Multiple Parameter Optimization for Maximization of Pectinase Production by Rhizopus sp. C4 under Solid State Fermentation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Substrates and Chemicals
2.2. Source of Fungal Inoculum
2.3. Solid State Fermentation
2.4. Polygalacturonase (PG) Activity
2.5. Experimental Design for Optimization
2.6. Model Validation
3. Results and Discussion
3.1. Cultural Characteristics of Fungal Isolate
3.2. Quantitative Estimation of Pectinase
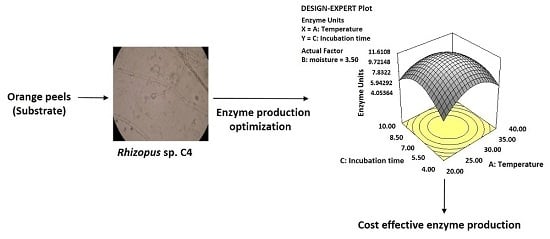
3.3. Optimization of Culture Conditions Using Response Surface Methodology
3.4. Validation of the Model
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Huang, Y.S.; Ho, S.C. Polymethoxy flavones are responsible for the anti-inflammatory activity of citrus fruit peel. Food Chem. 2010, 119, 868–873. [Google Scholar] [CrossRef]
- Martín, M.A.; Siles, J.A.; Chica, A.F.; Martín, A. Biomethanization of orange peel waste. Bioresour. Technol. 2010, 101, 8993–8999. [Google Scholar] [CrossRef] [PubMed]
- Anwar, F.; Naseer, R.; Bhanger, M.I.; Ashraf, S.; Talpur, F.N.; Aladedunye, F.A. Physico-chemical characteristics of citrus seeds and seed oils from Pakistan. J. Am. Oil Chem. Soc. 2008, 85, 321–330. [Google Scholar] [CrossRef]
- Palit, S.; Banerjee, R. Optimization of extraction parameters for recovery of α- amylase from the fermented bran of Bacillus circulans GRS313. Braz. Arch. Biol. Technol. 2001, 44, 107–111. [Google Scholar] [CrossRef]
- Oszmianski, J.; Wojdyło, A.; Kolniak, J. Effect of pectinase treatment on extraction of antioxidant phenols from pomace, for the production of pure enriched cloudy apple juices. Food Chem. 2011, 127, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Pedrolli, D.B.; Gomes, E.; Monti, R.; Cano-Carmona, E. Studies on productivity and characterization of polygalacturonase from Aspergillusgiganteus submerged culture using citrus pectin and orange waste. Appl. Biochem. Biotechnol. 2008, 144, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Kumar, Y.S.; Varakumar, S.; Reddy, O.V.S. Production and optimization of polygalacturonase from mango (Mangiferaindica L.) peel using Fusariummoniliforme in solid state fermentation. World J. Microbiol. Biotechnol. 2010, 26, 1973–1980. [Google Scholar] [CrossRef]
- Mamma, D.; Kourtoglou, E.; Christakopoulos, P. Fungal multienzyme production on industrial by-products of the citrus-processing industry. Bioresour. Technol. 2008, 99, 2373–2383. [Google Scholar] [CrossRef] [PubMed]
- Tari, C.; Gögus, N.; Tokatli, F. Optimization of biomass, pellet size and polygalacturonase production by Aspergillussojae ATCC 20235 using response surface methodology. Enzym. Microb. Technol. 2007, 40, 1108–1116. [Google Scholar] [CrossRef]
- Botella, C.; Diaz, A.; de Ory, I.; Webb, C.; Blandino, A. Xylanase and pectinase production by Aspergillus awamori on grape pomace in solid state fermentation. Process Biochem. 2007, 42, 98–101. [Google Scholar] [CrossRef]
- Kuhad, R.C.; Kapoor, M.; Rustagi, R. Enhanced production of an alkaline pectinase from Streptomyces sp. RCK-SC by whole-cell immobilization and solid-state cultivation. World J. Microbiol. Biotechnol. 2004, 20, 257–263. [Google Scholar] [CrossRef]
- De Gregorio, A.; Mandalari, G.; Arena, N.; Nucita, F.; Tripodo, M.M. SCP and crude pectinase production by slurry-state fermentation of lemon pulps. Bioresour. Technol. 2002, 83, 89–94. [Google Scholar] [CrossRef]
- Silva, D.; Martins, E.S.; Silva, R.; Gomes, E. Pectinase production by Penicilliumviridicatum rfc3 by solid state fermentation using agricultural wastes and agro-industrial by-products. Braz. J. Microbiol. 2002, 33, 318–324. [Google Scholar] [CrossRef]
- Castilho, L.R.; Alves, T.L.M.; Medronho, R.A. Recovery of pectolytic enzymes produced by solid state culture of Aspergillusniger. Process Biochem. 1999, 34, 181–186. [Google Scholar] [CrossRef]
- Gupta, S.; Kapoor, M.; Sharma, K.K.; Nair, L.M.; Kuhad, R.C. Production and recovery of an alkaline exo-polygalacturonase from Bacillus subtilis RCK under solid-state fermentation using statistical approach. Bioresour. Technol. 2008, 99, 937–945. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.K.; Babu, I.S.; Rao, G.H. Process optimization for citric acid production from raw glycerol using response surface methodology. Indian J. Biotechnol. 2008, 7, 496–501. [Google Scholar]
- Sharma, D.C.; Satyanarayana, T. Production and application of pectinolytic enzymes of Sporotrichum thermophile and Bacillus pumilus. In Biotechnological Approaches for Sustainable Development; Reddy, M.S., Khanna, S., Eds.; Allied Publishers Pvt. Ltd: New Delhi, India, 2004; pp. 164–169. [Google Scholar]
- He, G.Q.; Kong, Q.; Ding, L.X. Response surface methodology for optimizing the fermentation medium of Clostridium butyricum. Lett. Appl. Microbiol. 2004, 39, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Fernández, D.E.; Rodríguez-León, J.A.; de Carvalho, J.C.; Sturm, W.; Soccol, C.R. The behavior of kinetic parameters in production of pectinase and xylanase by solid-state fermentation. Bioresour. Technol. 2011, 102, 10657–10662. [Google Scholar] [CrossRef] [PubMed]
- Breed, R.S.; Murray, E.G.D.; Smith, N.R. Bergey’s Manual of Determinative Bacteriology, 7th ed.; The Williams and Wilkins Co., American Society of Microbiology: Baltimore, MD, USA, 1957. [Google Scholar]
- Miller, G.L. Use of dinitrisosalicilic acid reagent for determination of reducing sugars. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Myer, R.; Montgomery, D.C. Response Surface Methodology; John Wiley and Sons Inc.: New York, NY, USA, 2002. [Google Scholar]
- Patil, S.R.; Dayanand, A. Optimization of process for the production of fungal pectinases from deseeded sunflower head in submerged and solid state conditions. Bioresour. Technol. 2006, 97, 2340–2344. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, D.R.; Soni, S.K.; Tewari, R. Enhanced production of pectinase by Bacillus sp. DT7 using solid state fermentation. Bioresour. Technol. 2003, 88, 251–254. [Google Scholar] [CrossRef]
- Silva, D.; Tokuioshi, K.; da Silva Martins, E.; Da Silva, R.; Gomes, E. Production of pectinase by solid-state fermentation with Pencilliumviridicatum RFC3. Process Biochem. 2005, 40, 2885–2889. [Google Scholar] [CrossRef]
- Castilho, L.R.; Medronho, R.A.; Alves, T.L.M. Production and extraction of pectinase obtained by solid state fementation of agroindustrial residues with Aspergillusniger. Bioresour. Technol. 2000, 71, 45–50. [Google Scholar] [CrossRef]
- Yasmeen, Q.; Asgher, M.; Sheikh, M.A.; Nawaz, H. Optimization of ligninolytic enzymes production through response surface methodology. BioResource 2013, 8, 944–968. [Google Scholar] [CrossRef]
- Pandey, A.; Ashakumari, L.; Selvakumar, P.; Vijayalakshami, K.S. Influence of water activity on growth and activity of Aspergillusniger for glucoamylase production in solid state fermentation. World J. Microbiol. Biotechnol. 1994, 10, 485–486. [Google Scholar] [CrossRef] [PubMed]
- Vaithanomasat, P.; Apiwatanapiwat, W.; Petchoy, O.; Chedchant, J. Production of ligninolytic enzymes by white-rot fungus Datronia sp. KAPI0039 and their application for reactive dye removal. Int. J. Chem. Eng. 2010, 2010, 162504:1–162504:6. [Google Scholar]
- Lee, C.K.; Darah, I.; Ibrahim, C.O. Production and optimization of cellulose enzyme using Aspergillus niger USM AI 1 and comparison with Trichodermareesei via solid state fermentation system. Biotechnol. Res. Int. 2011, 2011, 658493:1–658493:6. [Google Scholar] [CrossRef] [PubMed]
- Zadrazil, F.; Karma, D.N.; Isikuemhen, O.S.; Schuchardt, F. Bioconversion of lignocellulosic into ruminant feed with white rot fungi. J. Appl. Anim. Res. 1999, 10, 105–124. [Google Scholar] [CrossRef]
- Tripathi, M.; Mishra, K.A.S.; Mishra, A.S.; Vaithiyanathan, S.; Prasad, R.; Jakhmola, R.C. Selection white rot basidiomycetes for bioconversion of mustard (Brassica compestris) straw under SSF into energy substrate for rumen microorganism. Lett. Appl. Microbiol. 2008, 46, 364–370. [Google Scholar] [CrossRef] [PubMed]
- Uma Maheswar Rao, J.L.; Satyanarayana, T. Statistical optimization of a high maltose-forming, hyperthermostable and Ca2+-independent amylase production by an extreme thermophile Geobacillus thermoleovorans using response surface methodology. J. Appl. Microbiol. 2003, 95, 712–718. [Google Scholar] [CrossRef] [PubMed]
- Moyo, S.; Gashe, B.A.; Collison, E.K.; Mpuchane, S. Optimizing growth condition for the pectinolytic activity of Kluyveromyceswickerhamii by using response surface methodology. Int. J. Food Microbiol. 2003, 15, 87–100. [Google Scholar] [CrossRef]
- Maciel, M.D.H.C.; Herculano, P.N.; Porto, T.S.; Teixeira, M.F.S.; Moreira, K.A.; de Souza-Motta, C.M. Production and partial characterization of pectinase from forage palm by Aspergillusniger URM4645. Afr. J. Biotechnol. 2011, 10, 2469–2475. [Google Scholar]



| Factors | Unit | Symbols | Actual Level Coded Factor | |
|---|---|---|---|---|
| −1 | 1 | |||
| Temperature | °C | A | 20 | 40 |
| Moisture | % | B | 1:1 | 1:5 |
| Incubation Time | Days | C | 4 | 12 |
| Run | A:Temperature | B:moisture | C:Incubation Time | Enzyme Units (IU/mL) | |
|---|---|---|---|---|---|
| Experimental Values | Predicted Value | ||||
| 1 | 40.00 | 1:5.00 | 10.00 | 2.90 | 2.62 |
| 2 | 30.00 | 1: 3.50 | 7.00 | 11.63 | 11.61 |
| 3 | 30.00 | 1:0.98 | 7.00 | 3.40 | 3.43 |
| 4 | 30.00 | 1:3.50 | 12.05 | 1.93 | 2.60 |
| 5 | 20.00 | 1:2.00 | 10.00 | 2.11 | 2.00 |
| 6 | 40.00 | 1:2.00 | 4.00 | 3.50 | 3.75 |
| 7 | 30.00 | 1:3.50 | 7.00 | 11.63 | 11.61 |
| 8 | 13.18 | 1:3.50 | 7.00 | 2.33 | 2.43 |
| 9 | 30.00 | 1:3.50 | 7.00 | 11.63 | 11.61 |
| 10 | 30.00 | 1:6.02 | 7.00 | 5.07 | 5.60 |
| 11 | 20.00 | 1:5.00 | 10.00 | 5.81 | 5.16 |
| 12 | 40.00 | 1:2.00 | 10.00 | 0.90 | 0.47 |
| 13 | 40.00 | 1:5.00 | 4.00 | 3.49 | 3.18 |
| 14 | 20.00 | 1:2.00 | 4.00 | 2.11 | 2.02 |
| 15 | 30.00 | 1:3.50 | 7.00 | 11.63 | 11.61 |
| 16 | 46.82 | 1:3.50 | 7.00 | 1.70 | 1.96 |
| 17 | 30.00 | 1:3.50 | 7.00 | 11.63 | 11.61 |
| 18 | 20.00 | 1:5.00 | 4.00 | 2.40 | 2.43 |
| 19 | 30.00 | 1:3.50 | 1.95 | 3.17 | 3.06 |
| Source | Sum of Squares | DF | Mean Value | F Square | Prob> F |
|---|---|---|---|---|---|
| Model | 300.42 | 9 | 33.38 | 170.45 | <0.0001 * |
| A | 0.53 | 1 | 0.53 | 2.72 | 0.1332 |
| B | 5.66 | 1 | 5.66 | 28.88 | 0.0004 * |
| C | 0.25 | 1 | 0.25 | 1.30 | 0.2835 |
| A2 | 148.07 | 1 | 148.07 | 756.08 | <0.0001 * |
| B2 | 85.90 | 1 | 85.90 | 438.65 | <0.0001 * |
| C2 | 131.55 | 1 | 131.55 | 671.73 | <0.0001 * |
| AB | 0.50 | 1 | 0.50 | 2.55 | 0.1445 |
| AC | 5.44 | 1 | 5.44 | 27.80 | 0.0005 * |
| BC | 3.67 | 1 | 3.67 | 18.75 | 0.0019 * |
| Residual | 1.76 | 9 | 0.20 | ||
| Lack of Fit | 1.76 | 5 | 0.35 | ||
| Pure Error | 0.000 | 4 | 0.000 | ||
| Cor Total | 302.18 | 18 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Handa, S.; Sharma, N.; Pathania, S. Multiple Parameter Optimization for Maximization of Pectinase Production by Rhizopus sp. C4 under Solid State Fermentation. Fermentation 2016, 2, 10. https://doi.org/10.3390/fermentation2020010
Handa S, Sharma N, Pathania S. Multiple Parameter Optimization for Maximization of Pectinase Production by Rhizopus sp. C4 under Solid State Fermentation. Fermentation. 2016; 2(2):10. https://doi.org/10.3390/fermentation2020010
Chicago/Turabian StyleHanda, Shweta, Nivedita Sharma, and Shruti Pathania. 2016. "Multiple Parameter Optimization for Maximization of Pectinase Production by Rhizopus sp. C4 under Solid State Fermentation" Fermentation 2, no. 2: 10. https://doi.org/10.3390/fermentation2020010