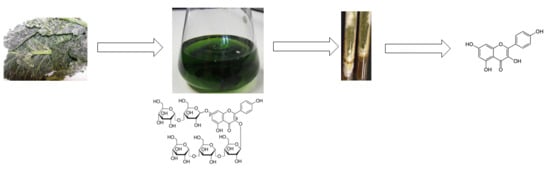
Bioconversion of Kaempferol and Quercetin Glucosides from Plant Sources Using Rhizopus spp.
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Material
4.1.1. Plant Material
4.1.2. Chemicals and Reagents
4.1.3. Fungal Strains and Preparation of the Inoculum
4.2. Experimental Procedure
4.2.1. Preparation of Extract
4.2.2. Fermentation Process
4.3. Analytical Methods
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Cartea, M.E.; Francisco, M.; Soengas, P.; Velasco, P. Phenolic compounds in brassica vegetables. Molecules 2010, 16, 251–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hollman, P.C.H.; Katan, M.B. Dietary flavonoids: Intake, health effects and bioavailability. Food Chem. Toxicol. 1999, 37, 937–942. [Google Scholar] [CrossRef]
- Pérez-Gregorio, R.M.; García-Falcón, M.S.; Simal-Gándara, J.; Rodrigues, A.S.; Almeida, D.P.F. Identification and quantification of flavonoids in traditional cultivars of red and white onions at harvest. J. Food Compos. Anal. 2010, 23, 592–598. [Google Scholar] [CrossRef]
- Hakkinen, S.H.; Karenlamp, S.O.; Heinonen, I.M.; Mykkanen, H.M.; Torronen, A.R. Content of the flavonols quercetin, myricetin, and kaempferol in 25 edible berries. J. Agric. Food Chem. 1999, 47, 2774–2779. [Google Scholar] [CrossRef]
- Erlun, I. Review of the flavonoids quercetin, hesperetin and naringenin. Dietary sources, bioactivities, bioavailability, and epidemiology. Nutr. Res. 2004, 24, 851–874. [Google Scholar] [CrossRef]
- Nile, S.H.; Park, S.W. Edible berries: Bioactive components and their effect on human health. Nutrition 2014, 30, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Mateos, A.; Heiss, C.; Borges, G.; Crozier, A. Berry (poly)phenols and cardiovascular health. J. Agric. Food Chem. 2014, 62, 3842–3851. [Google Scholar] [CrossRef] [PubMed]
- Fukumoto, L.R.; Mazza, G. Assessing antioxidant and prooxidant activities of phenolic compounds. J. Agric. Food Chem. 2000, 48, 3597–3604. [Google Scholar] [CrossRef]
- Zheng, Z.; Shetty, K. Solid-state bioconversion of phenolics from cranberry pomace and role of Lentinus edodes β-glucosidase. J. Agric. Food Chem. 2000, 48, 895–900. [Google Scholar] [CrossRef]
- Wu, Z.; Song, L.; Huang, D. Food grade fungal stress on germinating peanut seeds induced phytoalexins and enhanced polyphenolic antioxidants. J. Agric. Food Chem. 2011, 59, 5993–6003. [Google Scholar] [CrossRef]
- Hur, S.J.; Lee, S.Y.; Kim, Y.C.; Choi, I.; Kim, G.B. Effect of fermentation on the antioxidant activity in plant-based foods. Food Chem. 2014, 160, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Y.; Yu, H.S.; Feng, B.; Kang, L.P.; Pang, X.; Xiong, C.Q.; Zhao, Y.; Li, C.M.; Zhang, Y.; Ma, B.P. Selective hydrolysis of flavonoid glycosides by Curvularia lunata. Chin. J. Nat. Med. 2013, 11, 684–689. [Google Scholar] [CrossRef]
- Lin, S.; Zhu, Q.; Wen, L.; Yang, B.; Jiang, G.; Gao, H.; Chen, F.; Jiang, Y. Production of quercetin, kaempferol and their glycosidic derivatives from the aqueous-organic extracted residue of litchi pericarp with Aspergillus awamori. Food Chem. 2014, 145, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Rho, H.S.; Kim, D.H.; Chang, I.S. Enzymatic preparation of kaempferol from green tea seed and Its antioxidant activity. J. Agric. Food Chem. 2006, 54, 2951–2956. [Google Scholar] [CrossRef] [PubMed]
- Yadav, V.; Yadav, P.K.; Yadav, S.; Yadav, K.D.S. α-l-Rhamnosidase: A review. Process Biochem. 2010, 45, 1226–1235. [Google Scholar] [CrossRef]
- Cho, K.M.; Hong, S.Y.; Math, R.K.; Lee, J.H.; Kambiranda, D.M.; Kim, J.M.; Islam, S.M.A.; Yun, M.G.; Cho, J.J.; Lim, W.J.; et al. Biotransformation of phenolics (isoflavones, flavanols and phenolic acids) during the fermentation of cheonggukjang by Bacillus pumilus HY1. Food Chem. 2009, 114, 413–419. [Google Scholar] [CrossRef]
- Cho, K.M.; Lee, J.H.; Yun, H.D.; Ahn, B.Y.; Kim, H.; Seo, W.T. Changes of phytochemical constituents (isoflavones, flavanols, and phenolic acids) during cheonggukjang soybeans fermentation using potential probiotics Bacillus subtilis CS90. J. Food Compos. Anal. 2011, 24, 402–410. [Google Scholar] [CrossRef]
- Lee, S.H.; Seo, M.H.; Oh, D.K. Deglycosylation of isoflavones in isoflavone-rich soy germ flour by Aspergillus oryzae KACC 40247. J. Agric. Food Chem. 2013, 61, 12101–12110. [Google Scholar] [CrossRef]
- Di Gioia, D.; Strahsburger, E.; Lopez de Lacey, A.M.; Bregola, V.; Marotti, I.; Aloisio, I.; Biavati, B.; Dinelli, G. Flavonoid bioconversion in Bifidobacterium pseudocatenulatum B7003: A potential probiotic strain for functional food development. J. Funct. Foods 2014, 7, 671–679. [Google Scholar] [CrossRef] [Green Version]
- Oka, T.; Simpson, F.J.; Krishnamurty, H.G. Degradation of rutin by Aspergillus flavus. Studies on specificity, inhibition, and possible reaction mechanism of quercetinase. Can. J. Microbiol. 1972, 18, 493–508. [Google Scholar] [CrossRef]
- Lin, Y.T.; Hsiu, S.L.; Hou, Y.C.; Chen, H.Y.; Chao, P.D.L. Degradation of flavonoid aglycones by rabbit, rat and human fecal flora. Biol. Pharm. Bull. 2003, 26, 747–751. [Google Scholar] [CrossRef]
- Serra, A.; Macià, A.; Romero, M.P.; Reguant, J.; Ortega, N.; Motilva, M.J. Metabolic pathways of the colonic metabolism of flavonoids (flavonols, flavones and flavanones) and phenolic acids. Food Chem. 2012, 130, 383–393. [Google Scholar] [CrossRef]
- Huynh, T.N.; Smagghe, G.; Gonzales, G.B.; Van Camp, J.; Raes, K. Extraction and bioconversion of kaempferol metabolites from cauliflower outer leaves through fungal fermentation. Biochem. Eng. J. 2016, 116, 27–33. [Google Scholar] [CrossRef]
- Irimat, R. Fungal Bioconversion of Flavonoid Compounds in Cauliflower Waste as a Source of Bioactive Extracts. Master’s Thesis, Ghent University, Gent, Belgium, 2014. [Google Scholar]
- Taki, Y.; Ikeda, K.; Sato, C.; Yano, M.; Sato, T.; Konno, H. Production and characterization of β-glucosidase from Rhizopus oryzae MIBA348. J. Biol. Macromol. 2005, 5, 11–16. [Google Scholar]
- Yadav, G.; Singh, A.; Bhattacharya, P.; Yuvraj, J.; Banerjee, R. Comparative analysis of solid-state bioprocessing and enzymatic treatment of finger millet for mobilization of bound phenolics. Bioprocess. Biosyst. Eng. 2013, 36, 1563–1569. [Google Scholar] [CrossRef]
- Lee, I.H.; Chou, C.C. Distribution profiles of isoflavone isomers in black bean kojis prepared with various filamentous fungi. J. Agric. Food Chem. 2006, 54, 1309–1314. [Google Scholar] [CrossRef]
- Lee, I.H.; Hung, Y.H.; Chou, C.C. Solid-state fermentation with fungi to enhance the antioxidative activity, total phenolic and anthocyanin contents of black bean. Int. J. Food Microbiol. 2008, 121, 150–156. [Google Scholar] [CrossRef]
- Marotti, M.; Piccaglia, R. Characterization of flavonoids in different cultivars of onion (Allium cepa L.). J. Food Sci. 2002, 67, 1229–1232. [Google Scholar] [CrossRef]
- Rodríguez Galdón, B.; Rodríguez Rodríguez, E.M.; Díaz Romero, C. Flavonoids in onion cultivars (Allium cepa L.). J. Food Sci. 2008, 73, 599–605. [Google Scholar] [CrossRef]
- You, H.J.; Ahn, H.J.; Ji, G.E. Transformation of rutin to antiproliferative quercetin-3-glucoside by Aspergillus niger. J. Agric. Food Chem. 2010, 58, 10886–10892. [Google Scholar] [CrossRef]
- Battaglia, E.; Benoit, I.; van den Brink, J.; Wiebenga, A.; Coutinho, P.; Henrissat, B.; de Vries, R. Carbohydrate-active enzymes from the zygomycete fungus Rhizopus oryzae: A highly specialized approach to carbohydrate degradation depicted at genome level. BMC Genom. 2011, 12. [Google Scholar] [CrossRef]
- Gesell, M.; Hammer, E.; Mikolasch, A.; Schauer, F. Oxidation and ring cleavage of dibenzofuran by the filamentous fungus Paecilomyces lilacinus. Arch. Microbiol. 2004, 182, 51–59. [Google Scholar] [CrossRef]
- Schlueter, R.; Röder, A.; Czekalski, N.; Gliesche, D.; Mikolasch, A.; Schauer, F. Novel mechanisms of biotransformation of p-tert-amylphenol by bacteria and fungi with special degradation abilities and simultaneous detoxification of the disinfectant. Appl. Microbiol. Biotechnol. 2013, 98, 373–384. [Google Scholar] [CrossRef]
- Slana, M.; Žigon, D.; Makovec, T.; Lenasi, H. The response of filamentous fungus Rhizopus nigricans to flavonoids. J. Basic Microbiol. 2011, 51, 433–441. [Google Scholar] [CrossRef]
- Olsen, H.; Aaby, K.; Borge, G.I.A. Characterisation and quantificiaton of flavonoids and hydroxycinnamic acids in curly kale (Brassica oleracea L. Convar. acephala Var; sabellica) by HPLC-DAD-ESI-MSn. J. Agric. Food Chem. 2009, 57, 2816–2825. [Google Scholar] [CrossRef]
- Gonzales, G.B.; Raes, K.; Coelus, S.; Struijs, K.; Smagghe, G.; Van Camp, J. Ultra(high)-pressure liquid chromatography–electrospray ionization-time-of-flight-ion mobility-high definition mass spectrometry for the rapid identification and structural characterization of flavonoid glycosides from cauliflower waste. J. Chromatogr. 2014, 1323, 39–48. [Google Scholar] [CrossRef]




| Extract | Rt (min) | m/z Values | Identity | |
|---|---|---|---|---|
| Exact Mass | Fragments | |||
| Cauliflower Outer Leaves | 18.15 | 609 | 609,284 | Kaempferol-3-O-diglucoside |
| 21.58 | 447 | 447,284 | Kaempferol-3-O-glucoside | |
| 26.91 | 285 | 285 | Kaempferol | |
| Onion | 16.57 | 625 | 625,463,300 | Quercetin-3-O-glucoside-7-O-glucoside |
| 19.42 | 463 | 463,300 | Quercetin-3-O-glucoside | |
| 24.29 | 301 | 301 | Quercetin | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huynh, N.T.; Smagghe, G.; Gonzales, G.B.; Van Camp, J.; Raes, K. Bioconversion of Kaempferol and Quercetin Glucosides from Plant Sources Using Rhizopus spp. Fermentation 2018, 4, 102. https://doi.org/10.3390/fermentation4040102
Huynh NT, Smagghe G, Gonzales GB, Van Camp J, Raes K. Bioconversion of Kaempferol and Quercetin Glucosides from Plant Sources Using Rhizopus spp. Fermentation. 2018; 4(4):102. https://doi.org/10.3390/fermentation4040102
Chicago/Turabian StyleHuynh, Nguyen Thai, Guy Smagghe, Gerard Bryan Gonzales, John Van Camp, and Katleen Raes. 2018. "Bioconversion of Kaempferol and Quercetin Glucosides from Plant Sources Using Rhizopus spp." Fermentation 4, no. 4: 102. https://doi.org/10.3390/fermentation4040102