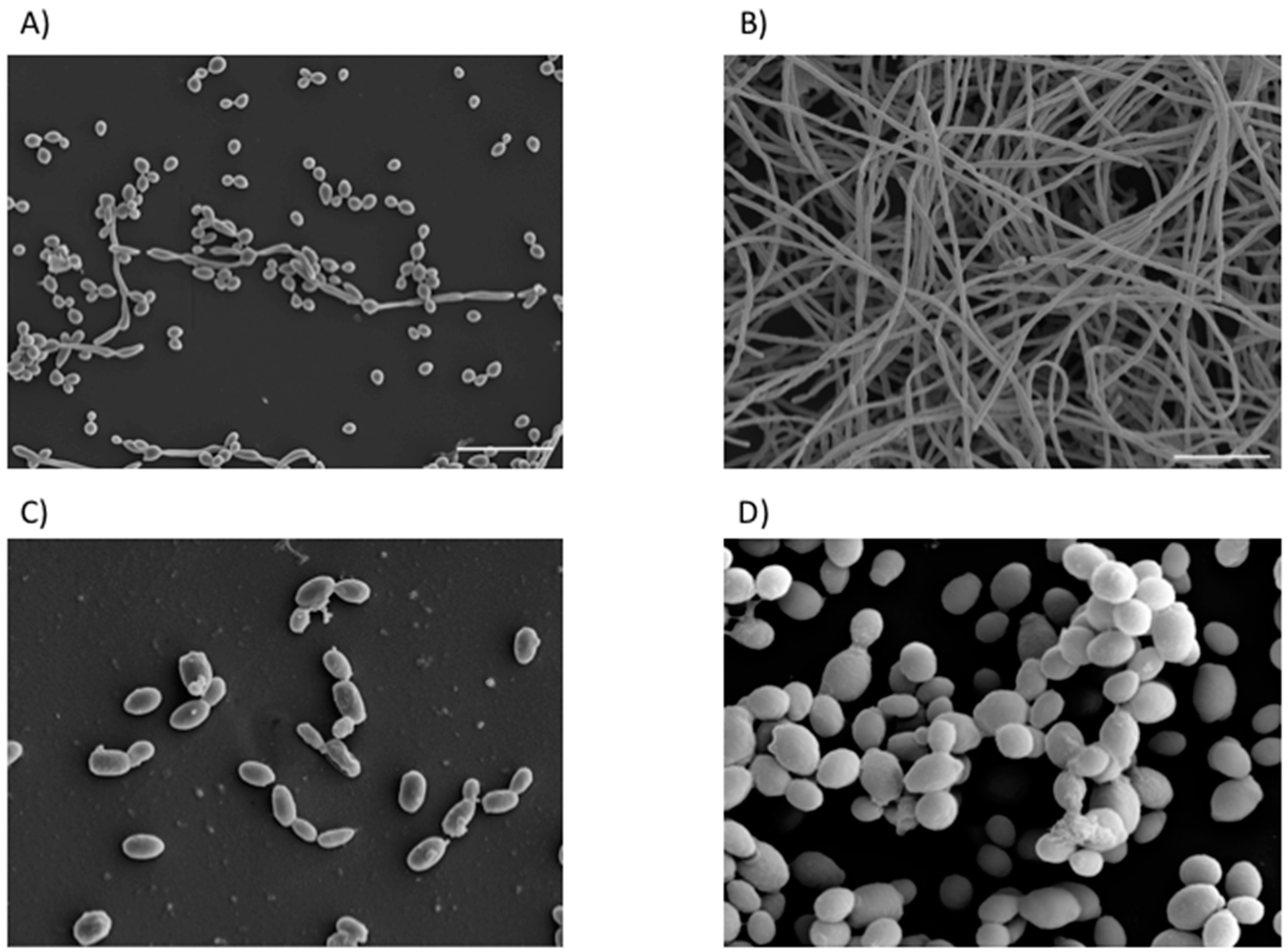
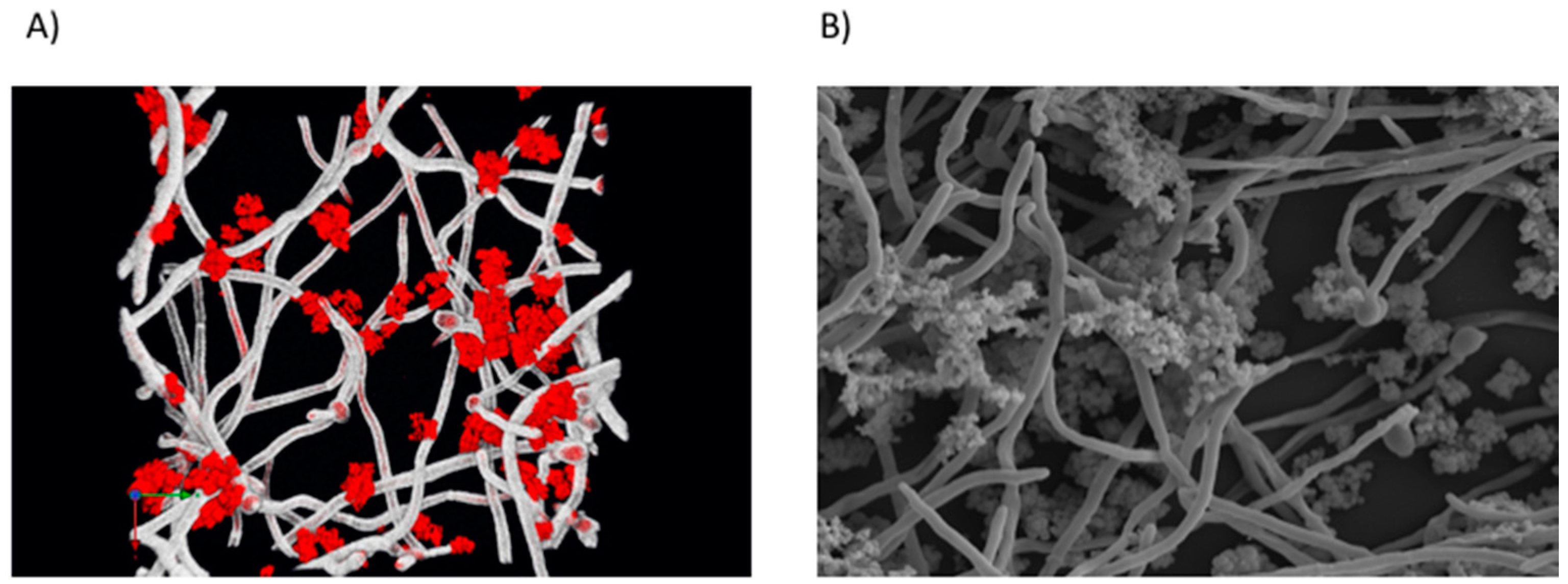
| Location | Fungi | Bacteria | Reference |
|---|---|---|---|
| Mucosal | |||
| Oral cavity | C. albicans, C. glabrata | Streptococcus mutans, Streptococcus gordonii, Porphomonas gingivalis, Staphylococcus aureus | [27,28,29,30,31] |
| Respiratory tract | C. albicans | Pseudomonas aeruginosa | [32,33] |
| Gastrointestinal tract | C. albicans | Enterococcus faecalis, Clostridium difficle | [34,35] |
| Vagina | C. albicans | Lactobacillus spp. | [36] |
| Wounds | C. albicans, C. auris | Pseudomonas aeruginosa, Staphylococcus aureus | [37,38] |
| Device-related | |||
| Denture | C. albicans, C. glabrata | Lactobacillus spp. | [39,40] |
| Voice prosthesis | C. albicans, C. tropicalis | Rothia dentocariosa | [41,42] |
| Artificial heart valves | C. albicans | Staphylococcus aureus, Staphylococcus epidermidis | [43,44] |
| Vascular catheter | C. albicans, C. auris | Staphylococcus aureus, Staphylococcus epidermidis | [45,46,47] |
| Urinary catheter | C. albicans, C. auris | Escherichia coli | [48,49] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kean, R.; Delaney, C.; Rajendran, R.; Sherry, L.; Metcalfe, R.; Thomas, R.; McLean, W.; Williams, C.; Ramage, G. Gaining Insights from Candida Biofilm Heterogeneity: One Size Does Not Fit All. J. Fungi 2018, 4, 12. https://doi.org/10.3390/jof4010012
Kean R, Delaney C, Rajendran R, Sherry L, Metcalfe R, Thomas R, McLean W, Williams C, Ramage G. Gaining Insights from Candida Biofilm Heterogeneity: One Size Does Not Fit All. Journal of Fungi. 2018; 4(1):12. https://doi.org/10.3390/jof4010012
Chicago/Turabian StyleKean, Ryan, Christopher Delaney, Ranjith Rajendran, Leighann Sherry, Rebecca Metcalfe, Rachael Thomas, William McLean, Craig Williams, and Gordon Ramage. 2018. "Gaining Insights from Candida Biofilm Heterogeneity: One Size Does Not Fit All" Journal of Fungi 4, no. 1: 12. https://doi.org/10.3390/jof4010012




