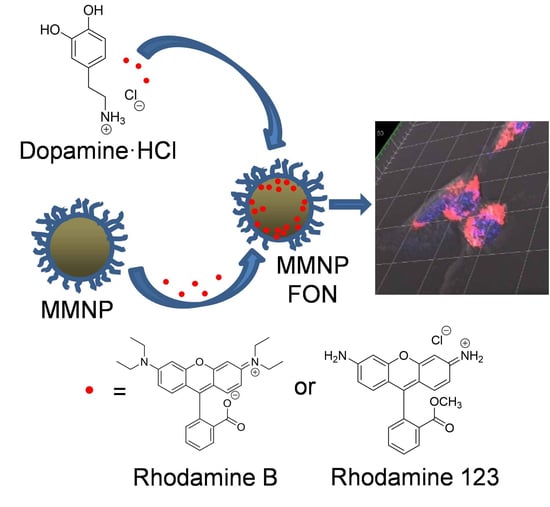
Size Control and Fluorescence Labeling of Polydopamine Melanin-Mimetic Nanoparticles for Intracellular Imaging
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Nanoparticle Synthesis and Modification
2.2.1. MMNP Synthesis
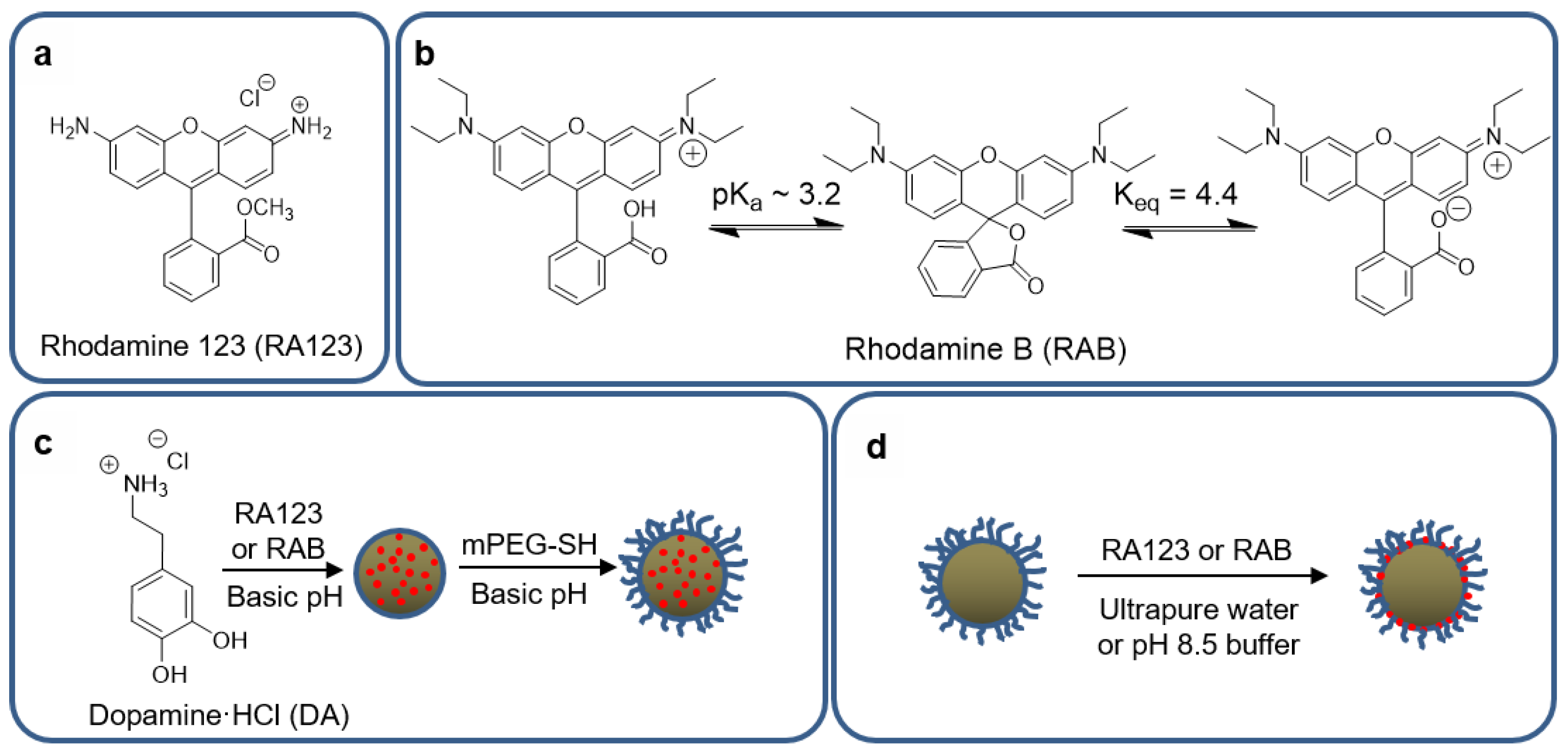
2.2.2. In Situ Modification of MMNPs with Rhodamine B or Rhodamine 123
2.2.3. PEGylation of MMNPs
2.2.4. Post-Functionalization of MMNP@PEG with Rhodamine B or Rhodamine 123
2.2.5. Fluorophore Release Testing
2.3. Nanoparticle Characterization
2.3.1. Extinction Coefficient Calculation
2.3.2. Dynamic Light Scattering and Zeta Potential Analysis
2.3.3. Electron Microscopy Imaging
2.3.4. Spectroscopic Characterization
2.3.5. X-ray Photoelectron Spectroscopy Characterization
2.4. In Vitro Uptake and Cytocompatibility Evaluation
2.4.1. MMNP@PEG Cytocompatibility Study
2.4.2. MMNP@PEG@RA123 Uptake Study
2.5. Statistical Analysis
3. Results and Discussion
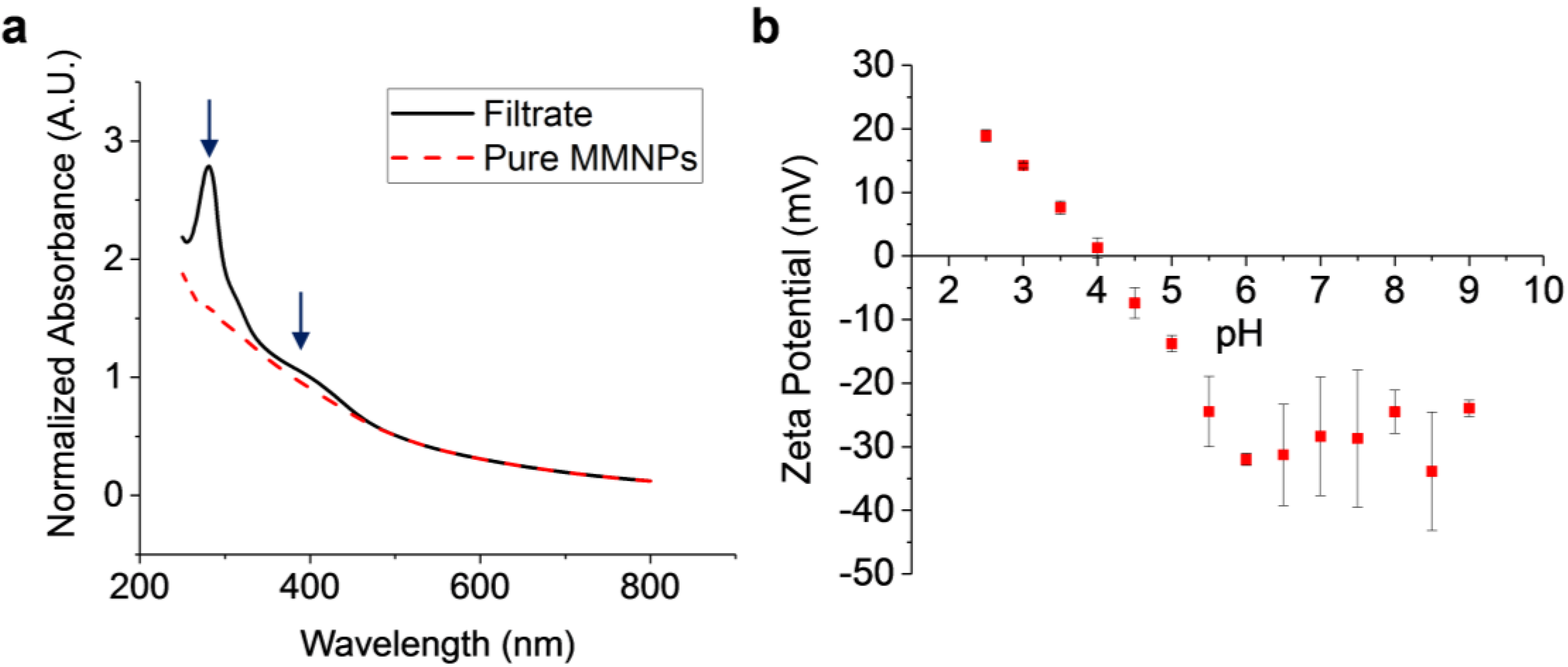
3.1. MMNP Formation via Dopamine Autoxidation, Comparisons with Melanin, and Characterization of Surface Charge
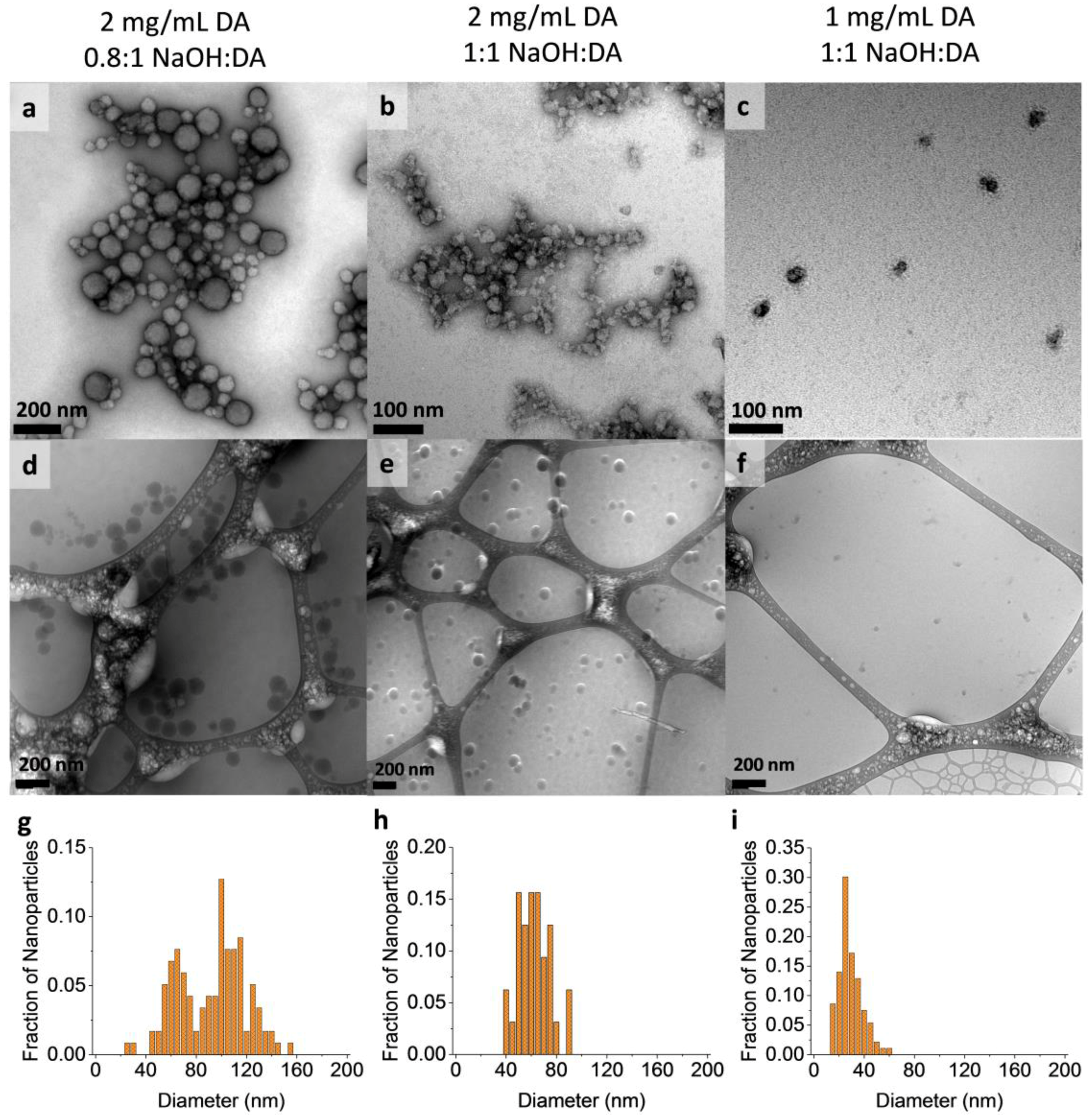
3.2. MMNP Size Control
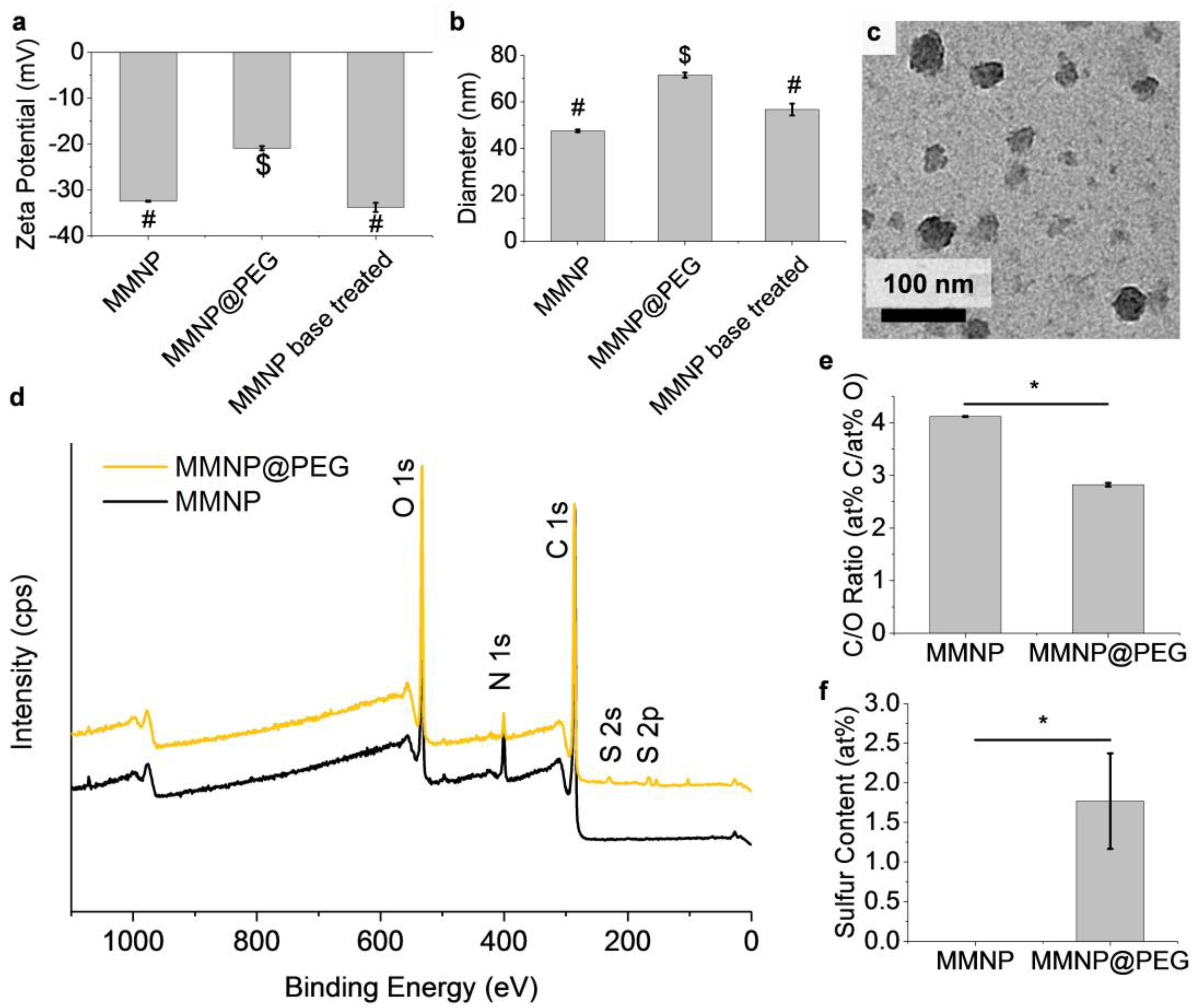
3.3. PEGylation to Produce MMNP@PEG
3.4. Stability of MMNP@PEG
3.5. Fluorescence Functionalization
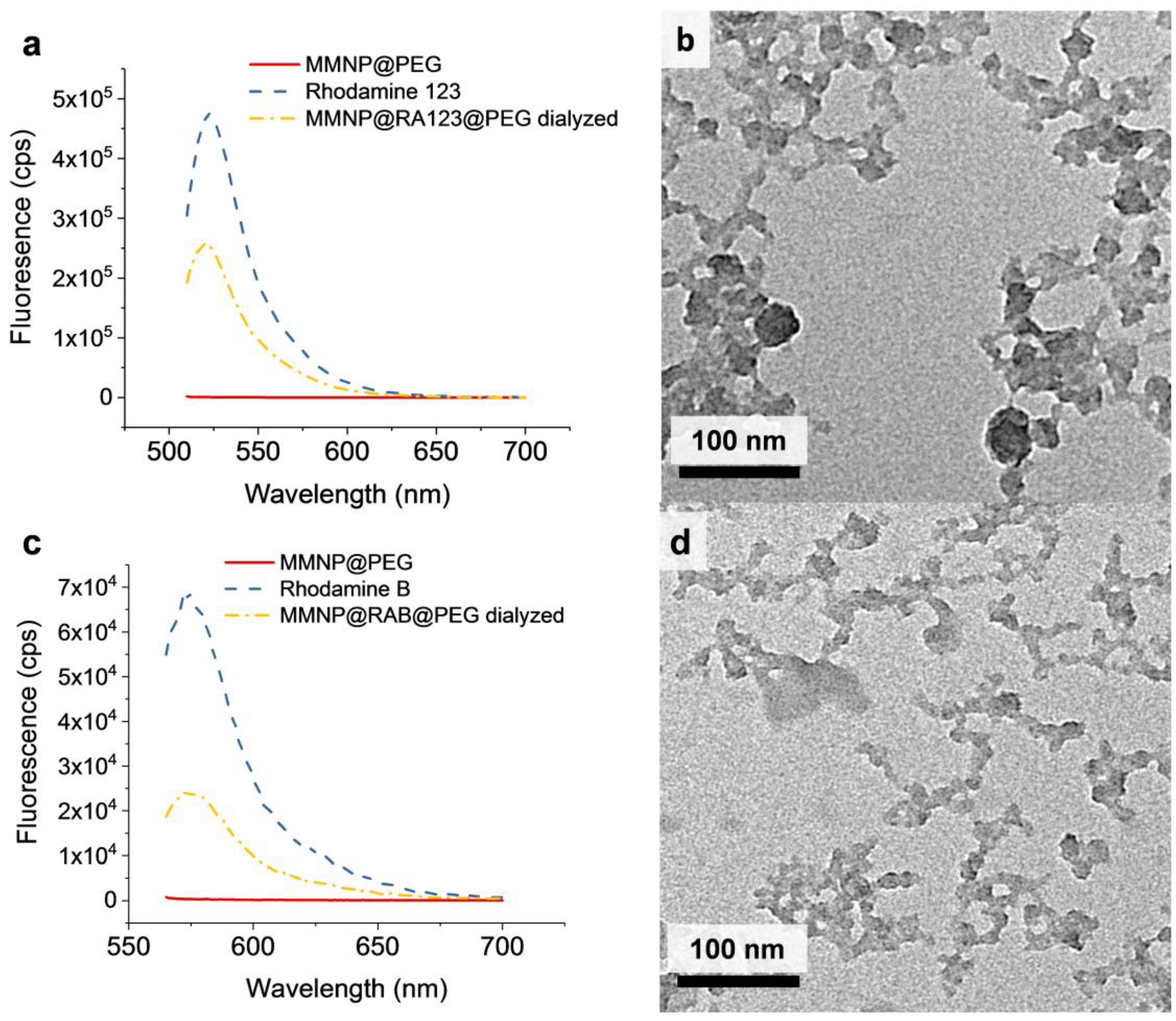
3.5.1. In situ Incorporation
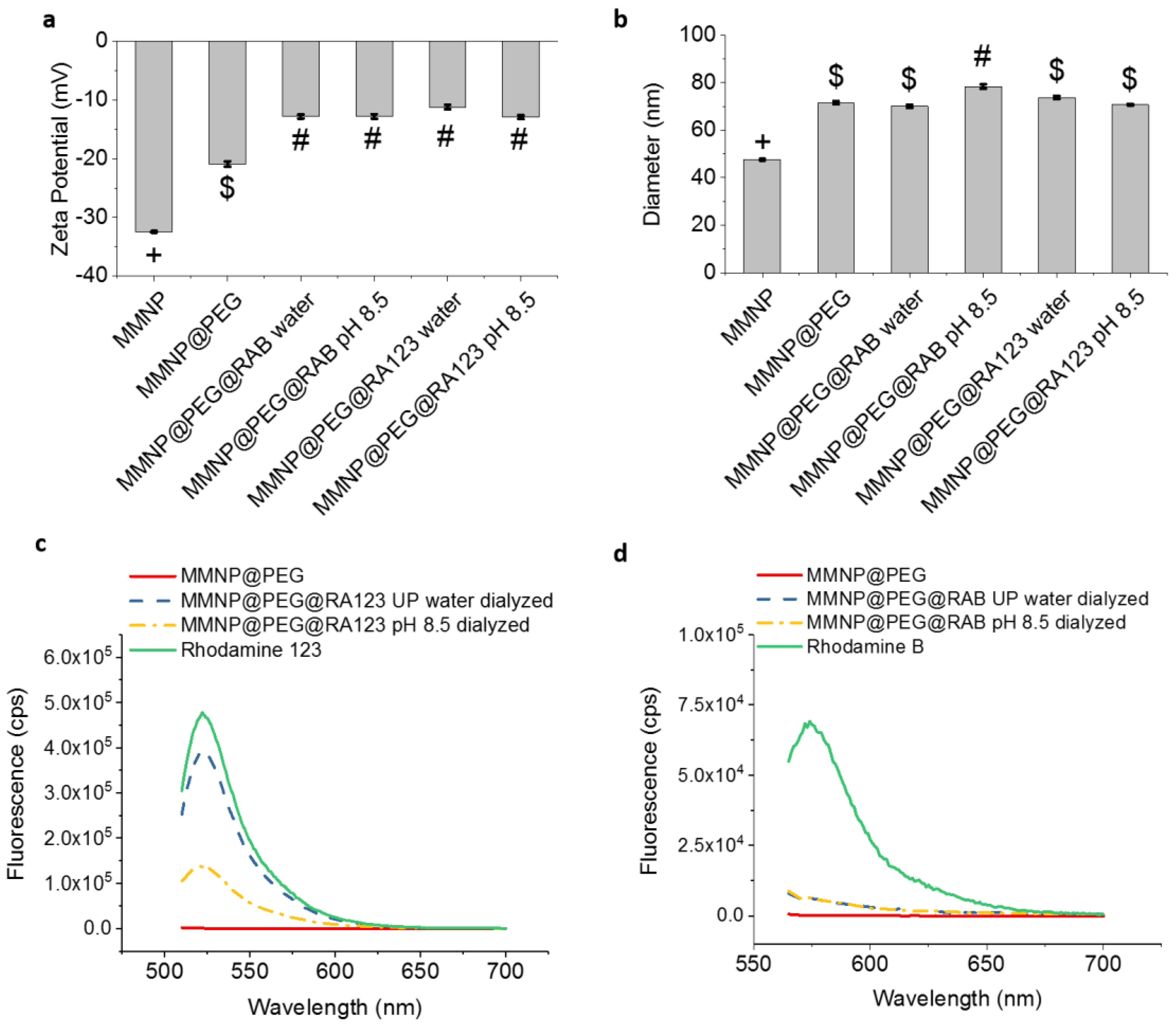
3.5.2. Post-Functionalization
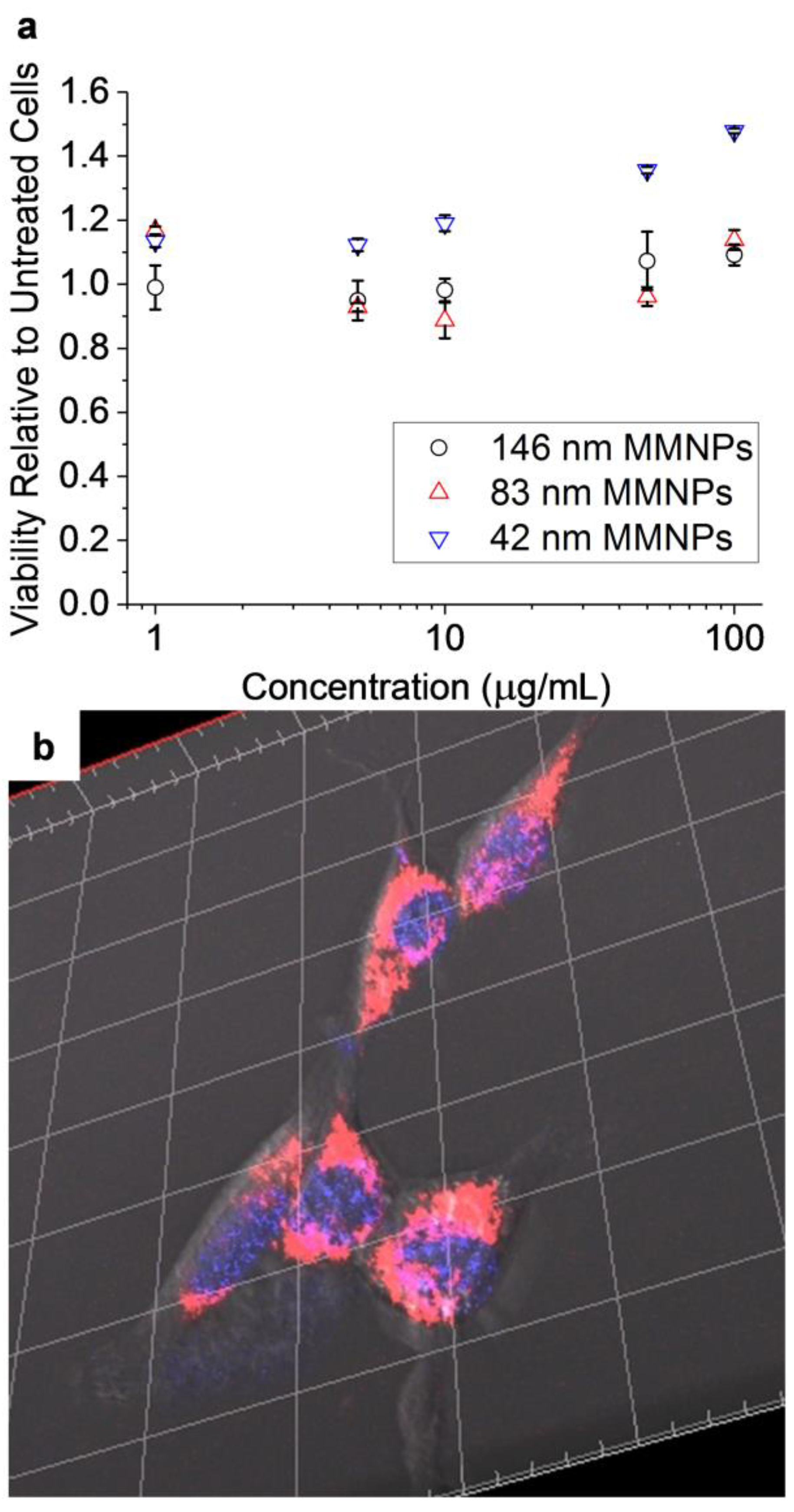
3.6. In Vitro Cytocompatibility of MMNP@PEG and Imaging of MMNP@PEG@RA123
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References and Note
- Petros, R.A.; DeSimone, J.M. Strategies in the design of nanoparticles for therapeutic applications. Nat. Rev. Drug Discov. 2010, 9, 615–627. [Google Scholar] [CrossRef] [PubMed]
- Peer, D.; Karp, J.M.; Hong, S.; FaroKhzad, O.C.; Margalit, R.; Langer, R. Nanocarriers as an emerging platform for cancer therapy. Nat. Nanotechnol. 2007, 2, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Heath, J.R.; Davis, M.E. Nanotechnology and cancer. Ann. Rev. Med. 2008, 59, 251–265. [Google Scholar] [CrossRef] [PubMed]
- Gu, F.X.; Karnik, R.; Wang, A.Z.; Alexis, F.; Levy-Nissenbaum, E.; Hong, S.; Langer, R.S.; Farokhzad, O.C. Targeted nanoparticles for cancer therapy. Nano Today 2007, 2, 14–21. [Google Scholar] [CrossRef]
- De, M.; Ghosh, P.S.; Rotello, V.M. Applications of nanoparticles in biology. Adv. Mater. 2008, 20, 4225–4241. [Google Scholar] [CrossRef]
- Davis, M.E.; Chen, Z.; Shin, D.M. Nanoparticle therapeutics: An emerging treatment modality for cancer. Nat. Rev. Drug Discov. 2008, 7, 771–782. [Google Scholar] [CrossRef] [PubMed]
- Byrne, J.D.; Betancourt, T.; Brannon-Peppas, L. Active targeting schemes for nanoparticle systems in cancer therapeutics. Adv. Drug Deliv. Rev. 2008, 60, 1615–1626. [Google Scholar] [CrossRef] [PubMed]
- Suhair, S.; Rania, H.; Heba, A.-H.; Ola, T. Synergistic Interplay of medicinal chemistry and formulation strategies in nanotechnology—From drug discovery to nanocarrier design and development. Curr. Top. Med. Chem. 2017, 17, 1451–1468. [Google Scholar] [CrossRef]
- Wilhelm, S.; Tavares, A.J.; Dai, Q.; Ohta, S.; Audet, J.; Dvorak, H.F.; Chan, W.C.W. Analysis of nanoparticle delivery to tumours. Nat. Rev. Mater. 2016, 1, 16014. [Google Scholar] [CrossRef]
- Tsoi, K.M.; MacParland, S.A.; Ma, X.Z.; Spetzler, V.N.; Echeverri, J.; Ouyang, B.; Fadel, S.M.; Sykes, E.A.; Goldaracena, N.; Kaths, J.M.; et al. Mechanism of hard-nanomaterial clearance by the liver. Nat. Mater. 2016, 15, 1212–1221. [Google Scholar] [CrossRef] [PubMed]
- Reisch, A.; Klymchenko, A.S. Fluorescent polymer nanoparticles based on dyes: Seeking brighter tools for bioimaging. Small 2016, 12, 1968–1992. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.S.; Chiu, D.T. Soft fluorescent nanomaterials for biological and biomedical imaging. Chem. Soc. Rev. 2015, 44, 4699–4722. [Google Scholar] [CrossRef] [PubMed]
- Ju, K.-Y.; Lee, Y.; Lee, S.; Park, S.B.; Lee, J.-K. Bioinspired polymerization of dopamine to generate melanin-like nanoparticles having an excellent free-radical-scavenging property. Biomacromolecules 2011, 12, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-T.; Martin-Martinez, F.J.; Jung, G.S.; Buehler, M.J. Polydopamine and eumelanin molecular structures investigated with ab initio calculations. Chem. Sci. 2017. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Na, Y.S.; Choi, S.; Song, I.T.; Kim, W.Y.; Lee, H. Non-covalent self-assembly and covalent polymerization co-contribute to polydopamine formation. Adv. Funct. Mater. 2012, 22, 4711–4717. [Google Scholar] [CrossRef]
- D’Ischia, M.; Wakamatsu, K.; Cicoira, F.; Di Mauro, E.; Garcia-Borron, J.C.; Commo, S.; Galván, I.; Ghanem, G.; Kenzo, K.; Meredith, P.; et al. Melanins and melanogenesis: From pigment cells to human health and technological applications. Pigment Cell Melanoma Res. 2015, 28, 520–544. [Google Scholar] [CrossRef]
- Meredith, P.; Sarna, T. The physical and chemical properties of eumelanin. Pigment Cell Res. 2006, 19, 572–594. [Google Scholar] [CrossRef] [PubMed]
- Bustamante, J.; Bredeston, L.; Malanga, G.; Mordoh, J. Role of melanin as a scavenger of active oxygen species. Pigment Cell Res. 1993, 6, 348–353. [Google Scholar] [CrossRef] [PubMed]
- Moses, D.N.; Mattoni, M.A.; Slack, N.L.; Waite, J.H.; Zok, F.W. Role of melanin in mechanical properties of Glycera jaws. Acta Biomater. 2006, 2, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Dellatore, S.M.; Miller, W.M.; Messersmith, P.B. Mussel-inspired surface chemistry for multifunctional coatings. Science 2007, 318, 426–430. [Google Scholar] [CrossRef] [PubMed]
- Lynge, M.E.; van der Westen, R.; Postma, A.; Stadler, B. Polydopamine—A nature-inspired polymer coating for biomedical science. Nanoscale 2011, 3, 4916–4928. [Google Scholar] [CrossRef] [PubMed]
- Howes, P.D.; Chandrawati, R.; Stevens, M.M. Colloidal nanoparticles as advanced biological sensors. Science 2014, 346. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ai, K.; Liu, J.; Deng, M.; He, Y.; Lu, L. Dopamine-melanin colloidal nanospheres: An efficient near-infrared photothermal therapeutic agent for in vivo cancer therapy. Adv. Mater. 2013, 25, 1353–1359. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Yang, L.; Lin, M.-F.; Ma, J.; Lu, X.; Lee, P.S. Polydopamine spheres as active templates for convenient synthesis of various nanostructures. Small 2013, 9, 596–603. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Li, Y.; Hu, Z.; Yue, X.; Proetto, M.T.; Jones, Y.; Gianneschi, N.C. Mimicking melanosomes: Polydopamine nanoparticles as artificial microparasols. ACS Cent. Sci. 2017. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.-H.; Yu, C.-J.; Yang, Y.-C.; Tseng, W.-L. Formation of fluorescent polydopamine dots from hydroxyl radical-induced degradation of polydopamine nanoparticles. Phys. Chem. Chem. Phys. 2015, 17, 15124–15130. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, S.; Xu, L.; Feng, L.; Ji, Y.; Tao, L.; Li, S.; Wei, Y. Biocompatible polydopamine fluorescent organic nanoparticles: Facile preparation and cell imaging. Nanoscale 2012, 4, 5581–5584. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yan, Y.; Müllner, M.; van Koeverden, M.P.; Noi, K.F.; Zhu, W.; Caruso, F. Engineering fluorescent poly(dopamine) capsules. Langmuir 2014, 30, 2921–2925. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zuo, F.; Liao, Z.; Qin, Z.; Du, S.; Zhao, Z. Mussel-inspired one-pot synthesis of a fluorescent and water-soluble polydopamine–Polyethyleneimine copolymer. Macromol. Rapid Commun. 2015, 36, 909–915. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Ji, J.; Zhang, X.; Zhang, X.; Yang, B.; Deng, F.; Li, Z.; Wang, K.; Yang, Y.; Wei, Y. Self-polymerization of dopamine and polyethyleneimine: Novel fluorescent organic nanoprobes for biological imaging applications. J. Mater. Chem. B 2015, 3, 3476–3482. [Google Scholar] [CrossRef]
- Chan, W.C.W.; Nie, S. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science 1998, 281, 2016–2018. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH image to ImageJ: 25 years of image analysis. Nat Meth 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Repetto, G.; del Peso, A.; Zurita, J.L. Neutral red uptake assay for the estimation of cell viability/cytotoxicity. Nat. Protoc. 2008, 3, 1125–1131. [Google Scholar] [CrossRef] [PubMed]
- Bisaglia, M.; Mammi, S.; Bubacco, L. Kinetic and structural analysis of the early oxidation products of dopamine: Analysis of the interactions with α-synuclein. J. Biol. Chem. 2007, 282, 15597–15605. [Google Scholar] [CrossRef] [PubMed]
- Herlinger, E.; Jameson, R.F.; Linert, W. Spontaneous autoxidation of dopamine. J. Chem. Soc. Perkin Trans. 2 1995, 259–263. [Google Scholar] [CrossRef]
- Sarna, T.; Swartz, H.A. The physical properties of melanins. In The Pigmentary System; Blackwell Publishing Ltd.: Hoboken, NJ, USA, 2007; pp. 311–341. [Google Scholar]
- Sarna, T.; Sealy, R.C. Photoinduced oxygen consumption in melanin systems. Action spectra and quantum yields for eumelanin and synthetic melanin. Photochem. Photobiol. 1984, 39, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Ball, V. Impedance spectroscopy and zeta potential titration of dopa-melanin films produced by oxidation of dopamine. Colloids Surf. A 2010, 363, 92–97. [Google Scholar] [CrossRef]
- Stetefeld, J.; McKenna, S.A.; Patel, T.R. Dynamic light scattering: A practical guide and applications in biomedical sciences. Biophys. Rev. 2016, 8, 409–427. [Google Scholar] [CrossRef] [PubMed]
- In the mushroom regime of PEG packing at the MMNP surface, the diameter increase (∆d) would be expected to be double the Flory radius (2 × Rf). The Flory radius is Rf = αn3/5, where n is the number monomers per polymer chain, and α is the segmental length of one monomer [41]. For PEG, α = 0.35 nm, and 5 kDa PEG has n ≈ 113 monomers per chain, resulting in Rf = 6.0 nm and ∆d = 12.0 nm.
- Allen, C.; Dos Santos, N.; Gallagher, R.; Chiu, G.N.C.; Shu, Y.; Li, W.M.; Johnstone, S.A.; Janoff, A.S.; Mayer, L.D.; Webb, M.S.; et al. Controlling the physical behavior and biological performance of liposome formulations through use of surface grafted poly(ethylene glycol). Biosci. Rep. 2002, 22, 225–250. [Google Scholar] [CrossRef] [PubMed]
- Perry, J.L.; Reuter, K.G.; Kai, M.P.; Herlihy, K.P.; Jones, S.W.; Luft, J.C.; Napier, M.; Bear, J.E.; DeSimone, J.M. PEGylated PRINT nanoparticles: The impact of PEG density on protein binding, macrophage association, biodistribution, and pharmacokinetics. Nano Lett. 2012, 12, 5304–5310. [Google Scholar] [CrossRef] [PubMed]
- Gref, R.; Lück, M.; Quellec, P.; Marchand, M.; Dellacherie, E.; Harnisch, S.; Blunk, T.; Müller, R.H. ‘Stealth’ corona-core nanoparticles surface modified by polyethylene glycol (PEG): Influences of the corona (PEG chain length and surface density) and of the core composition on phagocytic uptake and plasma protein adsorption. Colloids Surf. B 2000, 18, 301–313. [Google Scholar] [CrossRef]
- Fang, C.; Shi, B.; Pei, Y.Y.; Hong, M.H.; Wu, J.; Chen, H.Z. In vivo tumor targeting of tumor necrosis factor-α-loaded stealth nanoparticles: Effect of MePEG molecular weight and particle size. Eur. J. Pharm. Sci. 2006, 27, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Bazile, D.; Prud’homme, C.; Bassoullet, M.T.; Marlard, M.; Spenlehauer, G.; Veillard, M. Stealth Me. PEG-PLA nanoparticles avoid uptake by the mononuclear phagocytes system. J. Pharm. Sci. 1995, 84, 493–498. [Google Scholar] [CrossRef] [PubMed]
- Walkey, C.D.; Olsen, J.B.; Guo, H.; Emili, A.; Chan, W.C. Nanoparticle size and surface chemistry determine serum protein adsorption and macrophage uptake. J. Am. Chem. Soc. 2012, 134, 2139–2147. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Y.; Yuan, Y.; Liu, C.; Tao, X.; Shan, X.; Xu, F. In vitro macrophage uptake and in vivo biodistribution of PLA-PEG nanoparticles loaded with hemoglobin as blood substitutes: Effect of PEG content. J. Mater. Sci. Mater. Med. 2009, 20, 1881–1891. [Google Scholar] [CrossRef] [PubMed]
- Liebscher, J.; Mrówczyński, R.; Scheidt, H.A.; Filip, C.; Hădade, N.D.; Turcu, R.; Bende, A.; Beck, S. Structure of polydopamine: A never-ending story? Langmuir 2013, 29, 10539–10548. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Kim, K.C.; Lee, B.; Chen, Z.; Noda, S.; Jang, S.S.; Lee, S.W. Self-polymerized dopamine as an organic cathode for Li- and Na-ion batteries. Energy Environ. Sci. 2017, 10, 205–215. [Google Scholar] [CrossRef]
- Lee, H.; Rho, J.; Messersmith, P.B. Facile conjugation of biomolecules onto surfaces via mussel adhesive protein inspired coatings. Adv. Mater. 2009, 21, 431–434. [Google Scholar] [CrossRef] [PubMed]
- Hinckley, D.A.; Seybold, P.G. A spectroscopic/thermodynamic study of the rhodamine B lactone ⇌ zwitterion equilibrium. Spectrochim. Acta Part A 1988, 44, 1053–1059. [Google Scholar] [CrossRef]
- Mchedlov-Petrossyan, N.O.; Vodolazkaya, N.A.; Doroshenko, A.O. Ionic equilibria of fluorophores in organized solutions: The influence of micellar microenvironment on protolytic and photophysical properties of rhodamine B. J. Fluoresc. 2003, 13, 235–248. [Google Scholar] [CrossRef]
- Butcher, K.J.; Hurst, J. Aromatic amines as nucleophiles in the Bargellini reaction. Tetrahedron Lett. 2009, 50, 2497–2500. [Google Scholar] [CrossRef]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amin, D.R.; Sugnaux, C.; Lau, K.H.A.; Messersmith, P.B. Size Control and Fluorescence Labeling of Polydopamine Melanin-Mimetic Nanoparticles for Intracellular Imaging. Biomimetics 2017, 2, 17. https://doi.org/10.3390/biomimetics2030017
Amin DR, Sugnaux C, Lau KHA, Messersmith PB. Size Control and Fluorescence Labeling of Polydopamine Melanin-Mimetic Nanoparticles for Intracellular Imaging. Biomimetics. 2017; 2(3):17. https://doi.org/10.3390/biomimetics2030017
Chicago/Turabian StyleAmin, Devang R., Caroline Sugnaux, King Hang Aaron Lau, and Phillip B. Messersmith. 2017. "Size Control and Fluorescence Labeling of Polydopamine Melanin-Mimetic Nanoparticles for Intracellular Imaging" Biomimetics 2, no. 3: 17. https://doi.org/10.3390/biomimetics2030017