The Plant Defensin NaD1 Enters the Cytoplasm of Candida albicans via Endocytosis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Protein Source
2.2. Fungal Strains
2.3. Confocal Microscopy
2.4. Flow Cytometry
2.5. Growth Inhibition Assays
3. Results
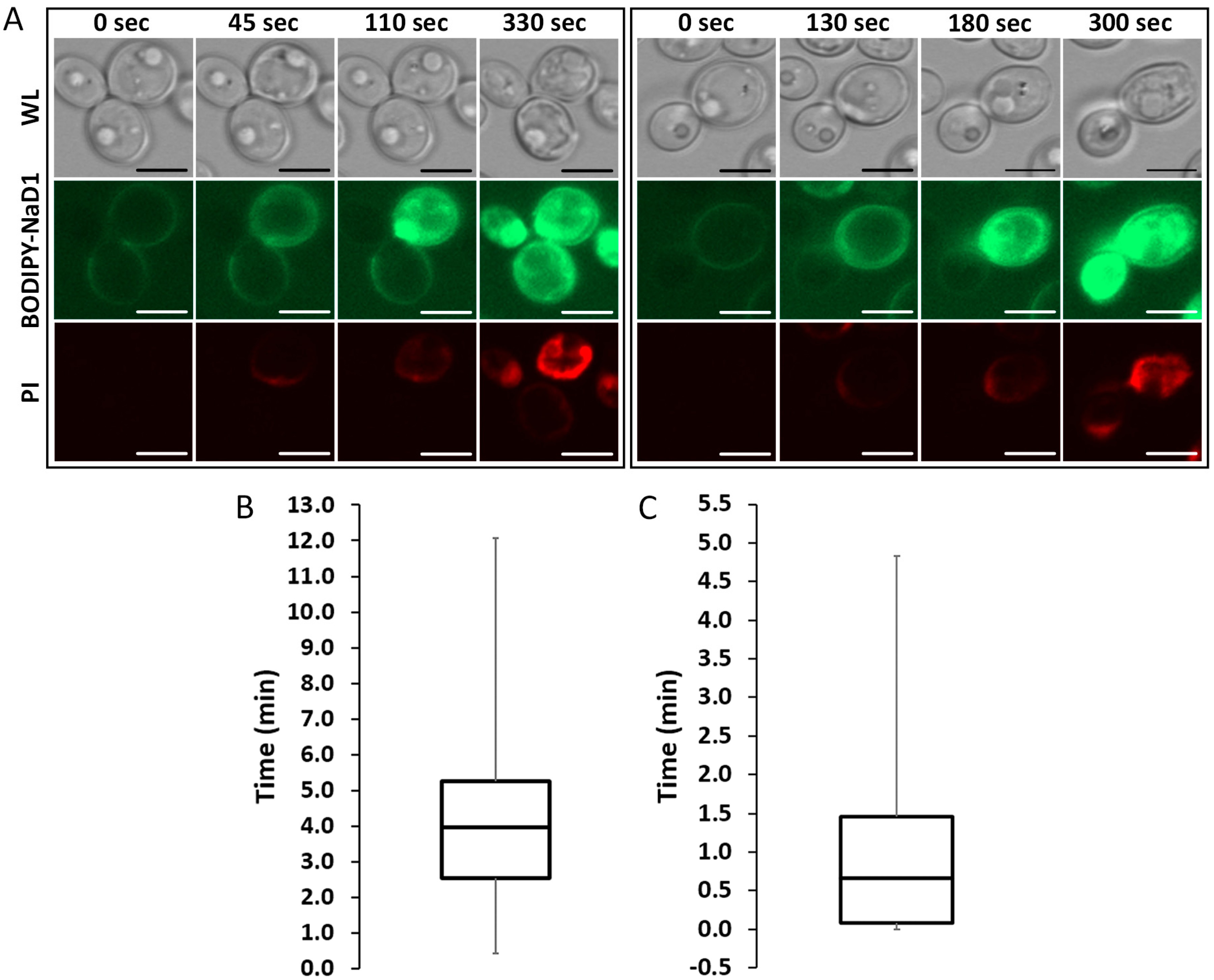
3.1. NaD1 Uptake into C. albicans Cells Occurs in Three Stages
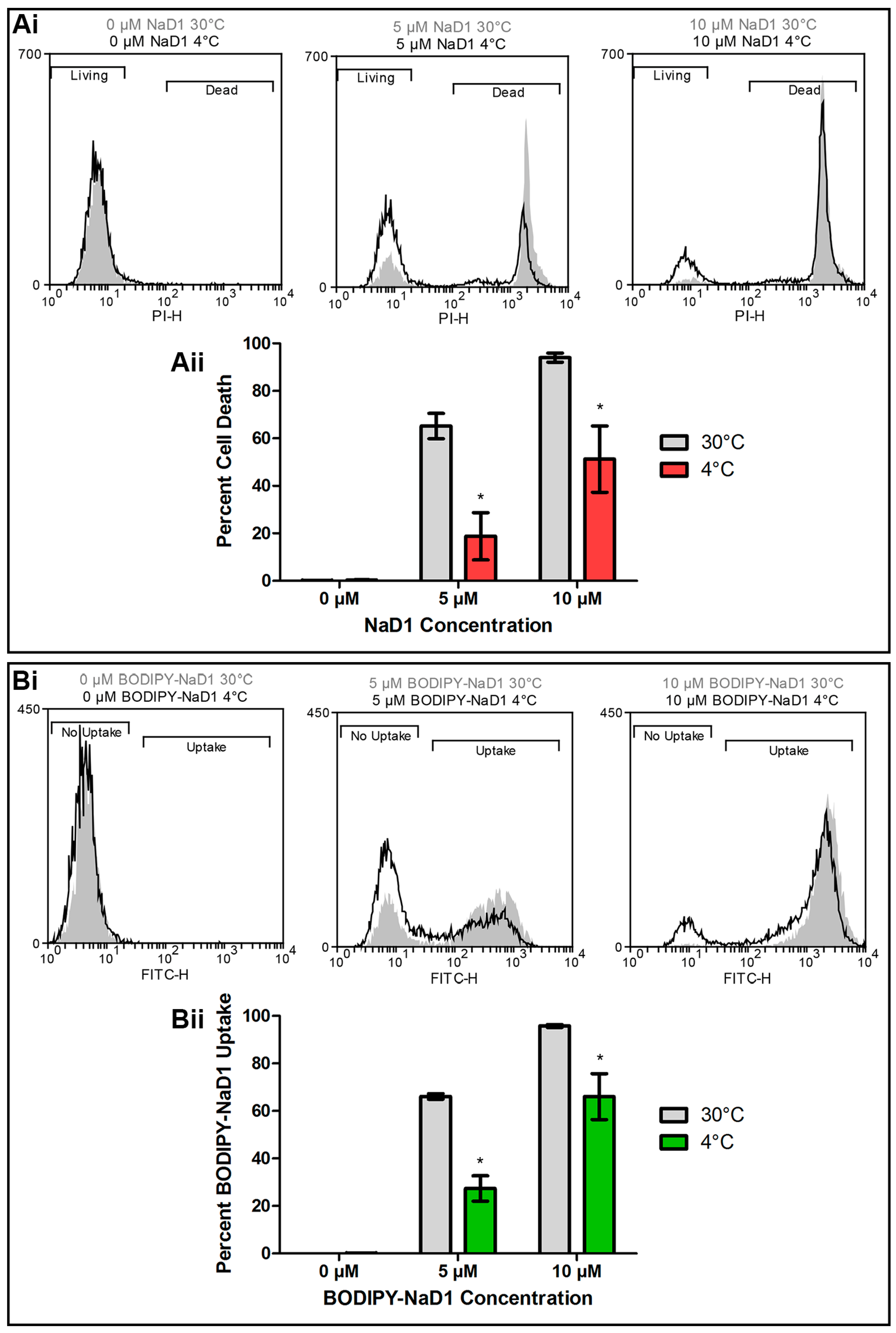
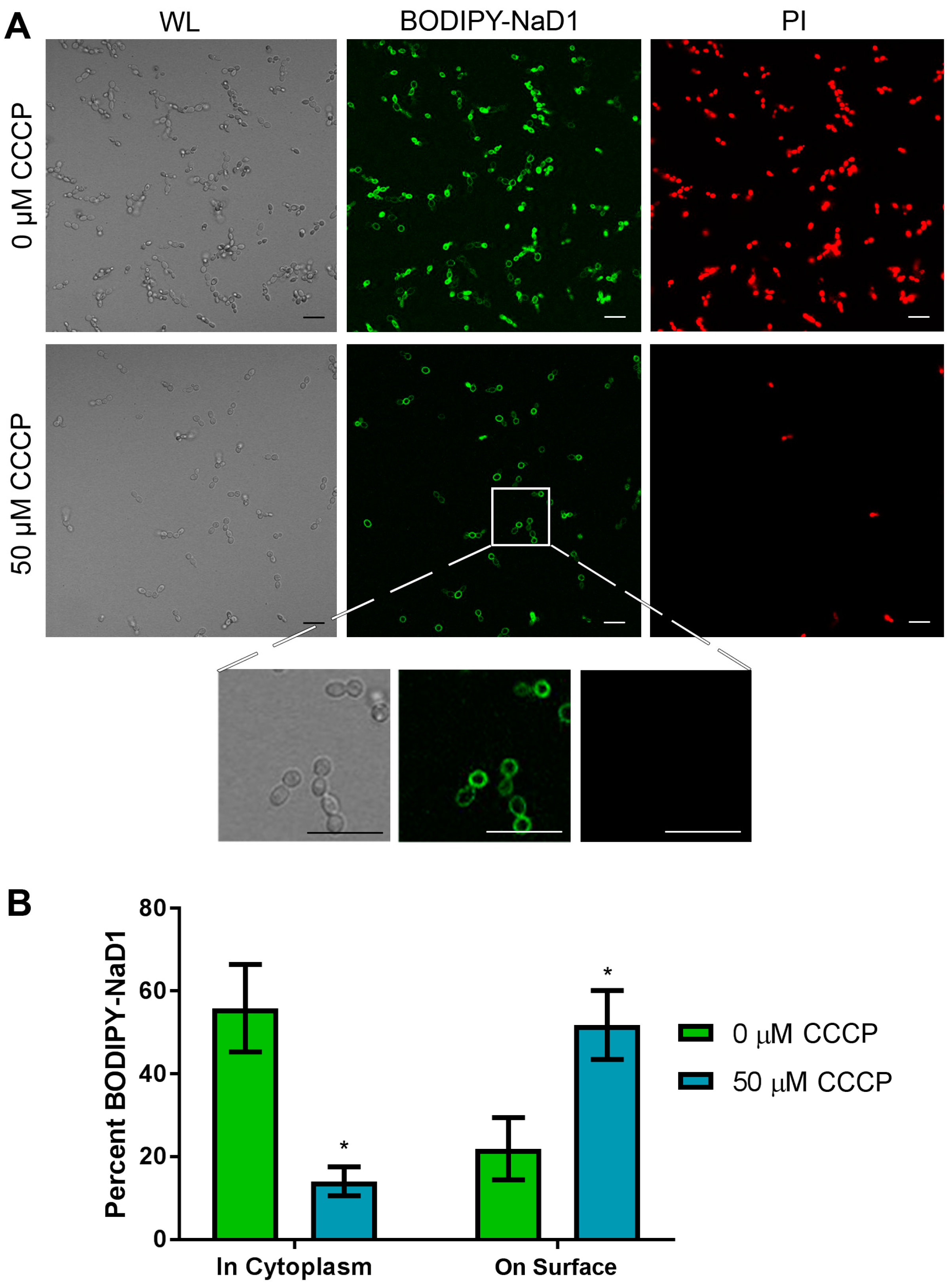
3.2. NaD1’s Antifungal Activity and Uptake into Fungal Cells is Reduced at 4 °C and after Treatment with CCCP
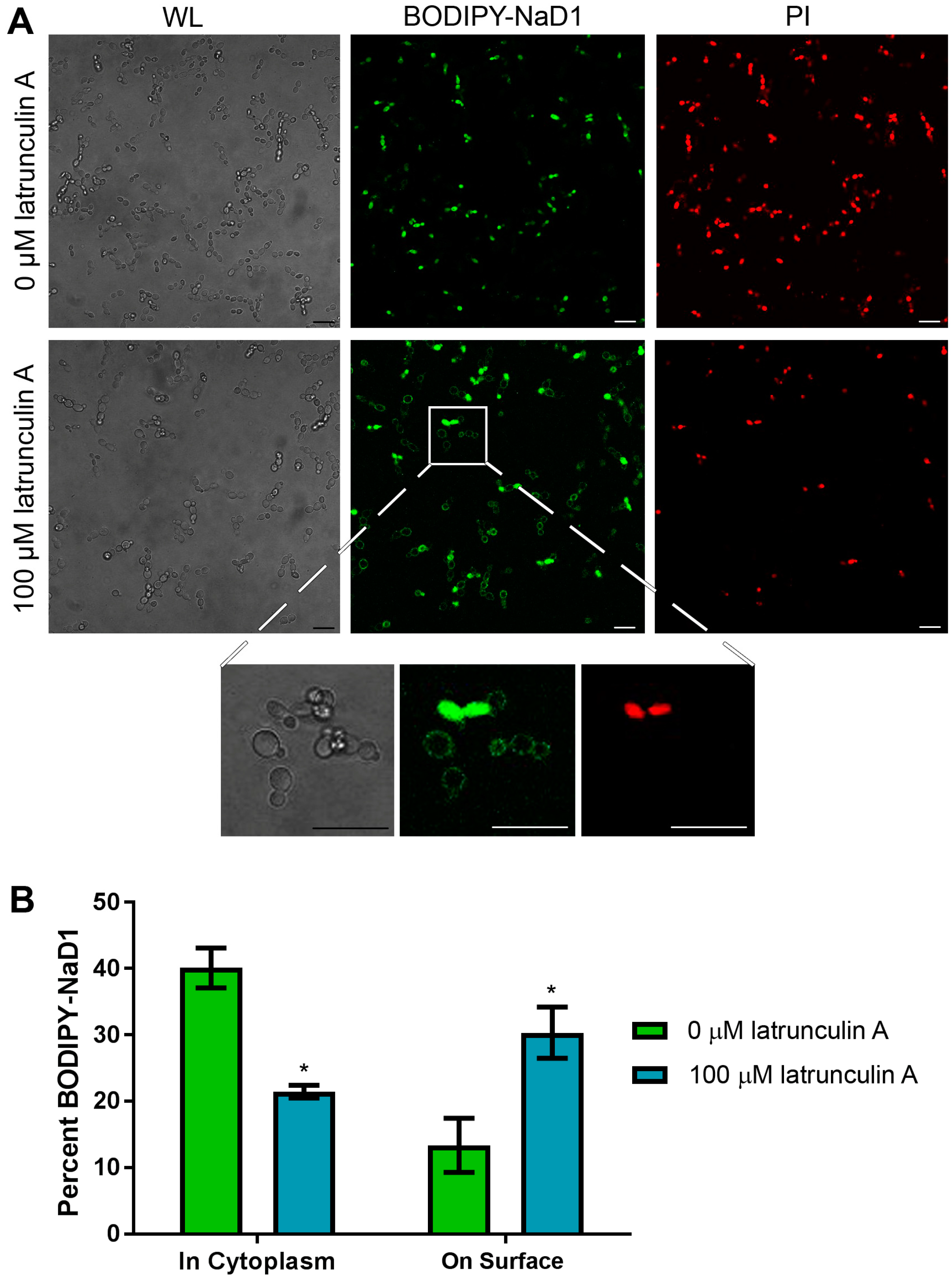
3.3. NaD1 Uptake and NaD1 Induced Cell Death are Reduced in the Presence of the Actin Assembly Inhibitor Latrunculin A
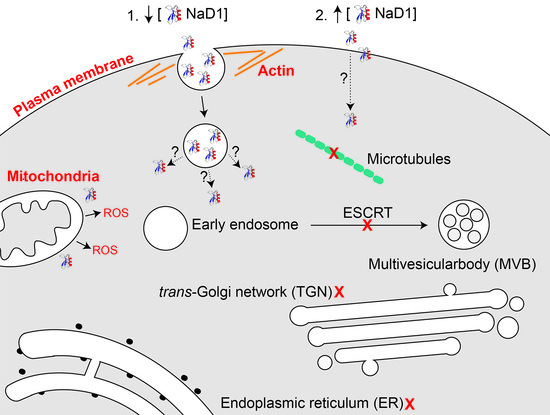
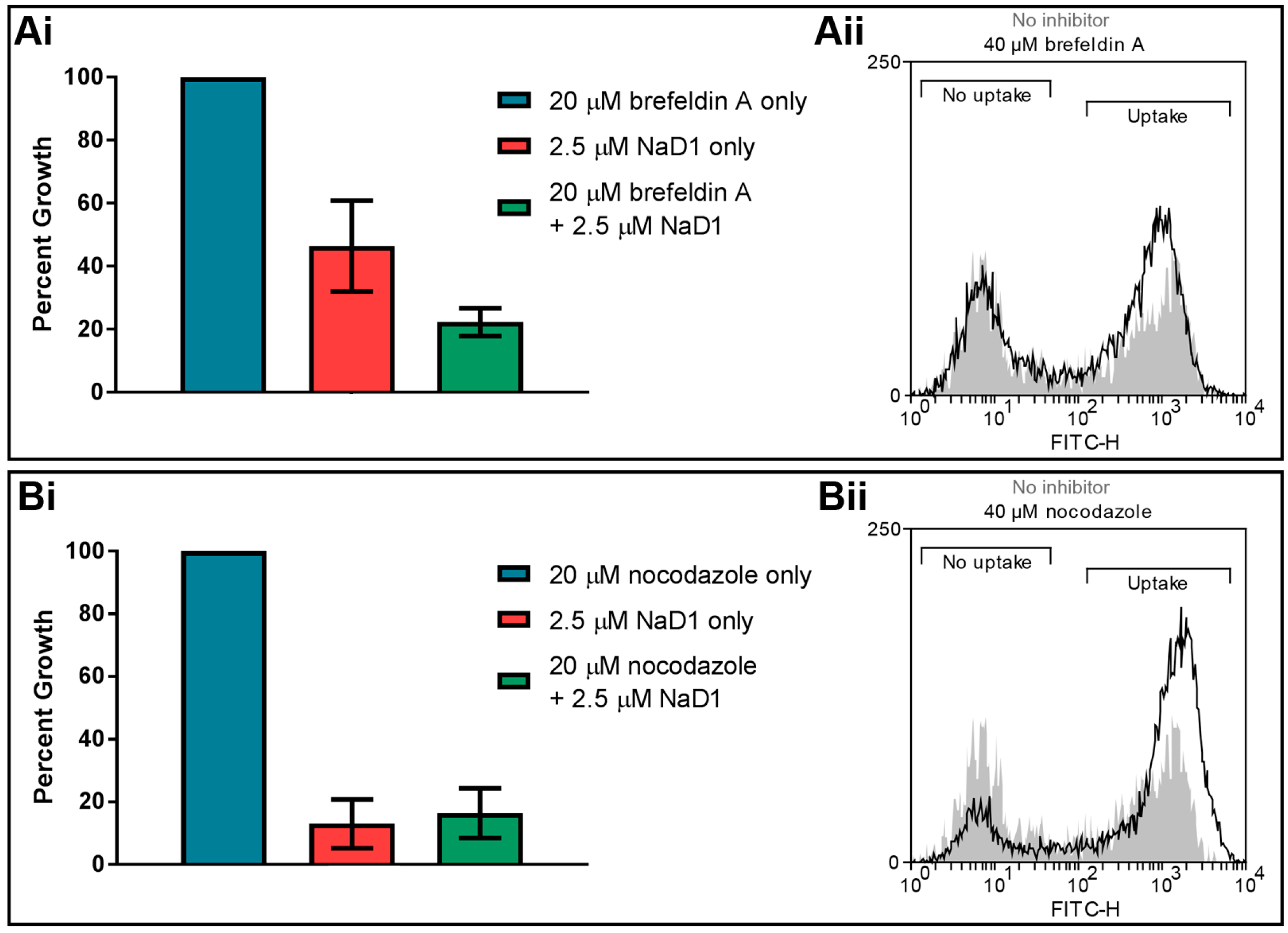
3.4. Internal Protein Transport Is not Required for NaD1 Activity
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Fisher, M.C.; Henk, D.A.; Briggs, C.J.; Brownstein, J.S.; Madoff, L.C.; McCraw, S.L.; Gurr, S.J. Emerging fungal threats to animal, plant and ecosystem health. Nature 2012, 484, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Strange, R.N.; Scott, P.R. Plant disease: A threat to global food security. Annu. Rev. Phytopathol. 2005, 43, 83–116. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.D.; Denning, D.W.; Gow, N.A.R.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012, 4, 165rv113. [Google Scholar] [CrossRef] [PubMed]
- Latge, J.P.; Calderone, R. Host-microbe interactions: Fungi invasive human fungal opportunistic infections. Curr. Opin. Microbiol. 2002, 5, 355–358. [Google Scholar] [CrossRef]
- Wanke, B.; Lazéra, M.D.S.; Nucci, M. Fungal infections in the immunocompromised host. Clin. Haematol. 2000, 95, 153–158. [Google Scholar] [CrossRef]
- Sanglard, D. Resistance of human fungal pathogens to antifungal drugs. Curr. Opin. Microbiol. 2002, 5, 379–385. [Google Scholar] [CrossRef]
- De Lucca, A.J.; Walsh, T.J. Antifungal peptides: Novel therapeutic compounds against emerging pathogens. Antimicrob. Agents Chemother. 1999, 43, 1–11. [Google Scholar] [PubMed]
- Jenssen, H.; Hamill, P.; Hancock, R.E. Peptide antimicrobial agents. Clin. Microbiol. Rev. 2006, 19, 491–511. [Google Scholar] [CrossRef] [PubMed]
- Oren, Z.; Shai, Y. Mode of action of linear amphipathic α-helical antimicrobial peptides. Pept. Sci. 1998, 47, 451–463. [Google Scholar] [CrossRef]
- Nicolas, P. Multifunctional host defense peptides: Intracellular-targeting antimicrobial peptides. FEBS J. 2009, 276, 6483–6496. [Google Scholar] [CrossRef] [PubMed]
- Lay, F.T.; Anderson, M.A. Defensins-Components of the innate immune system in plants. Curr. Protein Pept. Sci. 2005, 6, 85–101. [Google Scholar] [CrossRef] [PubMed]
- Van der Weerden, N.L.; Lay, F.T.; Anderson, M.A. The plant defensin, NaD1, enters the cytoplasm of Fusarium oxysporum hyphae. J. Biol. Chem. 2008, 283, 14445–14452. [Google Scholar] [CrossRef] [PubMed]
- Hayes, B.M.E.; Bleackley, M.R.; Wiltshire, J.L.; Anderson, M.A.; Traven, A.; van der Weerden, N.L. Identification and mechanism of action of the plant defensin NaD1 as a new member of the antifungal drug arsenal against Candida albicans. Antimicrob. Agents Chemother. 2013, 57, 3667–3675. [Google Scholar] [CrossRef] [PubMed]
- Van der Weerden, N.L.; Hancock, R.E.W.; Anderson, M.A. Permeabilization of fungal hyphae by the plant defensin NaD1 occurs through a cell wall-dependent process. J. Biol. Chem. 2010, 285, 37513–37520. [Google Scholar] [CrossRef] [PubMed]
- El-Mounadi, K.; Islam, K.T.; Hernández-Ortiz, P.; Read, N.D.; Shah, D.M. Antifungal mechanisms of a plant defensin MtDef4 are not conserved between the ascomycete fungi Neurospora crassa and Fusarium graminearum. Mol. Microbiol. 2016, 100, 542–559. [Google Scholar] [CrossRef] [PubMed]
- Payne, J.A.E.; Bleackley, M.R.; Lee, T.-H.; Shafee, T.M.A.; Poon, I.K.H.; Hulett, M.D.; Aguilar, M.-I.; van der Weerden, N.L.; Anderson, M.A. The plant defensin NaD1 introduces membrane disorder through a specific interaction with the lipid, phosphatidylinositol 4,5 bisphosphate. BBA Biomembr. 2016, 1858, 1099–1109. [Google Scholar] [CrossRef] [PubMed]
- Heytler, P.G.; Prichard, W.W. A new class of uncoupling agents—Carbonyl cyanide phenylhydrazones. Biochem. Biophys. Res. Commun. 1962, 7, 272–275. [Google Scholar] [CrossRef]
- Oberparleiter, C.; Kaiserer, L.; Haas, H.; Ladurner, P.; Andratsch, M.; Marx, F. Active internalization of the Penicillium chrysogenum antifungal protein PAF in sensitive Aspergilli. Antimicrob. Agents Chemother. 2003, 47, 3598–3601. [Google Scholar] [CrossRef] [PubMed]
- Ayscough, K.R.; Stryker, J.; Pokala, N.; Sanders, M.; Crews, P.; Drubin, D.G. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 1997, 137, 399–416. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; Donaldson, J.G. Search for inhibitors of endocytosis: Intended specificity and unintended consequences. Cell. Logist. 2012, 2, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Chadha, S.; Saraswat, D.; Bajwa, J.S.; Li, R.A.; Conti, H.R.; Edgerton, M. Histatin 5 uptake by Candida albicans utilizes polyamine transporters Dur3 and Dur31 proteins. J. Biol. Chem. 2011, 286, 43748–43758. [Google Scholar] [CrossRef] [PubMed]
- Tati, S.; Jang, W.S.; Li, R.; Kumar, R.; Puri, S.; Edgerton, M. Histatin 5 resistance of Candida glabrata can be reversed by insertion of Candida albicans polyamine transporter-encoding genes DUR3 and DUR31. PLoS ONE 2013, 8, e61480. [Google Scholar] [CrossRef] [PubMed]
- Deshayes, S.; Morris, M.C.; Divita, G.; Heitz, F. Cell-penetrating peptides: Tools for intracellular delivery of therapeutics. Cell. Mol. Life Sci. 2005, 62, 1839–1849. [Google Scholar] [CrossRef] [PubMed]
- Lindgren, M.E.; Hällbrink, M.M.; Elmquist, A.M.; Langel, U. Passage of cell-penetrating peptides across a human epithelial cell layer in vitro. Biochem. J. 2004, 377, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Fungal Genetics Stock Center. Available online: http://www.fgsc.net/.
- Davis, D.; Edwards, J.E.; Mitchell, A.P.; Ibrahim, A.S. Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect. Immun. 2000, 68, 5953–5959. [Google Scholar] [CrossRef] [PubMed]
- Davis, D.A.; Bruno, V.M.; Loza, L.; Filler, S.G.; Mitchell, A.P. Candida albicans Mds3p, a conserved regulator of pH responses and virulence identified through insertional mutagenesis. Genetics 2002, 162, 1573–1581. [Google Scholar] [PubMed]
- Xu, W.; Smith, F.J.; Subaran, R.; Mitchell, A.P. Multivesicular body-ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans. Mol. Biol. Cell 2004, 15, 5528–5537. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Meth. 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Broekaert, W.F.; Terras, F.R.G.; Cammue, B.P.A.; Vanderleyden, J. An automated quantitative assay for fungal growth inhibition. FEMS Microbiol. Lett. 1990, 69, 55–59. [Google Scholar] [CrossRef]
- Iacopetta, B.J.; Morgan, E.H. The kinetics of transferrin endocytosis and iron uptake from transferrin in rabbit reticulocytes. J. Biol. Chem. 1983, 258, 9108–9115. [Google Scholar] [PubMed]
- Munn, A.L. Molecular requirements for the internalisation step of endocytosis: Insights from yeast. BBA Mol. Basis Dis. 2001, 1535, 236–257. [Google Scholar] [CrossRef]
- Lippincott-Schwartz, J.; Yuan, L.; Tipper, C.; Amherdt, M.; Orci, L.; Klausner, R.D. Brefeldin A’s effects on endosomes, lysosomes, and the TGN suggest a general mechanism for regulating organelle structure and membrane traffic. Cell 1991, 67, 601–616. [Google Scholar] [CrossRef]
- Wood, S.A.; Park, J.E.; Brown, W.J. Brefeldin A causes a microtubule-mediated fusion of the trans-Golgi network and early endosomes. Cell 1991, 67, 591–600. [Google Scholar] [CrossRef]
- Apodaca, G. Endocytic traffic in polarized epithelial cells: Role of the actin and microtubule cytoskeleton. Traffic 2001, 2, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Matteoni, R.; Kreis, T.E. Translocation and clustering of endosomes and lysosomes depends on microtubules. J. Biol. Chem. 1987, 105, 1253–1265. [Google Scholar] [CrossRef]
- Henne, W.M.; Buchkovich, N.J.; Emr, S.D. The ESCRT pathway. Dev. Cell 2011, 21, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Saksena, S.; Sun, J.; Chu, T.; Emr, S.D. ESCRTing proteins in the endocytic pathway. Trends Biochem. Sci. 2007, 32, 561–573. [Google Scholar] [CrossRef] [PubMed]
- Lobo, D.S.; Pereira, I.B.; Fragel-Madeira, L.; Medeiros, L.N.; Cabral, L.M.; Faria, J.; Bellio, M.; Campos, R.C.; Linden, R.; Kurtenbach, E. Antifungal Pisum sativum defensin 1 interacts with Neurospora crassa cyclin F related to the cell cycle. Biochemistry 2007, 46, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Sagaram, U.S.; El-Mounadi, K.; Buchko, G.W.; Berg, H.R.; Kaur, J.; Pandurangi, R.S.; Smith, T.J.; Shah, D.M. Structural and functional studies of a phosphatidic acid-binding antifungal plant defensin MtDef4: Identification of an RGFRRR motif governing fungal cell entry. PLoS ONE 2013, 8, e82485. [Google Scholar] [CrossRef] [PubMed]
- Cools, T.L.; Vriens, K.; Struyfs, C.; Verbandt, S.; Ramada, M.H.S.; Brand, G.D.; Bloch, C.; Koch, B.; Traven, A.; Drijfhout, J.W.; et al. The antifungal plant defensin HsAFP1 is a phosphatidic acid-interacting peptide inducing membrane permeabilization. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Jang, W.S.; Bajwa, J.S.; Sun, J.N.; Edgerton, M. Salivary histatin 5 internalization by translocation, but not endocytosis, is required for fungicidal activity in Candida albicans. Mol. Microbiol. 2010, 77, 354–370. [Google Scholar] [CrossRef] [PubMed]
- Gyurko, C.; Lendenmann, U.; Troxler, R.F.; Oppenheim, F.G. Candida albicans mutants deficient in respiration are resistant to the small cationic salivary antimicrobial peptide histatin 5. Antimicrob. Agents Chemother. 2000, 44, 348–354. [Google Scholar] [CrossRef] [PubMed]
- Bleackley, M.R.; Wiltshire, J.L.; Perrine-Walker, F.; Vasa, S.; Burns, R.L.; van der Weerden, N.L.; Anderson, M.A. Agp2p, the plasma membrane transregulator of polyamine uptake, regulates the antifungal activities of the plant defensin NaD1 and other cationic peptides. Antimicrob. Agents Chemother. 2014, 58, 2688–2698. [Google Scholar] [CrossRef] [PubMed]
- Aouida, M.; Texeira, M.R.; Thevelein, J.M.; Poulin, R.; Ramotar, D. Agp2, a member of the yeast amino acid permease family, positively regulates polyamine transport at the transcriptional level. PLoS ONE 2013, 8, e65717. [Google Scholar] [CrossRef]
- Ordonez, S.R.; Amarullah, I.H.; Wubbolts, R.W.; Veldhuizen, E.J.A.; Haagsman, H.P. Fungicidal mechanisms of cathelicidins LL-37 and CATH-2 revealed by live-cell imaging. Antimicrob. Agents Chemother. 2014, 58, 2240–2248. [Google Scholar] [CrossRef] [PubMed]
- Den Hertog, A.; van Marle, J.; van Veen, H.; van’t Hof, W.; Bolscher, J.; Veerman, E.; Nieuw Amerongen, A. Candidacidal effects of two antimicrobial peptides: Histatin 5 causes small membrane defects, but LL-37 causes massive disruption of the cell membrane. Biochem. J. 2005, 388, 689–695. [Google Scholar] [CrossRef] [PubMed]
- Cuesta-Marbán, Á.; Botet, J.; Czyz, O.; Cacharro, L.M.; Gajate, C.; Hornillos, V.; Delgado, J.; Zhang, H.; Amat-Guerri, F.; Acuña, A.U.; et al. Drug uptake, lipid rafts, and vesicle trafficking modulate resistance to an anticancer lysophosphatidylcholine analogue in yeast. J. Biol. Chem. 2013, 288, 8405–8418. [Google Scholar] [CrossRef] [PubMed]

| IC50 (µM) | |||||||
|---|---|---|---|---|---|---|---|
| Wild type (DAY286) | vps2Δ | vps23Δ | vps24Δ | vps28Δ | vps36Δ | snf7Δ | bro1Δ |
| 1.8 ± 0.49 | 1.8 ± 0.40 | 1.8 ± 0.43 | 2.1 ± 0.33 | 1.6 ± 0.33 | 1.7 ± 0.28 | 1.9 ± 0.45 | 2.1 ± 0.35 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hayes, B.M.E.; Bleackley, M.R.; Anderson, M.A.; Van der Weerden, N.L. The Plant Defensin NaD1 Enters the Cytoplasm of Candida albicans via Endocytosis. J. Fungi 2018, 4, 20. https://doi.org/10.3390/jof4010020
Hayes BME, Bleackley MR, Anderson MA, Van der Weerden NL. The Plant Defensin NaD1 Enters the Cytoplasm of Candida albicans via Endocytosis. Journal of Fungi. 2018; 4(1):20. https://doi.org/10.3390/jof4010020
Chicago/Turabian StyleHayes, Brigitte M. E., Mark R. Bleackley, Marilyn A. Anderson, and Nicole L. Van der Weerden. 2018. "The Plant Defensin NaD1 Enters the Cytoplasm of Candida albicans via Endocytosis" Journal of Fungi 4, no. 1: 20. https://doi.org/10.3390/jof4010020