Alginate-Chitosan Hydrogels Containing shRNA Plasmid for Inhibition of CTNNB1 Expression in Breast Cancer Cells
Abstract
:1. Introduction
2. Results and Discussion
2.1. Preparation and Control of pshRNA-Loaded Alginate-Chitosan Hydrogels
2.2. Characterization of the Hydrogels
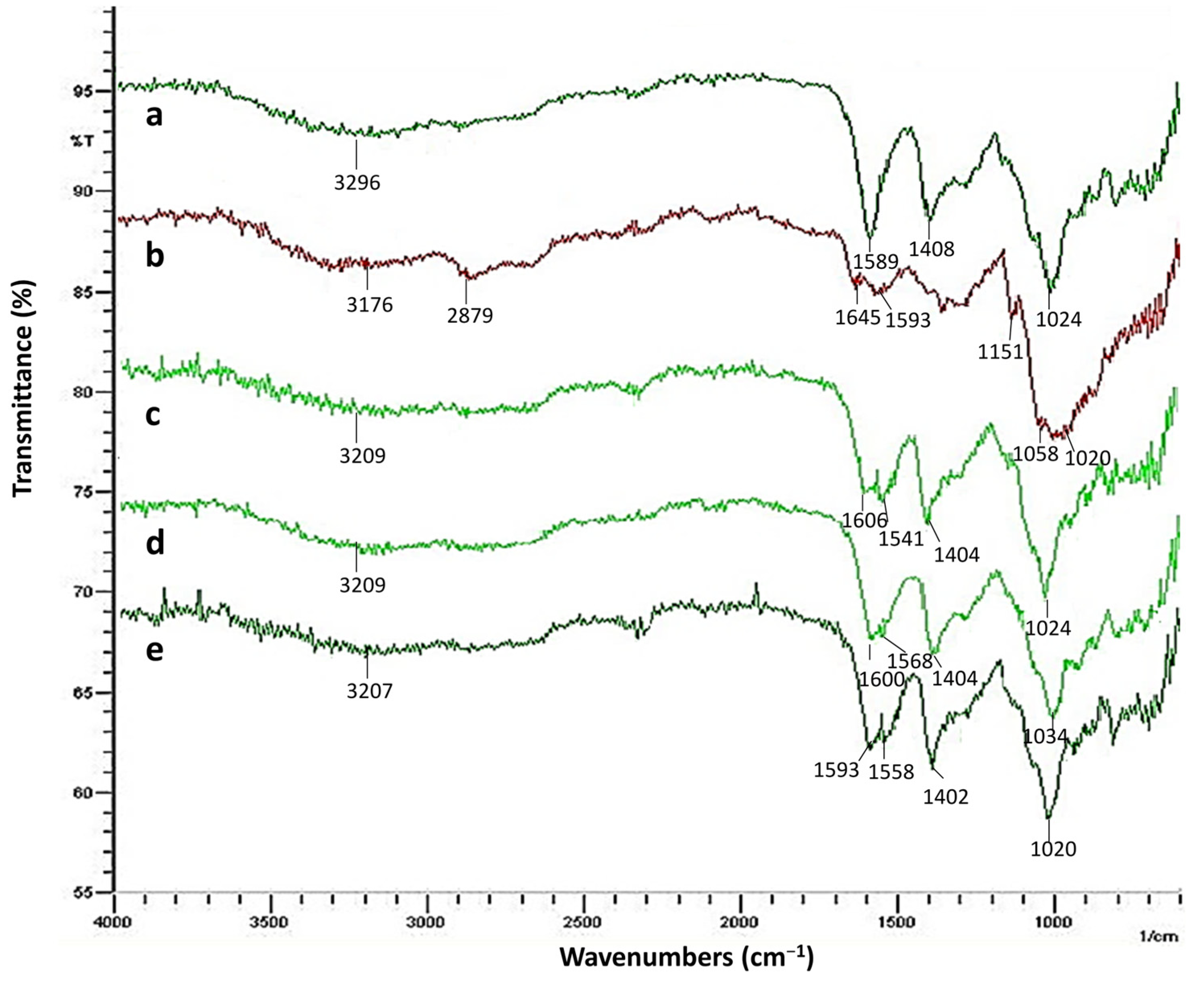
2.2.1. FTIR Analysis
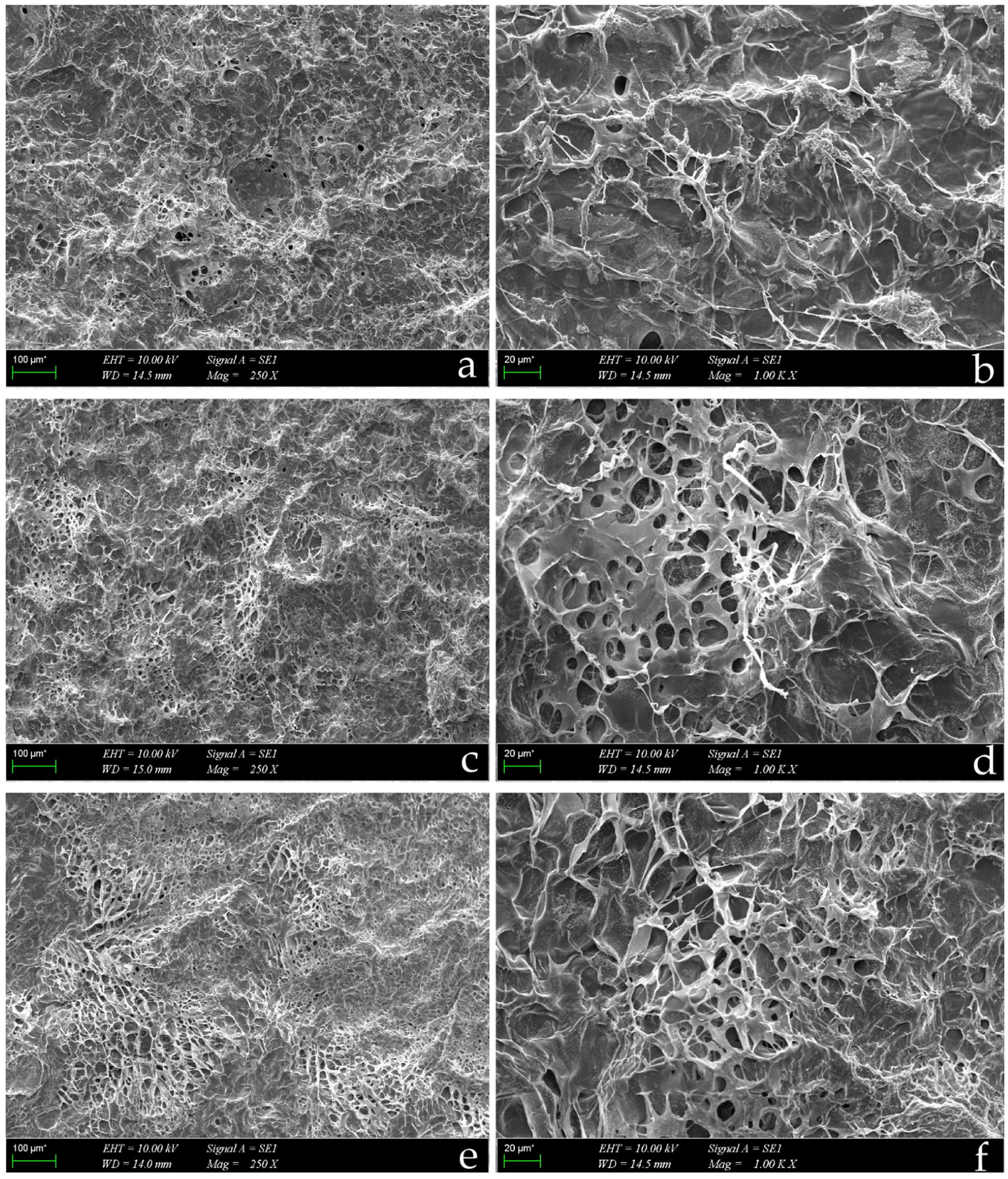
2.2.2. SEM Analysis
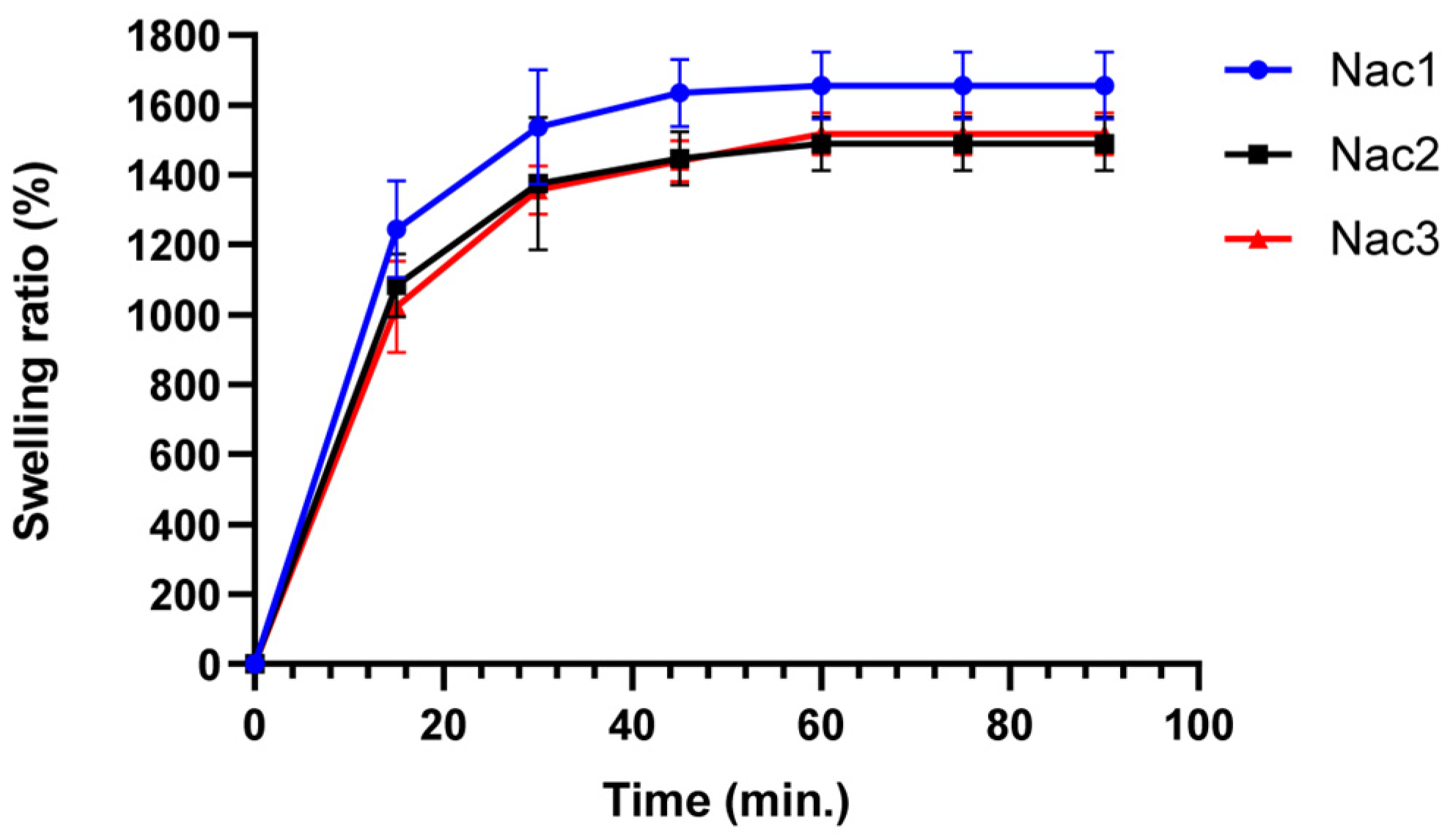
2.2.3. Swelling Properties
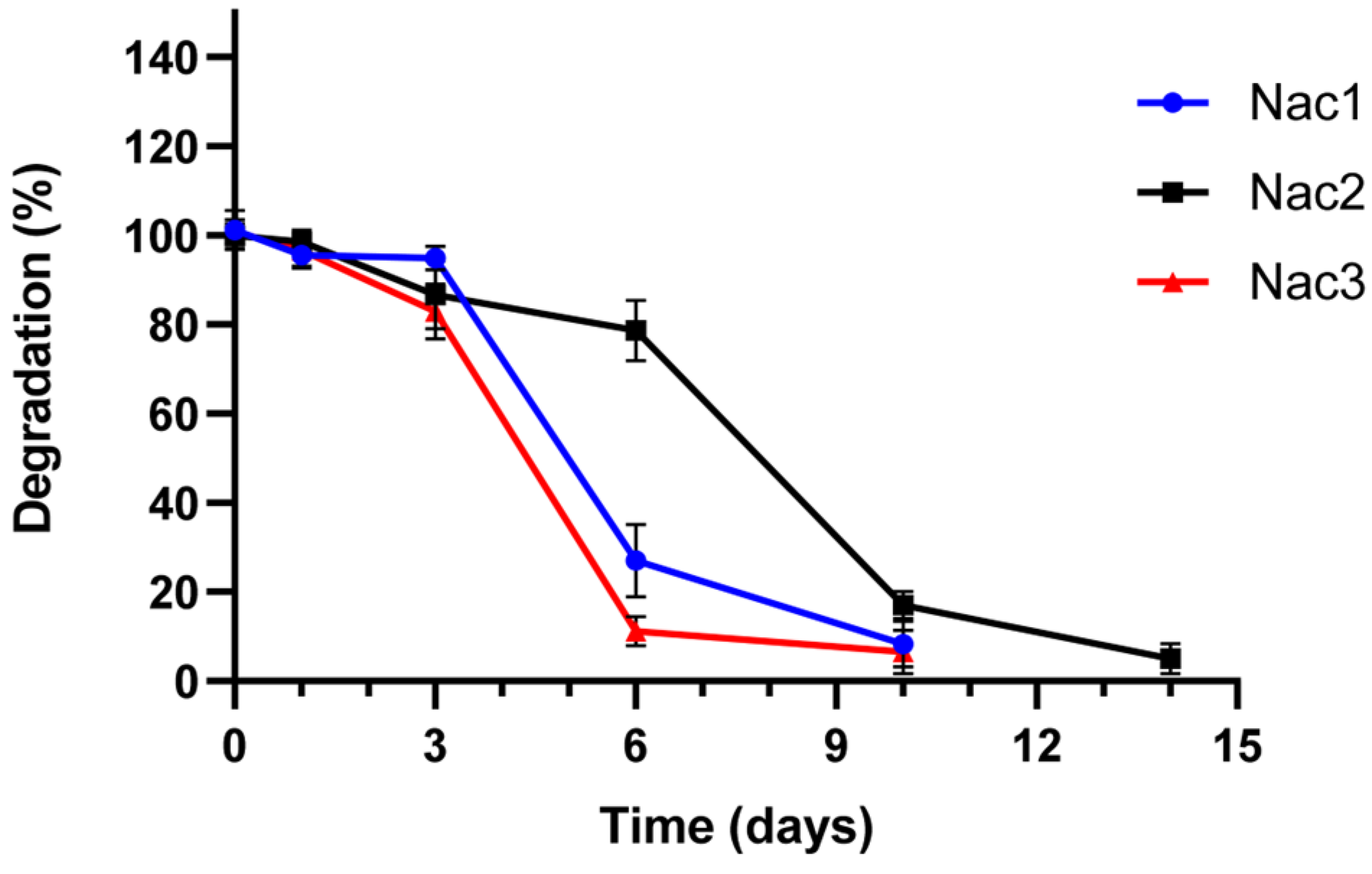
2.2.4. Degradation Properties
2.2.5. Serum Stability
2.2.6. Release Profiles
2.3. Cellular Uptake and CTNNB1 Protein Levels
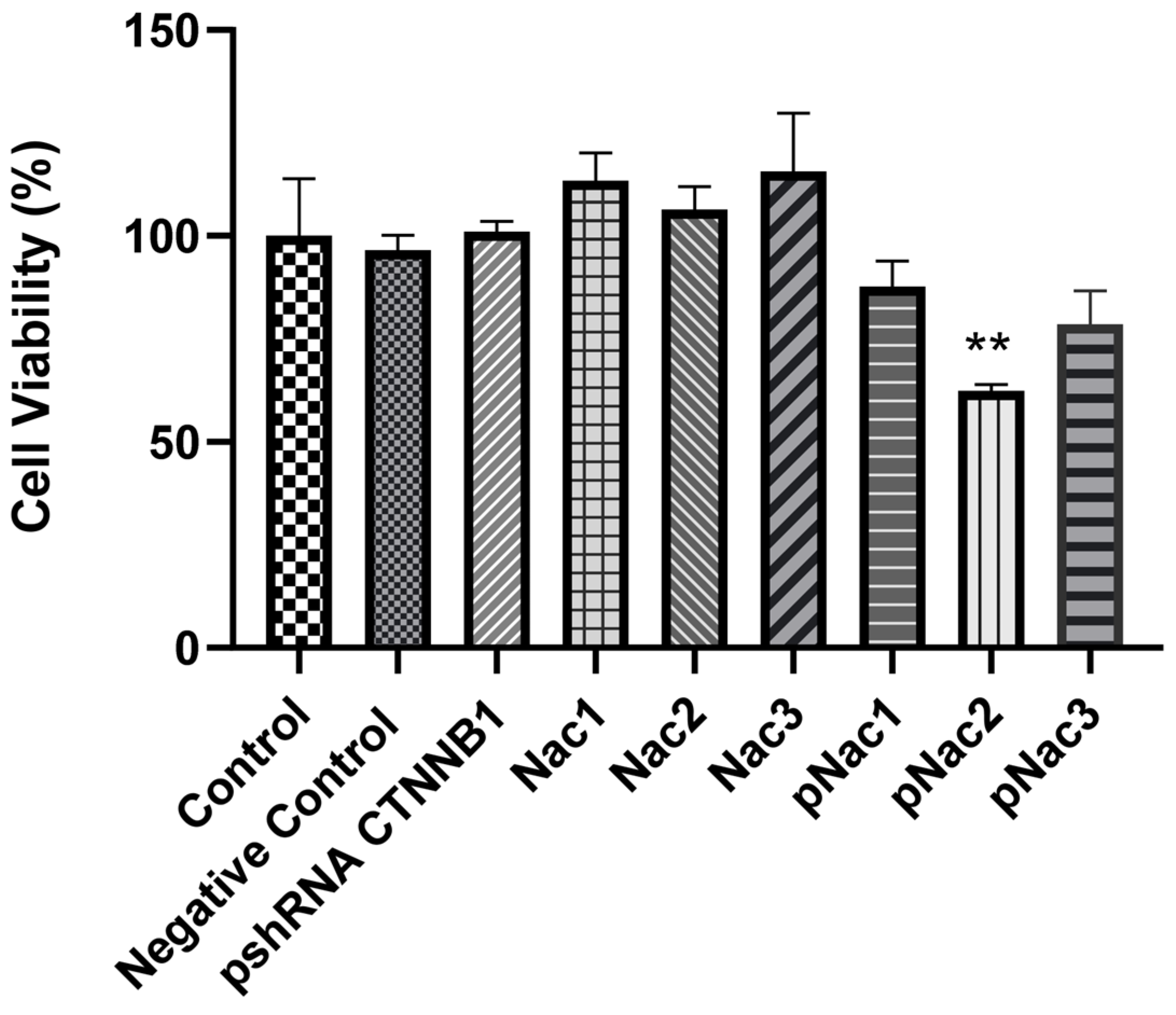
2.4. Cell Proliferation
3. Conclusions
4. Materials and Methods
4.1. Materials
4.2. Transformation and Isolation of shRNA Plasmid
4.3. Preparation of pshRNA-Loaded Alginate-Chitosan Hydrogels
4.4. Characterization of pshRNA-Loaded Alginate-Chitosan Hydrogels
4.4.1. FTIR Analysis
4.4.2. SEM Analysis
4.4.3. Swelling Study
4.4.4. In Vitro Degradation Assay
4.4.5. Serum Stability Assay
4.4.6. Release Study
4.5. In Vitro Transfection
4.6. Determination of CTNNB1 Expression
4.7. Cell Proliferation
4.8. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Harbeck, N.; Penault-Llorca, F.; Cortes, J.; Gnant, M.; Houssami, N.; Poortmans, P.; Ruddy, K.; Tsang, J.; Cardoso, F. Breast cancer. Nat. Rev. Dis. Prim. 2019, 5, 66. [Google Scholar] [CrossRef] [PubMed]
- Khramtsov, A.I.; Khramtsova, G.F.; Tretiakova, M.; Huo, D.; Olopade, O.I.; Goss, K.H. Wnt/β-catenin pathway activation is enriched in basal-like breast cancers and predicts poor outcome. Am. J. Pathol. 2010, 176, 2911–2920. [Google Scholar] [CrossRef] [PubMed]
- López-Knowles, E.; Zardawi, S.J.; McNeil, C.M.; Millar, E.K.; Crea, P.; Musgrove, E.A.; Sutherland, R.L.; O’Toole, S.A. Cytoplasmic localization of β-catenin is a marker of poor outcome in breast cancer patients. Cancer Epidemiol. Biomark. Prev. 2010, 19, 301–309. [Google Scholar] [CrossRef] [Green Version]
- Thompson, M.D.; Monga, S.P.S. WNT/β-catenin signaling in liver health and disease. Hepatology 2007, 45, 1298–1305. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.; Fan, S.; Ma, W.; Fan, P.; Wang, B.; Zhang, J.; Wang, H.; Tang, B.; Zhang, Q.; Yu, X.; et al. Roles of Wnt/β-catenin signaling in the gastric cancer stem cells proliferation and salinomycin treatment. Cell Death Dis. 2014, 5, e1039. [Google Scholar] [CrossRef] [Green Version]
- Abraham, S.C.; Reynolds, C.; Lee, J.H.; Montgomery, E.A.; Baisden, B.L.; Krasinskas, A.M.; Wu, T.T. Fibromatosis of the breast and mutations involving the APC/β-catenin pathway. Hum. Pathol. 2002, 33, 39–46. [Google Scholar] [CrossRef]
- Dahlman, J.E.; Kauffman, K.J.; Langer, R.; Anderson, D.G. Nanotechnology for in vivo targeted siRNA delivery. Adv. Genet. 2014, 88, 37–69. [Google Scholar] [CrossRef]
- Ramakrishnan, S. Hydrogel-siRNA for cancer therapy. Cancer Biol. Ther. 2011, 11, 849–851. [Google Scholar] [CrossRef] [Green Version]
- Charbe, N.B.; Amnerkar, N.D.; Ramesh, B.; Tambuwala, M.M.; Bakshi, H.A.; Aljabali, A.A.A.; Khadse, S.C.; Satheeshkumar, R.; Satija, S.; Metha, M.; et al. Small interfering RNA for cancer treatment: Overcoming hurdles in delivery. Acta Pharm. Sin. B 2020, 10, 2075–2109. [Google Scholar] [CrossRef]
- Gupta, N.; Schmitt, F.; Grebhardt, S.; Mayer, D. β-catenin is a positive regulator of estrogen receptor-α function in breast cancer cells. Cancers 2011, 3, 2990–3001. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Prosperi, J.R.; Choudhury, N.; Olopade, O.I.; Goss, K.H. β-catenin is required for the tumorigenic behavior of triple-negative breast cancer cells. PLoS ONE 2015, 10, e0117097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.S.; Garg, H.; Joshi, A.; Manjunath, N. Strategies for targeted nonviral delivery of siRNAs in vivo. Trends Mol. Med. 2009, 15, 491–500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- des Rieux, A.; Shikanov, A.; Shea, L.D. Fibrin hydrogels for non-viral vector delivery in vitro. J. Control. Release 2009, 136, 148–154. [Google Scholar] [CrossRef] [Green Version]
- Wieland, J.A.; Houchin-Ray, T.L.; Shea, L.D. Non-viral vector delivery from PEG-hyaluronic acid hydrogels. J. Control. Release 2007, 120, 233–241. [Google Scholar] [CrossRef] [Green Version]
- Naghizadeh, Z.; Karkhaneh, A.; Khojasteh, A. Self-crosslinking effect of chitosan and gelatin on alginate based hydrogels: Injectable in situ forming scaffolds. Mater. Sci. Eng. C 2018, 89, 256–264. [Google Scholar] [CrossRef]
- Baysal, K.; Aroguz, A.Z.; Adiguzel, Z.; Baysal, B.M. Chitosan/alginate crosslinked hydrogels: Preparation, characterization and application for cell growth purposes. Int. J. Biol. Macromol. 2013, 59, 342–348. [Google Scholar] [CrossRef]
- Cao, Y.; Tan, Y.F.; Wong, Y.S.; Liew, M.W.J.; Venkatraman, S. Recent advances in chitosan-based carriers for gene delivery. Mar. Drugs 2019, 17, 381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chuan, D.; Jin, T.; Fan, R.; Zhou, L.; Guo, G. Chitosan for gene delivery: Methods for improvement and applications. Adv. Colloid Interface Sci. 2019, 268, 25–38. [Google Scholar] [CrossRef]
- Curley, C.J.; Dolan, E.B.; Otten, M.; Hinderer, S.; Duffy, G.P.; Murphy, B.P. An injectable alginate/extra cellular matrix (ECM) hydrogel towards acellular treatment of heart failure. Drug Deliv. Transl. Res. 2018, 9, 1–13. [Google Scholar] [CrossRef]
- Salehi, M.; Bagher, Z.; Kamrava, S.K.; Ehterami, A.; Alizadeh, R.; Farhadi, M.; Falah, M.; Komeili, A. Alginate/chitosan hydrogel containing olfactory ectomesenchymal stem cells for sciatic nerve tissue engineering. J. Cell. Physiol. 2019, 234, 15357–15368. [Google Scholar] [CrossRef]
- Wei, L.; Tan, J.; Li, L.; Wang, H.; Liu, S.; Chen, J.; Weng, Y.; Liu, T. Chitosan/alginate hydrogel dressing loaded fgf/ve-cadherin to accelerate full-thickness skin regeneration and more normal skin repairs. Int. J. Mol. Sci. 2022, 23, 1249. [Google Scholar] [CrossRef]
- Deng, B.; Shen, L.; Wu, Y.; Shen, Y.; Ding, X.; Lu, S.; Jia, J.; Qian, J.; Ge, J. Delivery of alginate-chitosan hydrogel promotes endogenous repair and preserves cardiac function in rats with myocardial infarction. J. Biomed. Mater. Res. A 2015, 103, 907–918. [Google Scholar] [CrossRef] [PubMed]
- Rassu, G.; Salis, A.; Porcu, E.P.; Giunchedi, P.; Roldo, M.; Gavini, E. Composite chitosan/alginate hydrogel for controlled release of deferoxamine: A system to potentially treat iron dysregulation diseases. Carbohydr. Polym. 2016, 136, 1338–1347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khong, T.T.; Aarstad, O.A.; Skjåk-Bræk, G.; Draget, K.I.; Vårum, K.M. Gelling concept combining chitosan and alginate—Proof of principle. Biomacromolecules 2013, 14, 2765–2771. [Google Scholar] [CrossRef]
- Liu, Q.; Li, Q.; Xu, S.; Zheng, Q.; Cao, X. Preparation and properties of 3d printed alginate–chitosan polyion complex hydrogels for tissue engineering. Polymers 2018, 10, 664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emani, S.; Vangala, A.; Buonocore, F.; Yarandi, N.; Calabrese, G. Chitosan hydrogels cross-linked with trimesic acid for the delivery of 5-fluorouracil in cancer therapy. Pharmaceutics 2023, 15, 1084. [Google Scholar] [CrossRef] [PubMed]
- Rahmati, M.; Ehterami, A.; Saberani, R.; Abbaszadeh-Goudarzi, G.; Rezaei Kolarijani, N.; Khastar, H.; Garmabi, B.; Salehi, M. Improving sciatic nerve regeneration by using alginate/chitosan hydrogel containing berberine. Drug Deliv. Transl. Res. 2020, 11, 1983–1993. [Google Scholar] [CrossRef] [PubMed]
- Kopplin, G.; Lervik, A.; Draget, K.I.; Aachmann, F.L. Alginate gels crosslinked with chitosan oligomers—A systematic investigation into alginate block structure and chitosan oligomer interaction. RSC Adv. 2021, 11, 13780–13798. [Google Scholar] [CrossRef]
- Cheaburu, Y.; Yilmaz, O.; Kose, F.A.; Bibire, N. Chitosan-Graft-Poly(N-Isopropylacrylamide)/PVA cryogels as carriers for mucosal delivery of voriconazole. Polymers 2019, 11, 1432. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.J.; Park, S.J.; Kim, S.I. Swelling behavior of interpenetrating polymer network hydrogels composed of poly(vinyl alcohol) and chitosan. React. Funct. Polym. 2003, 55, 53–59. [Google Scholar] [CrossRef]
- Aied, A.; Greiser, U.; Pandit, A.; Wang, W. Polymer gene delivery: Overcoming the obstacles. Drug Discov. Today 2013, 18, 1090–1098. [Google Scholar] [CrossRef] [PubMed]
- Santos-Carballal, B.; Fernández Fernández, E.; Goycoolea, F. Chitosan in non-viral gene delivery: Role of structure, characterization methods, and insights in cancer and rare diseases therapies. Polymers 2018, 10, 444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dadsetan, M.; Szatkowski, J.P.; Shogren, K.L.; Yaszemski, M.J.; Maran, A. Hydrogel-mediated DNA delivery confers estrogenic response in nonresponsive osteoblast cells. J. Biomed. Mater. Res. A 2009, 91, 1170–1777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, S.; Wu, S.; Zhang, J.; Zhang, S.; Huang, Y.; Zhu, H.; Li, Y.; Qi, B. Controlled release of curcumin from gelatin hydrogels by the molecular-weight modulation of an oxidized dextran cross-linker. Food Chem. 2023, 418, 135966. [Google Scholar] [CrossRef] [PubMed]
- Zolfagharzadeh, V.; Ai, J.; Soltani, H.; Hassanzadeh, S.; Khanmohammadi, M. Sustain release of loaded insulin within biomimetic hydrogel microsphere for sciatic tissue engineering in vivo. Int. J. Biol. Macromol. 2023, 225, 687–700. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, M. Chitosan-alginate as scaffolding material for cartilage tissue engineering. J. Biomed. Mater. Res. A 2005, 75, 485–493. [Google Scholar] [CrossRef]
- Lara-Velazquez, M.; Alkharboosh, R.; Norton, E.S.; Ramirez-Loera, C.; Freeman, W.D.; Guerrero-Cazares, H.; Forte, A.J.; Quiñones-Hinojosa, A.; Sarabia-Estrada, R. Chitosan-based non-viral gene and drug delivery systems for brain cancer. Front. Neurol. 2020, 11, 740. [Google Scholar] [CrossRef]
- Zhang, D.; Fei, F.; Li, S.; Zhao, Y.; Yang, Z.; Qu, J.; Zhang, X.; Yin, Y.; Zhang, S. The role of β-catenin in the initiation and metastasis of TA2 mice spontaneous breast cancer. J. Cancer 2017, 8, 2114–2123. [Google Scholar] [CrossRef] [Green Version]
- Ashaie, M.A.; Islam, R.A.; Kamaruzman, N.I.; Ibnat, N.; Tha, K.K.; Chowdhury, E.H. Targeting cell adhesion molecules via carbonate apatite-mediated delivery of specific sirnas to breast cancer cells in vitro and in vivo. Pharmaceutics 2019, 11, 309. [Google Scholar] [CrossRef] [Green Version]
- Saygili, E.; Kaya, E.; Ilhan-Ayisigi, E.; Saglam-Metiner, P.; Alarcin, E.; Kazan, A.; Girgic, E.; Kim, Y.W.; Gunes, K.; Eren-Ozcan, G.G.; et al. An alginate-poly(acrylamide) hydrogel with TGF-β3 loaded nanoparticles for cartilage repair: Biodegradability, biocompatibility and protein adsorption. Int. J. Biol. Macromol. 2021, 172, 381–393. [Google Scholar] [CrossRef]
- Salva, E.; Akbuğa, J. In vitro silencing effect of chitosan nanoplexes containing siRNA expressing vector targeting VEGF in breast cancer cell lines. Pharmazie 2010, 65, 896–902. [Google Scholar] [PubMed]
- Gao, Y.; Ji, H.; Peng, L.; Gao, X.; Jiang, S. Development of PLGA-PEG-PLGA hydrogel delivery system for enhanced immunoreaction and efficacy of newcastle disease virus dna vaccine. Molecules 2020, 25, 2505. [Google Scholar] [CrossRef] [PubMed]

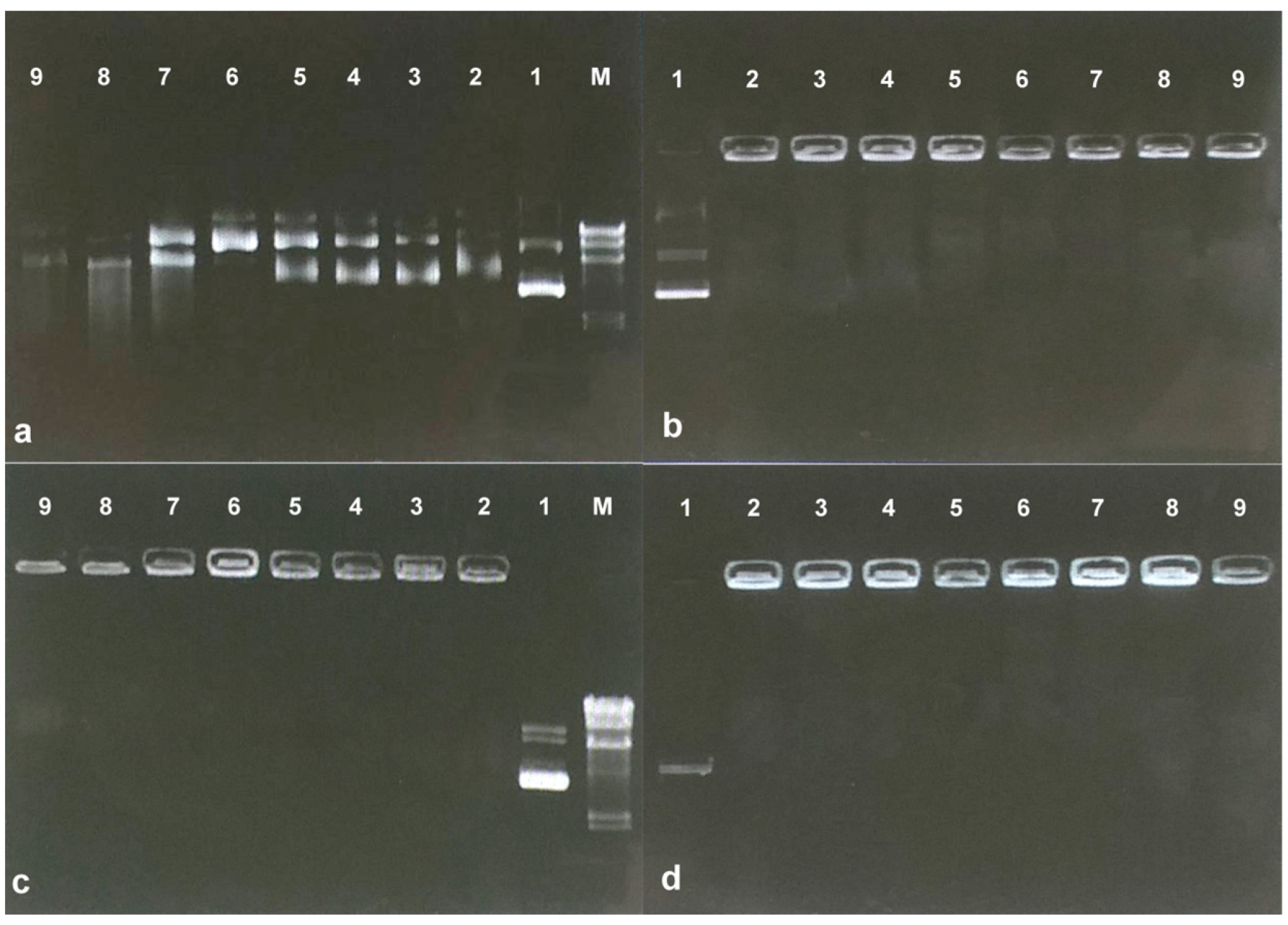



| Formulation | Sodium Alginate (%) | Chitosan (%) | pH |
|---|---|---|---|
| Nac1 | 2% | 2% | 5.59 ± 0.07 |
| Nac2 | 2.5% | 2.5% | 5.98 ± 0.05 |
| Nac3 | 3% | 3% | 6.21 ± 0.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cömez, B.; Özbaş, S. Alginate-Chitosan Hydrogels Containing shRNA Plasmid for Inhibition of CTNNB1 Expression in Breast Cancer Cells. Gels 2023, 9, 541. https://doi.org/10.3390/gels9070541
Cömez B, Özbaş S. Alginate-Chitosan Hydrogels Containing shRNA Plasmid for Inhibition of CTNNB1 Expression in Breast Cancer Cells. Gels. 2023; 9(7):541. https://doi.org/10.3390/gels9070541
Chicago/Turabian StyleCömez, Birnur, and Suna Özbaş. 2023. "Alginate-Chitosan Hydrogels Containing shRNA Plasmid for Inhibition of CTNNB1 Expression in Breast Cancer Cells" Gels 9, no. 7: 541. https://doi.org/10.3390/gels9070541





