A MYB Transcription Factor from Brassica juncea Regulates Purple Leaves in Pak Choi (Brassica campestris L. ssp. chinensis)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Identification and Genetic Analysis of the Purple-Leaf Phenotype
2.3. Bulked Segregant Analysis Sequence (BSA-Seq) for Primary Mapping
2.4. Molecular Marker Development
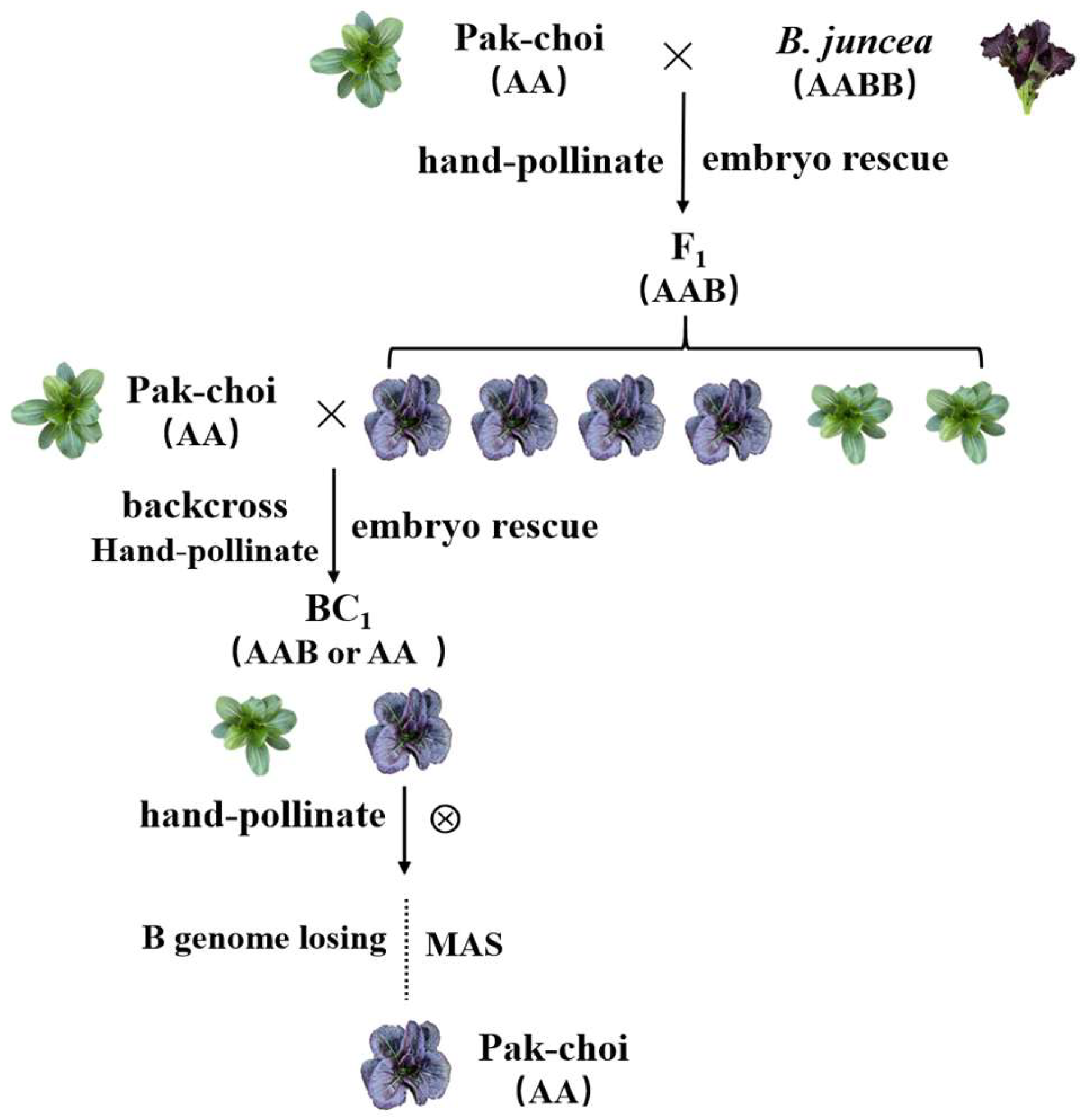
2.5. Embryo Rescue from Distant Hybridization
3. Results
3.1. Inheritance Pattern Analysis of the Purple-Leaf Phenotype in Pak Choi
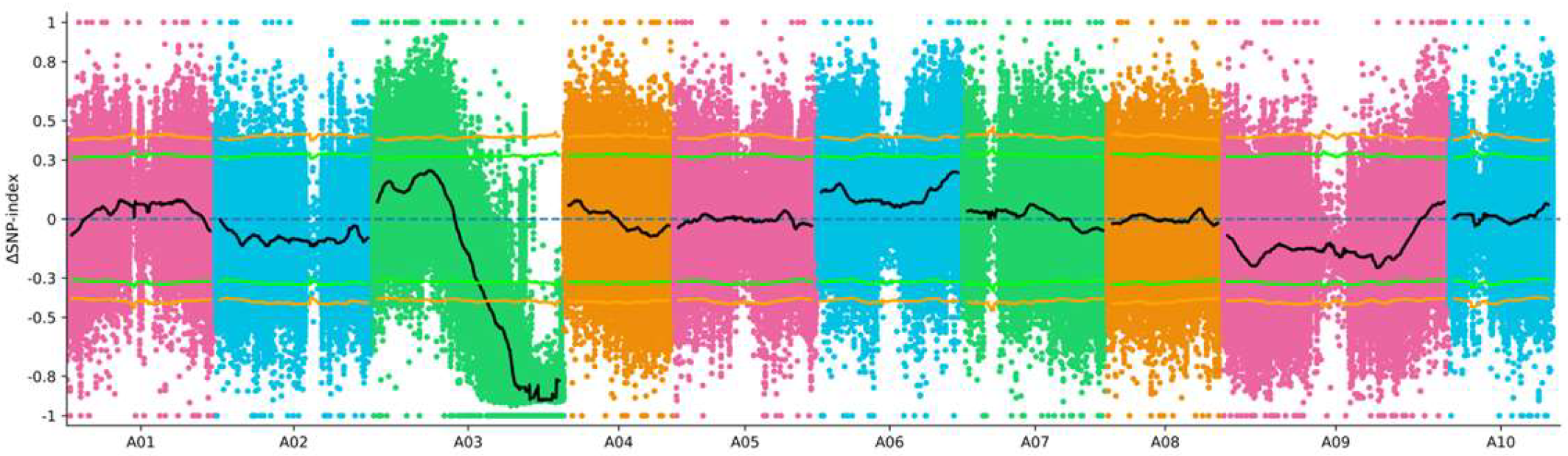
3.2. Primary Mapping of the Purple-Leaf Phenotype via BSA-Seq
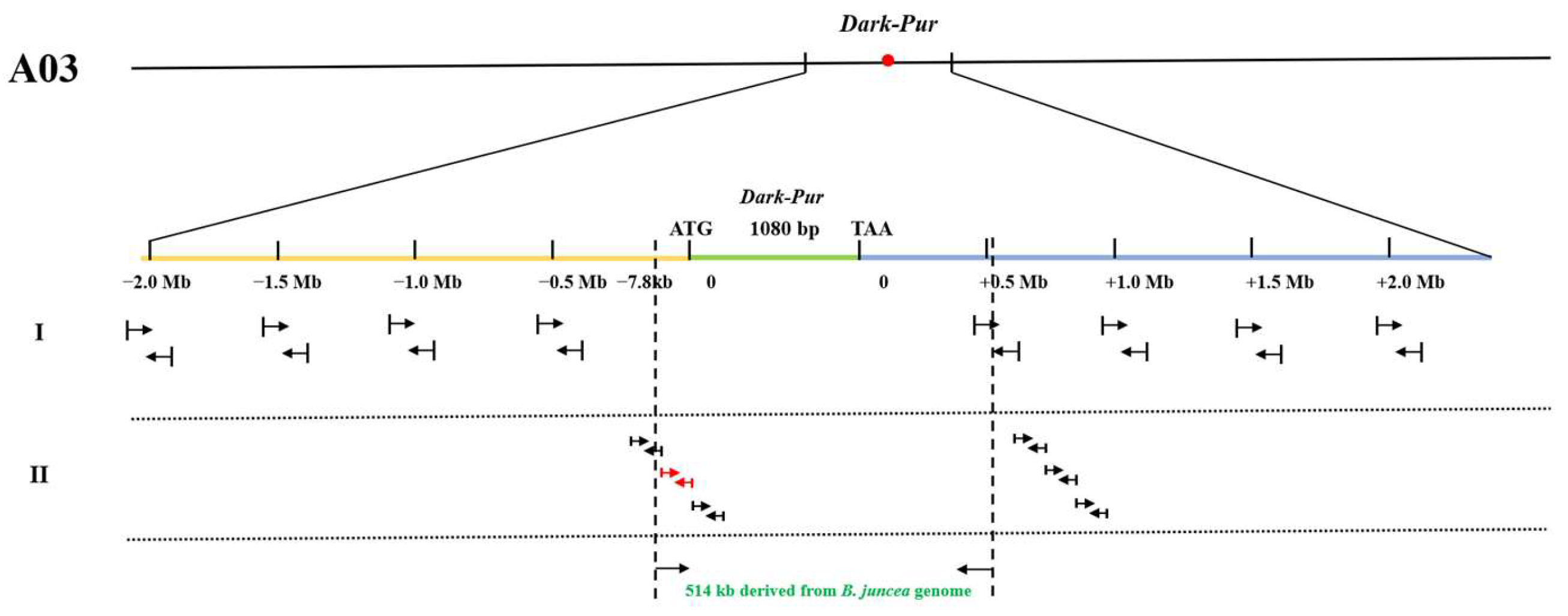
3.3. Fine Mapping and Candidate Gene Excavation for the Purple-Leaf Phenotype in Pak Choi
3.4. The Purple-Leaf Gene in Pak Choi Was Obtained from B. juncea through Distant Hybridization
3.5. A New Pak Choi Germplasm Line Was Created with Green Pak Choi × Purple B. juncea through Distant Hybridization
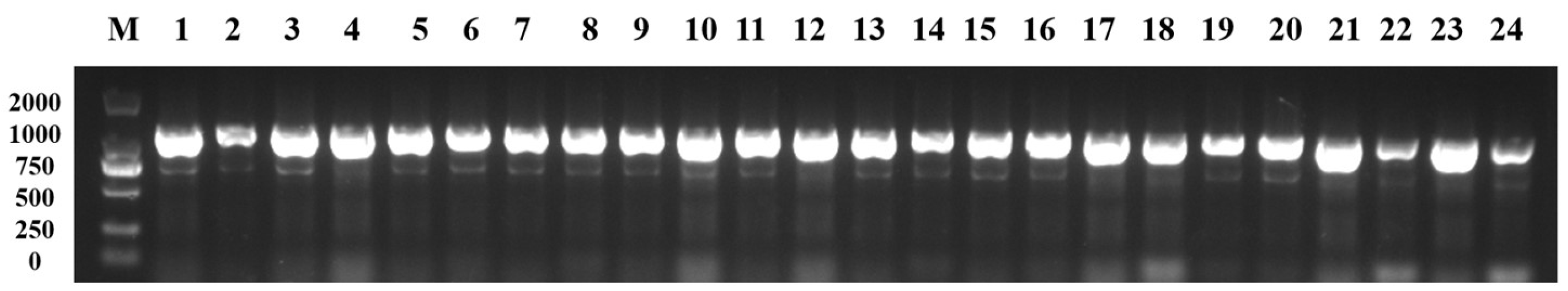
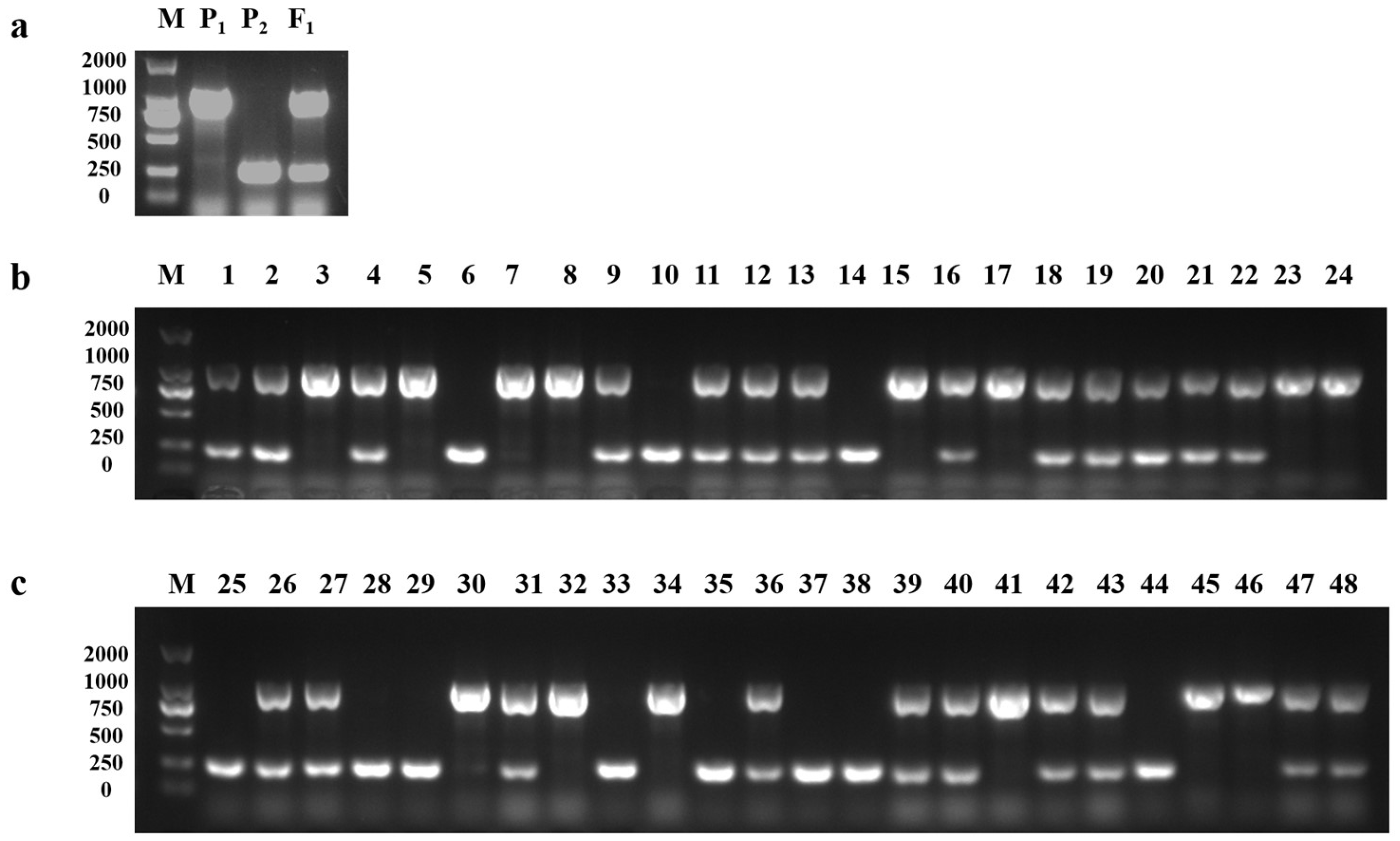
3.6. The Purple-Leaf Phenotype of 20 Pak Choi Varieties Were All Derived from B. juncea through Distant Hybridization
4. Discussion
4.1. MYB Transcription Factor Regulated the Purple Phenotype
4.2. Distant Hybridization Is a Very Effective Method for Creating New Germplasms
4.3. The Genomic Similarity of Brassica Crops Is the Basis of Distant Hybridization
4.4. The Purple-Leaf Phenotype of the Pak Choi Varieties Is Derived from Purple B. juncea through Distant Hybridization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Tan, C.; Chen, H.D.; Dai, G.Q.; Liu, Y.; Shen, W.J.; Wang, C.C.; Liu, D.; Liu, S.J.; Xu, S.Q.; Zhu, B.; et al. Identification and characterization of the gene BraANS.A03 associated with purple leaf color in pak choi (Brassica rapa L. ssp. chinensis). Planta 2023, 258, 14. [Google Scholar] [CrossRef]
- Liu, H.; Liu, Z.; Wu, Y.; Zheng, L.; Zhang, G. Regulatory Mechanisms of Anthocyanin Biosynthesis in Apple and Pear. Int. J. Mol. Sci. 2021, 22, 8441. [Google Scholar] [CrossRef]
- Nijveldt, R.J.; van Nood, E.; van Hoorn, D.E.C.; Boelens, P.G.; van Norren, K.; van Leeuwen, P.A.M. Flavonoids: A review of probable mechanisms of action and potential applications123. Am. J. Clin. Nutr. 2001, 74, 418–425. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Pang, Q.; Chen, X.; Li, T.; Fang, J.; LIN, S.; Jia, H. Transcriptome analysis of strawberry fruit in response to exogenous arginine. Planta 2020, 252, 82. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Zhang, M.D.; Wen, C.X.; Xie, X.L.; Tian, W.; Wen, S.Q.; Lu, R.K.; Liu, L.D. Integrated metabolomic and transcriptomic analysis of the anthocyanin regulatory networks in Salvia miltiorrhiza Bge. flowers. BMC Plant Biol. 2020, 20, 349. [Google Scholar] [CrossRef]
- Li, W.F.; Mao, J.; Yang, S.J.; Guo, Z.G.; Ma, Z.H.; Dawuda, M.M.; Zuo, C.W.; Chu, M.Y.; Chen, B.H. Anthocyanin accumulation correlates with hormones in the fruit skin of ‘Red Delicious’ and its four generation bud sport mutants. BMC Plant Biol. 2018, 18, 363. [Google Scholar] [CrossRef]
- Qiu, Z.; Wang, X.; Gao, J.; Guo, Y.; Huang, Z.; Du, Y. The tomato hoffman’s anthocyaninless gene encodes a bHLH transcription factor involved in anthocyanin biosynthesis that is developmentally regulated and induced by low temperatures. PLoS ONE 2016, 11, e0151067. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, H.; Fernandes, A.F.; Brás, N.; Mateus, N.; de Freitas, V.; Fernandes, I. Anthocyanins as Antidiabetic Agents—In Vitro and In Silico Approaches of Preventive and Therapeutic Effects. Molecules 2020, 25, 3813. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Hu, N.; Ding, C.; Zhang, Q.; Li, W.; Suo, Y.; Wang, H.; Bai, B.; Ding, C. In vitro and in vivo biological activities of anthocyanins from Nitraria tangutorun Bobr. fruits. Food Chem. 2016, 194, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Li, H.Y.; Deng, Z.Y.; Zhu, H.H.; Hu, C.L.; Liu, R.H.; Young, J.C.; Tsao, R. Highly pigmented vegetables: Anthocyanin compositions and their role in antioxidant activities. Food Res. Int. 2012, 46, 250–259. [Google Scholar] [CrossRef]
- Heng, S.P.; Cheng, Q.Q.; Zhang, T.; Liu, X.J.; Huang, H.; Yao, P.J.; Liu, Z.X.; Wan, Z.J.; Fu, T.D. Fine-mapping of the BjPur gene for purple leaf color in Brassica juncea. Theor. Appl. Genet. 2020, 133, 2989–3000. [Google Scholar] [CrossRef]
- Li, G.H.; Chen, H.C.; Liu, J.L.; Luo, W.L.; Xie, D.S.; Luo, S.B.; Wu, T.Q.; Akram, W.; Zhong, Y. A high-density genetic map developed by specific-locus amplified fragment (SLAF) sequencing and identification of a locus controlling anthocyanin pigmentation in stalk of Zicaitai (Brassica rapa L. ssp. chinensis var. purpurea). BMC Genom. 2019, 20, 343. [Google Scholar] [CrossRef]
- Zhuang, H.; Lou, Q.; Liu, H.; Han, H.; Wang, Q.; Tang, Z.; Ma, Y.; Wang, H. Differential Regulation of Anthocyanins in Green and Purple Turnips Revealed by Combined De Novo Transcriptome and Metabolome Analysis. Int. J. Mol. Sci. 2019, 20, 4387. [Google Scholar] [CrossRef]
- He, J.A.; Giusti, M.M. Anthocyanins: Natural colorants with health-promoting properties. Annu. Rev. Food Sci. Technol. 2010, 1, 163–187. [Google Scholar] [CrossRef]
- Castañeda-Ovando, A.; Pacheco-Hernández, M.D.; Páez-Hernández, M.E.; Rodríguez, J.A.; Galán-Vidal, C.A. Chemical studies of anthocyanins: A review. Food Chem. 2009, 113, 859–871. [Google Scholar] [CrossRef]
- Jeon, J.; Lim, C.J.; Kim, J.K.; Park, S.U. Comparative Metabolic Profiling of Green and Purple Pakchoi (Brassica rapa Subsp. Chinensis). Molecules 2018, 23, 1613. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Chen, G.P.; Dong, T.T.; Pan, Y.; Zhao, Z.P.; Tian, S.B.; Hu, Z.L. Anthocyanin Accumulation and Transcriptional Regulation of Anthocyanin Biosynthesis in Purple Bok-Choy (Brassica rapa var. chinensis). J. Agric. Food Chem. 2014, 62, 12366–12376. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Cui, Y.; Yao, Y.; An, L.; Bai, Y.; Li, X.; Yao, X.; Wu, K. Genome-wide identification of WD40 transcription factors and their regulation of the MYB-bHLH-WD40 (MBW) complex related to anthocyanin synthesis in Qingke (Hordeum vulgare L. var. nudum Hook. f.). BMC Genom. 2023, 24, 166. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Tikunov, Y.; Schouten, R.E.; Marcelis, L.F.M.; Visser, R.G.F.; Bovy, A. Anthocyanin Biosynthesis and Degradation Mechanisms in Solanaceous Vegetables: A Review. Front. Chem. 2018, 6, 52. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Li, H.; Huang, J.-R. Arabidopsis TT19 functions as a carrier to transport anthocyanin from the cytosol to tonoplasts. Mol. Plant 2012, 5, 387–400. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; He, Y.J.; Li, J.; Liu, Y.; Chen, H.Y. CBFs Function in Anthocyanin Biosynthesis by Interacting with MYB113 in Eggplant (Solanum melongena L.). Plant Cell Physiol. 2020, 61, 416–426. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, A.; Mendenhall, J.; Huo, Y.; Lloyd, A. TTG1 complex MYBs, MYB5 and TT2, control outer seed coat differentiation. Dev. Biol. 2009, 325, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.X.; Chiu, L.W.; Li, L. Transcriptional regulation of anthocyanin biosynthesis in red cabbage. Planta 2009, 230, 1141–1153. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, K.; Wu, J.; Guo, N.; Liang, J.L.; Wang, X.W.; Cheng, F. QTL-Seq and Sequence Assembly Rapidly Mapped the Gene BrMYBL2.1 for the Purple Trait in Brassica rapa. Sci. Rep. 2020, 10, 9. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.L.; Li, F.; Zhang, S.F.; Zhang, H.; Qian, W.; Li, P.R.; Zhang, S.J.; Sun, R.F. Mining for Candidate Genes in an Introgression Line by Using RNA Sequencing: The Anthocyanin Overaccumulation Phenotype in Brassica. Front. Plant Sci. 2016, 7, 1245. [Google Scholar] [CrossRef]
- Burdzinski, C.; Wendell, D.L. Mapping the Anthocyaninless (anl) Locus in Rapid-Cycling Brassica rapa (RBr) to Linkage Group R9. BMC Genet. 2007, 8, 64. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, L.; Gong, Z.; Hui, M. Screening RAPD markers linked to purple trait of Chinese cabbage and its chromosome location. Acta Bot. Boreali-Occident. Sin. 2008, 28, 901–906. [Google Scholar]
- Yu, Y.; Zhang, Y.; Zhang, D. SRAP Markers Linked to Purple Trait in Chinese Cabbage. Mol. Plant Breed. 2009, 7, 573–578. [Google Scholar] [CrossRef]
- Hayashi, K.; Matsumoto, S.; Tsukazaki, H.; Kondo, T.; Kubo, N.; Hirai, M. Mapping of a novel locus regulating anthocyanin pigmentation in Brassica rapa. Breed Sci. 2010, 60, 76–80. [Google Scholar] [CrossRef]
- Guo, N.; Wu, J.; Zheng, S.N.; Cheng, F.; Liu, B.; Liang, J.L.; Cui, Y.; Wang, X.W. Anthocyanin profile characterization and quantitative trait locus mapping in zicaitai (Brassica rapa L. ssp chinensis var. purpurea). Mol. Breed. 2015, 35, 11. [Google Scholar] [CrossRef]
- Zhang, S.J.; Li, P.R.; Qian, W.; Zhang, S.F.; Li, F.; Zhang, H.; Wang, X.W.; Sun, R.F. Mapping and expression profiling reveal an inserted fragment from purple mustard involved anthocyanin accumulation in Chinese cabbage. Euphytica 2016, 212, 83–95. [Google Scholar] [CrossRef]
- Liu, J.; Wang, W.; Zhang, D.; Yu, S.; Zhang, F.; Zhao, X.; Yu, Y.; Xu, J.; Lu, G. Primary Mapping of pur, a Gene Controlling Purple Leaf Color in Brassica rapa. Acta Agric. Boreali-Sin. 2013, 28, 49–53. [Google Scholar]
- Wang, W.H.; Zhang, D.S.; Yu, S.C.; Liu, J.; Wang, D.; Zhang, F.L.; Yu, Y.J.; Zhao, X.Y.; Lu, G.X.; Su, T.B. Mapping the BrPur gene for purple leaf color on linkage group A03 of Brassica rapa. Euphytica 2014, 199, 293–302. [Google Scholar] [CrossRef]
- Katche, E.; Quezada-Martinez, D.; Katche, E.I.; Vasquez-Teuber, P.; Mason, A.S. Interspecific Hybridization for Brassica Crop Improvement. Crop Breed. Genet. Genom. 2019, 1, e190007. [Google Scholar]
- Nagaharu, U. Genome-analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn. J. Bot. 1935, 7, 389–452. [Google Scholar]
- Li, J.X.; Rao, L.L.; Chen, L.P. Production and characterization of interspecific hybrids between tuber mustard (Brassica juncea) and red cabbage (Brassica oleracea) through embryo culture. Acta Hortic. 2016, 1127, 431–436. [Google Scholar] [CrossRef]
- Peterka, H.; Budahn, H.; Schrader, O.; Ahne, R.; Schütze, W. Transfer of resistance against the beet cyst nematode from radish (Raphanus sativus) to rape (Brassica napus) by monosomic chromosome addition. Theor. Appl. Genet. 2004, 109, 30–41. [Google Scholar] [CrossRef]
- Liu, Y.J.; Li, G.L.; Zhang, S.J.; Zhang, S.F.; Zhang, H.; Sun, R.F.; Li, F. Comprehensive transcriptome-metabolome analysis and evaluation of the Dark_Pur gene from Brassica juncea that controls the differential regulation of anthocyanins in Brassica rapa. Genes 2022, 13, 283. [Google Scholar] [CrossRef]
- Teng, S.; Keurentjes, J.; Bentsink, L.; Koornneef, M.; Smeekens, S. Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 2005, 139, 1840–1852. [Google Scholar] [CrossRef]
- Ramsay, N.A.; Glover, B.J. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005, 10, 63–70. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Han, T.; Lyu, L.; Li, W.; Wu, W. Research progress in understanding the biosynthesis and regulation of plant anthocyanins. Sci. Hortic. 2023, 321, 112374. [Google Scholar] [CrossRef]
- Wang, L.J.; Lu, W.X.; Ran, L.Y.; Dou, L.W.; Yao, S.; Hu, J.; Fan, D.; Li, C.F.; Luo, K.M. R2R3-MYB transcription factor MYB6 promotes anthocyanin and proanthocyanidin biosynthesis but inhibits secondary cell wall formation in Populus tomentosa. Plant J. 2019, 99, 733–751. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Gong, J.X.; Chen, K.L.; Yao, W.K.; Zhang, B.X.; Wang, J.Y.; Tian, S.T.; Liu, H.L.; Wang, Y.Q.; Liu, Y.L.; et al. A novel R3 MYB transcriptional repressor, MaMYBx, finely regulates anthocyanin biosynthesis in grape hyacinth. Plant Sci. 2020, 298, 12. [Google Scholar] [CrossRef] [PubMed]
- Naing, A.H.; Kim, C.K. Roles of R2R3-MYB transcription factors in transcriptional regulation of anthocyanin biosynthesis in horticultural plants. Plant Mol. Biol. 2018, 98, 1–18. [Google Scholar] [CrossRef]
- Walker, A.R.; Lee, E.; Bogs, J.; McDavid, D.A.J.; Thomas, M.R.; Robinson, S.P. White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 2007, 49, 772–785. [Google Scholar] [CrossRef]
- Wang, Z.G.; Meng, D.; Wang, A.D.; Li, T.L.; Jiang, S.L.; Cong, P.H.; Li, T.Z. The Methylation of the PcMYB10 Promoter Is Associated with Green-Skinned Sport in Max Red Bartlett Pear. Plant Physiol. 2013, 162, 885–896. [Google Scholar] [CrossRef]
- Song, H.; Yi, H.; Lee, M.; Han, C.T.; Lee, J.; Kim, H.; Park, J.I.; Nou, I.S.; Kim, S.J.; Hur, Y. Purple Brassica oleracea var. capitata F-rubra is due to the loss of BoMYBL2-1 expression. BMC Plant Biol. 2018, 18, 16. [Google Scholar] [CrossRef]
- Zhu, M.Z.; Yang, L.M.; Zhang, Y.Y.; Zhuang, M.; Ji, J.L.; Hou, X.L.; Li, Z.S.; Han, F.Q.; Fang, Z.Y.; Lv, H.H.; et al. Introgression of clubroot resistant gene into Brassica oleracea L. from Brassica rapa based on homoeologous exchange. Hortic. Res. 2022, 9, 16. [Google Scholar] [CrossRef]
- Zhang, L.; Li, X.; Chang, L.; Wang, T.; Liang, J.; Lin, R.; Wu, J.; Wang, X. Expanding the genetic variation of Brassica juncea by introgression of the Brassica rapa Genome. Hortic. Res. 2022, 9, uhab054. [Google Scholar] [CrossRef]
- Wei, Y.X.; Zhu, M.Z.; Qiao, H.Y.; Li, F.; Zhang, S.J.; Zhang, S.F.; Zhang, H.; Sun, R.F. Characterization of interspecific hybrids between flowering Chinese cabbage and broccoli. Sci. Hortic. 2018, 240, 552–557. [Google Scholar] [CrossRef]
- Zhang, L.; Zhu, Z.Y.; Chen, F.F.; Zhu, Y.Q.; Guo, X.C.; Fu, M.J.; Chen, J.H.; Wu, J.G.; Zhu, Z.J. Production and identification of x Brassicoraphanus distant hybrids between radish (Raphanus sativus L.) and kohlrabi (Brassica oleracea L. var. Caulorapa DC.). N. Z. J. Crop Hortic. Sci. 2023, 51, 341–354. [Google Scholar] [CrossRef]
- Zhao, C.J.; Xie, M.L.; Liang, L.B.; Yang, L.; Han, H.S.; Qin, X.R.; Zhao, J.X.; Hou, Y.; Dai, W.D.; Du, C.F.; et al. Genome-Wide Association Analysis Combined with Quantitative Trait Loci Mapping and Dynamic Transcriptome Unveil the Genetic Control of Seed Oil Content in Brassica napus L. Front. Plant Sci. 2022, 13, 16. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.W.; Wang, H.Z.; Wang, J.; Sun, R.F.; Wu, J.; Liu, S.Y.; Bai, Y.Q.; Mun, J.H.; Bancroft, I.; Cheng, F.; et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011, 43, 1035–1039. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.Y.; Liu, Y.M.; Yang, X.H.; Tong, C.B.; Edwards, D.; Parkin, I.A.P.; Zhao, M.X.; Ma, J.X.; Yu, J.Y.; Huang, S.M.; et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 2014, 5, 11. [Google Scholar] [CrossRef]
- Parkin, I.A.P.; Koh, C.; Tang, H.B.; Robinson, S.J.; Kagale, S.; Clarke, W.E.; Town, C.D.; Nixon, J.; Krishnakumar, V.; Bidwell, S.L.; et al. Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea. Genome Biol. 2014, 15, 18. [Google Scholar] [CrossRef]
- Chalhoub, B. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef]
- Priya, P.; Arun, J.; Bisht, N.C.; Padmaja, K.L.; Sarita, S.; Vibha, G.; Pradhan, A.K.; Deepak, P. Comparative mapping of Brassica juncea and Arabidopsis thaliana using Intron Polymorphism (IP) markers: Homoeologous relationships, diversification and evolution of the A, B and C Brassica genomes. BMC Genom. 2008, 9, 113. [Google Scholar]
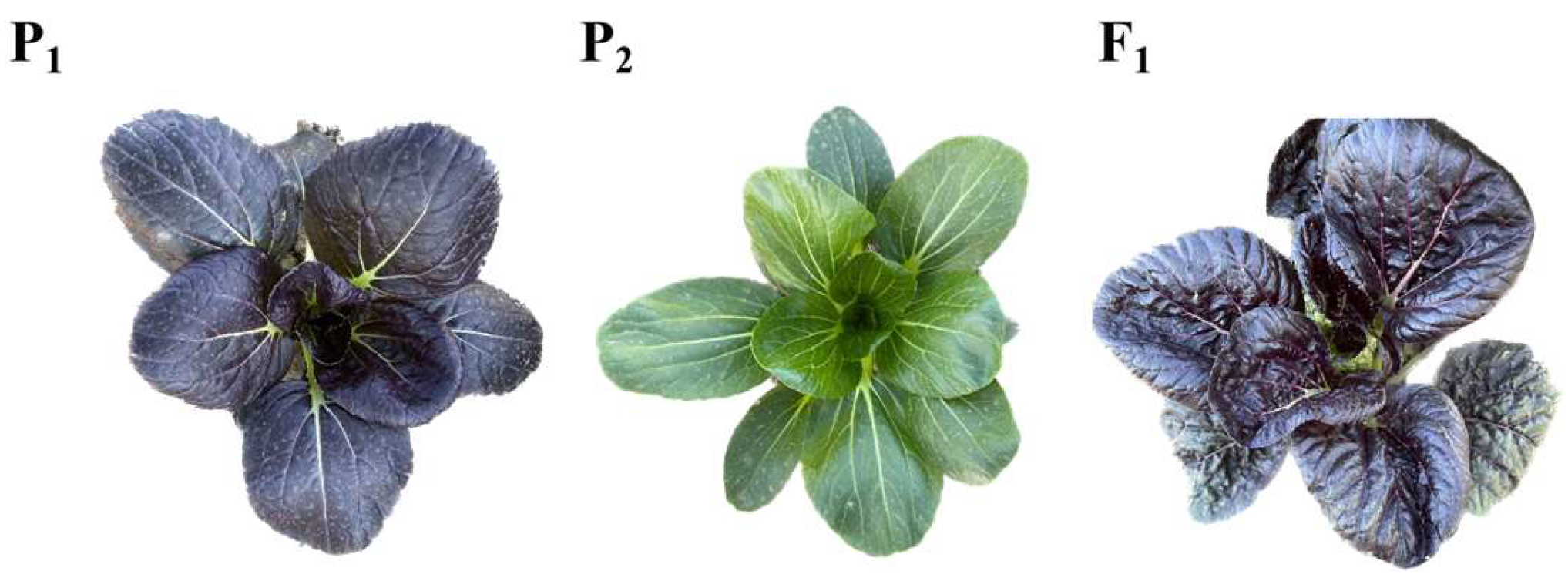


| Line | Purple Leaves | Green Leaves | Total | Expected Ratio | χ2 Test (Chi-Squared Test) |
|---|---|---|---|---|---|
| P1 | 10 | ||||
| P2 | 10 | ||||
| F1 (P1 × P2) | 10 | ||||
| F2 | 378 | 124 | 502 | 3:1 | χ2 = 0.024 < χ20.05,1 = 3.841 |
| BC1 (F1 × P2) | 102 | 107 | 209 | 1:1 | χ2 = 0.024 < χ20.05,1 = 3.841 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Zhu, Y.; Shi, B.; Zhang, S.; Zhang, S.; Zhang, H.; Sun, R.; Zhou, J.; Li, Z.; Li, G.; et al. A MYB Transcription Factor from Brassica juncea Regulates Purple Leaves in Pak Choi (Brassica campestris L. ssp. chinensis). Horticulturae 2024, 10, 276. https://doi.org/10.3390/horticulturae10030276
Wang X, Zhu Y, Shi B, Zhang S, Zhang S, Zhang H, Sun R, Zhou J, Li Z, Li G, et al. A MYB Transcription Factor from Brassica juncea Regulates Purple Leaves in Pak Choi (Brassica campestris L. ssp. chinensis). Horticulturae. 2024; 10(3):276. https://doi.org/10.3390/horticulturae10030276
Chicago/Turabian StyleWang, Xia, Yating Zhu, Bo Shi, Shujiang Zhang, Shifan Zhang, Hui Zhang, Rifei Sun, Jinyan Zhou, Ze Li, Guoliang Li, and et al. 2024. "A MYB Transcription Factor from Brassica juncea Regulates Purple Leaves in Pak Choi (Brassica campestris L. ssp. chinensis)" Horticulturae 10, no. 3: 276. https://doi.org/10.3390/horticulturae10030276





