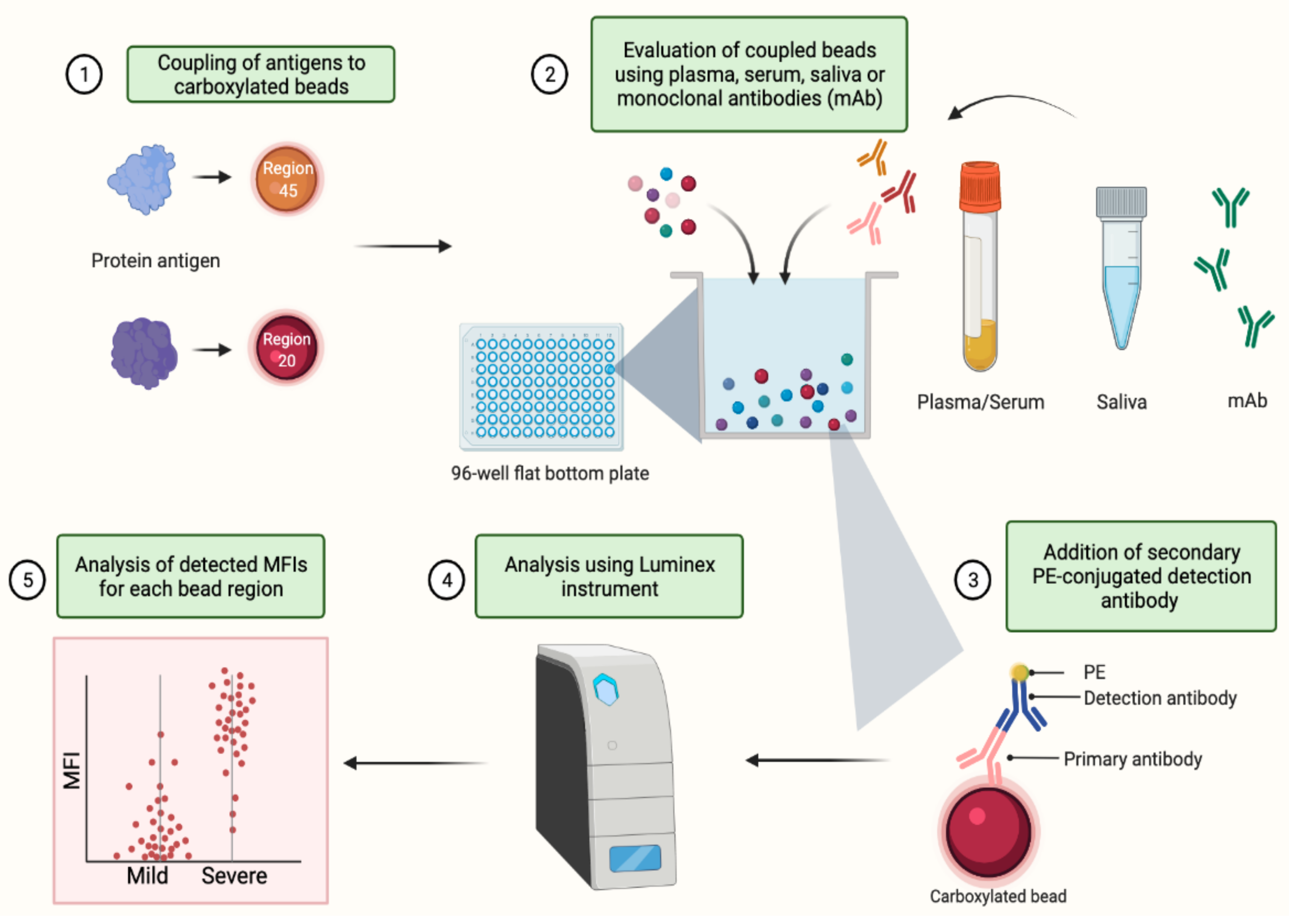
SARS-CoV-2 Multi-Antigen Serology Assay
Abstract
:1. Introduction
2. Experimental Design
2.1. Materials
- Phosphate-buffered saline (DPBS, pH 7.0–7.3 Gibco, Cat # 14190-144),
- Monobasic sodium phosphate buffer pH 6.0 (Sigma, St. Louis, MO, USA, Cat # S0751-500G),
- Sulfo-NHS (hydroxysulfosuccinimide sodium salt 98%, Sigma, St. Louis, MO, USA, Cat # 56485-250MG),
- EDC (N-Ethyl-N′-carbodiimide hydrochloride, Sigma, St. Louis, MO, USA, Cat # 03449-5G),
- Na-azide (Sodium azide, Sigma, St. Louis, MO, USA, Cat # S2002-25G),
- Tween-20 (Sigma, St. Louis, MO, USA Cat # P1379),
- MilliQ-water (MQ-H2O),
- PBT buffer (1× PBS, 1% BSA, 0.05% Tween-20; sterile filter the PBS-BSA prior to adding Tween-20),
- Bovine serum albumin (BSA, Sigma, St. Louis, MO, USA Cat # A7906-100G),
- Carboxylated microspheres (Bio-Plex Pro Magnetic COOH Beads, Bio-Rad, Hercules, CA, USA, 1 mL (1.25 × 107 bead/mL), Cat # MC100X01),
- R-PE AffiniPure F(ab’)2 Fragment, donkey anti-human IgG (Jackson Immuno Research, West Grove, PA, USA, Cat # 709-116-098),
- R-PE AffiniPure F(ab’)2 Fragment, donkey anti-human IgM (Jackson Immuno Research, West Grove, PA, USA, Cat # 709-116-073),
- Goat F(ab’)2 anti-human IgA-PE (Southern Biotech, Birmingham, AL, USA, Cat # 2052-09),
- Antigens (Table 1),
- Positive control (plasma or saliva from COVID-19 diagnosed convalescent individuals),
- Negative control (naïve control plasma or saliva collected from individuals before the start of the pandemic or SARS-CoV-2 negative),
- 96-well round-bottom plate, clear (Corning, Corning, NY, USA, Cat # 353077),
- 96-well flat-bottom plate, black (Greiner Bio-One, Kremsmunster, Austria, Cat # 655090),
- Microcentrifuge tubes (Eppendorf, Safe-Lock tubes, Cat # 0030 120.191),
- Falcon tubes (15 mL Polystyrene conical tube, Corning, Corning, NY, USA, Cat # 352095),
- Magpix Calibration Kit (Luminex, Austin, TX, USA, Cat # MPX-CAL-K25),
- Magpix Verification Kit (Luminex, Austin, TX, USA, Cat # MPX-PVER-K25), and
- Magpix Drive Fluid (Luminex, Austin, TX, USA, Cat # MPX-DF-4PK).
2.2. Equipment
- Magnetic plate washer (BioTek Instruments, Winooski, VT, USA, BioTek ELx50),
- Bio-Rad Sure Beads magnetic rack (Bio-Rad, Hercules, CA, USA, Cat # 1614916),
- Ratek Platform shaker (Ratek Instruments, Boronia, Australia, MPS1 Microtiter/PCR Plate Shaker),
- Vortex Sonicator (Branson 2200),
- BioSan Vortex V-1 plus,
- Table centrifuge (Eppendorf Centrifuge 5424), and
- MAGPIX® Multiplexing System (Millipore, Burlington, MA, USA, MAGPIX® System, Xponent software V.4.2).
3. Procedure
3.1. Optimization of Coupling SARS-CoV-2 and Control Proteins to Magnetic Beads
3.1.1. Microsphere Activation
- Sonicate magnetic microsphere stocks for 15 s followed by 10 s vortexing, before transferring 50 µL beads in the trial coupling phase (6.25 × 105 beads) and 200 µL beads for the bulk coupling phase (2.5 × 106 beads) to a microcentrifuge tube.
- Place the tubes in a magnetic separator rack and allow separation to occur for 30–60 s.
- While tubes still positioned in the rack, remove the supernatant, taking care not to disturb the beads.
- Remove the tubes from the rack and resuspend beads in MQ-H2O (100 µL for trial coupling and 200 µL for bulk coupling) by vortexing for 20 s.
- Place tube back into magnetic separator rack for 30–60 s and while positioned in the rack remove supernatant, taking care not to disturb the beads.
- Remove the tubes from the rack and resuspend in 100 mM monobasic sodium phosphate buffer, pH 6.2 (80 µL for trial coupling and 160 µL for bulk coupling) and vortex for 20 s.
- Add sulfo-NHS (50 mg/mL in MQ-H2O), mix by gentle vortex for 10 s and add EDC (50 mg/mL in MQ-H2O), mix again by gentle vortex for 10 s (10 µL each for trial and 20 µL for bulk coupling) and incubate while rotating in the dark for 20 min, to activate the beads.
CRITICAL STEP Sulfo-NHS and EDC should be made fresh on the day of use.
- After the incubation, place the tubes into a magnetic separator rack for 30–60 s and while positioned in the rack, remove supernatant, taking care not to disturb the beads.
- Remove the tubes from the rack and resuspend in PBS, pH 7.0–7.3 (250 µL for trial and 500 µL for bulk coupling), vortex for 20 s and place in magnetic separator for 30–60 s and remove the supernatant.
- Remove tubes from magnetic separator and continue the washing with PBS one more time (for a total of 2 washes).
- Resuspend the activated and washed beads in PBS (250 µL volume for trial couplings and 500 µL volume for bulk coupling), vortex for 20 s and put aside for the protein coupling step.
3.1.2. Protein Coupling to Microspheres
- Add the appropriate volume of each protein, which corresponds to the protein concentration being tested, to the activated beads and incubate while rotating in the dark for 2 h at room temperature.
PAUSE STEP Beads can also be left over night at 4 °C, on a rotator.
- Place tubes again into a magnetic separator rack and allow separation for 30–60 s and remove supernatant without disturbing the beads.
- Resuspend the beads in 500 µL PBS-TBN (PBS, pH 7.0–7.3 plus 0.02% Tween-20, 0.1% BSA and 0.05% Na-azide), vortex for 20 s and repeat the washing two more times (three washes in total).
- After the final removal of supernatant, resuspend the beads in PBS-TBN (125 µL for trial couplings and 500 µL for bulk couplings), vortex for 10 s. Note: Bead loss is minimal when using magnetic beads and the magnetic separator racks and thus counting of beads after coupling are not required. However, a loss in bead numbers could occur due to the various centrifugation steps, when using non-magnetic beads. If non-magnetic beads are used, counting of the bead numbers under the microscope after final resuspension is recommended to achieve consistent final bead-concentration.
PAUSE STEP Store the coupled beads at 4 °C in the dark.
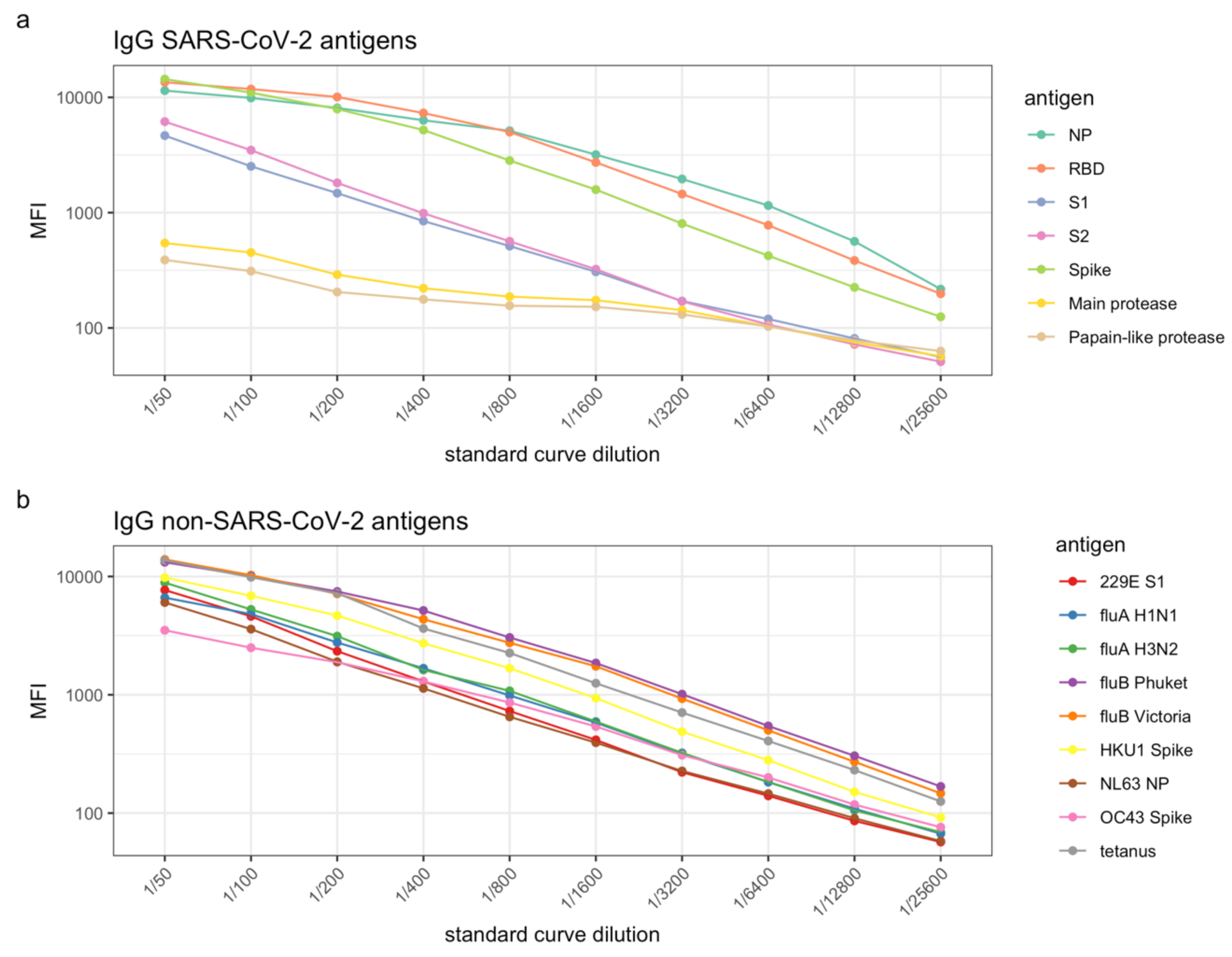
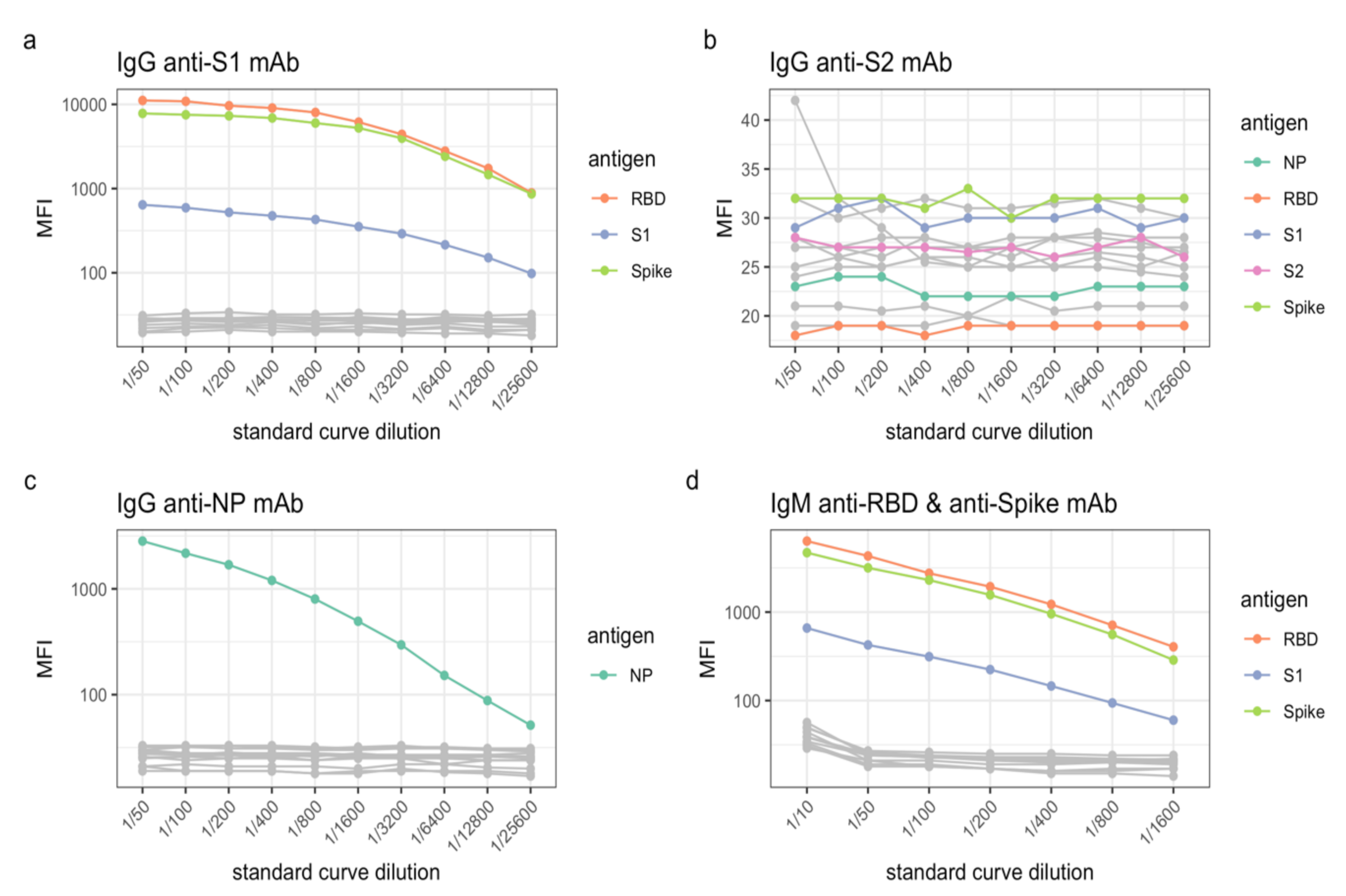
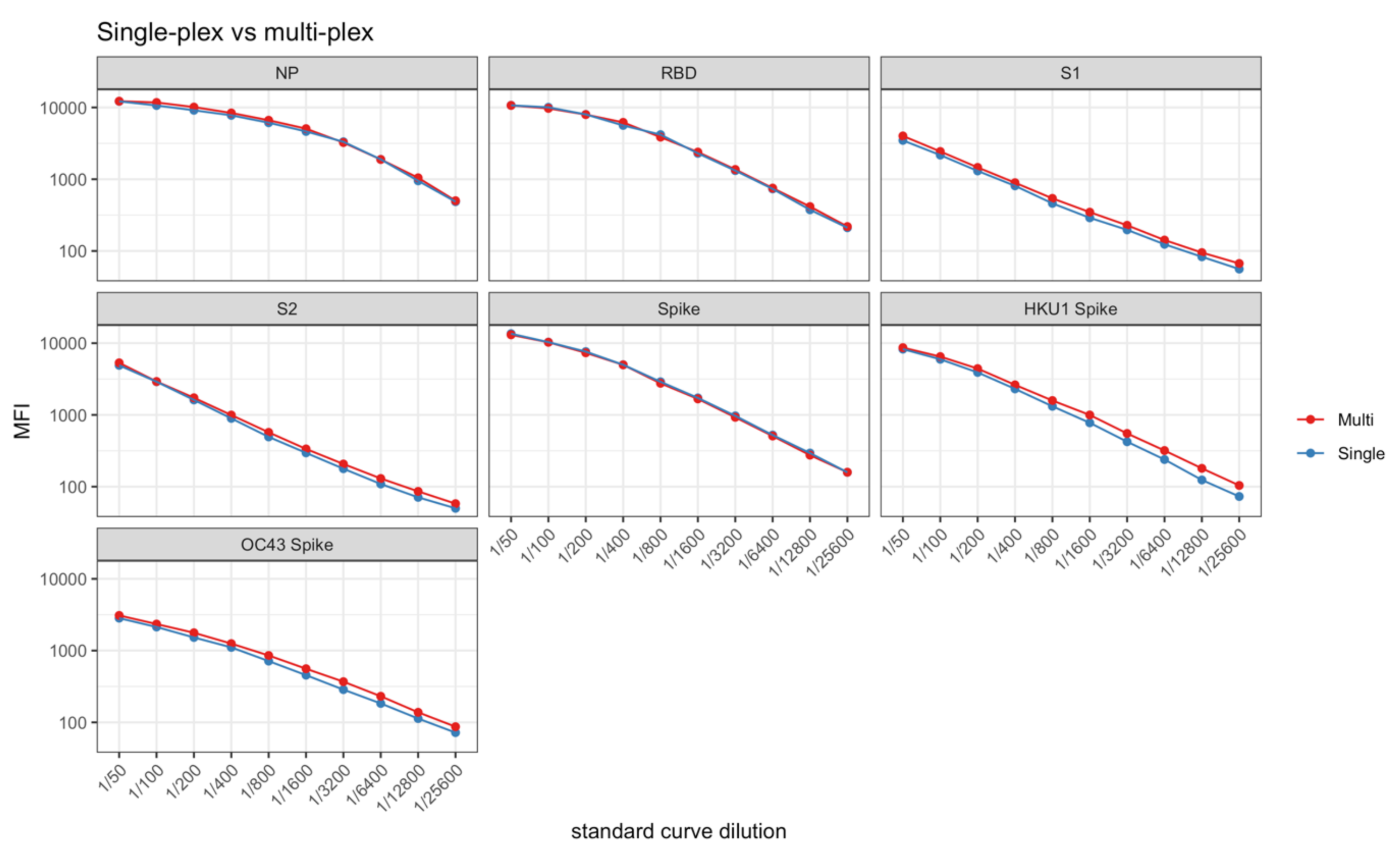
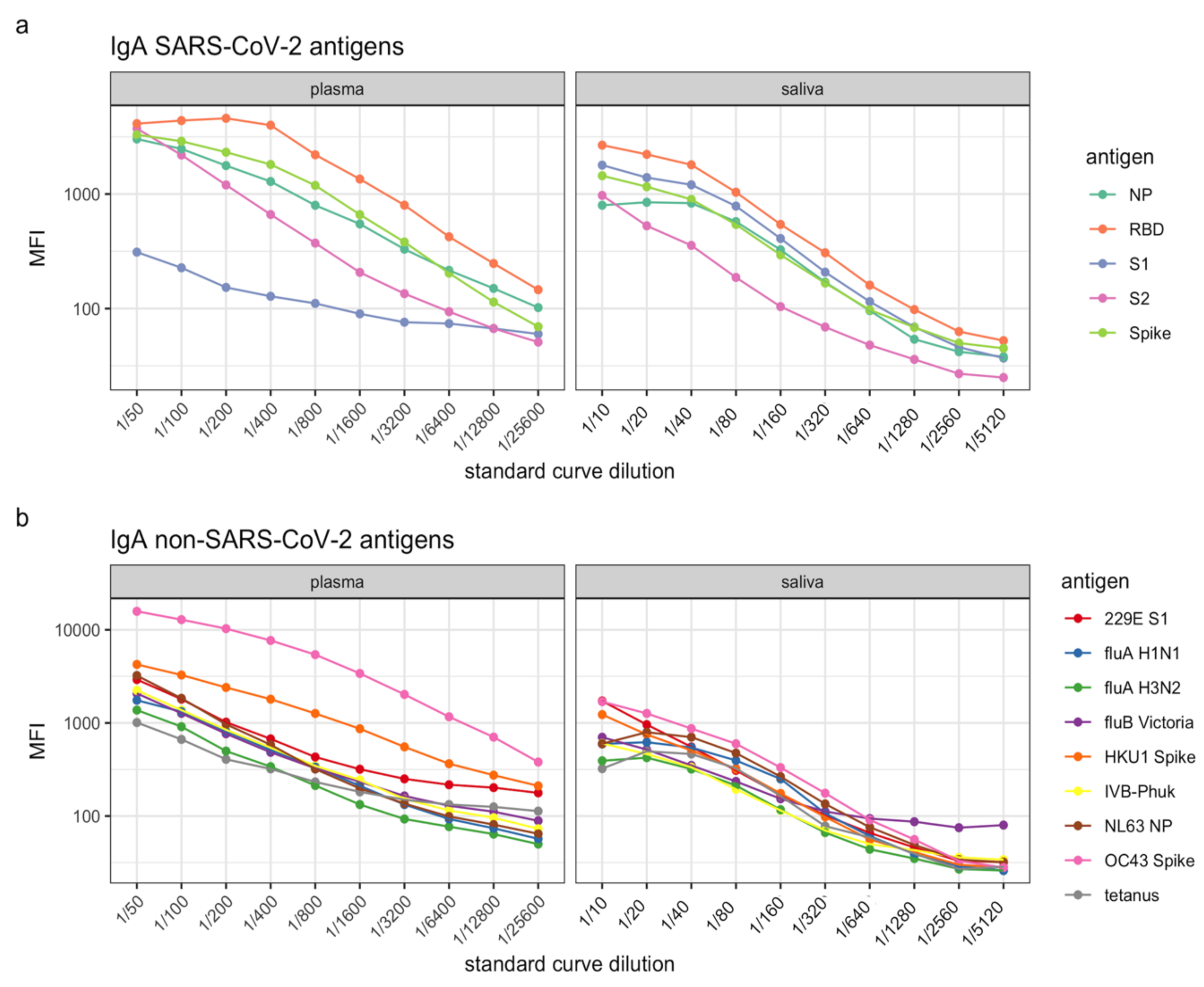
3.2. Evaluation of Capacity to Measure Antigen-Specific IgG, IgM and IgA Antibody Responses
- Saliva is pre-processed by centrifugation in a table centrifuge at 10,000× g for 15 min to remove cell pellets and debris. The saliva sample is then aliquoted before freezing at −80 °C until use.
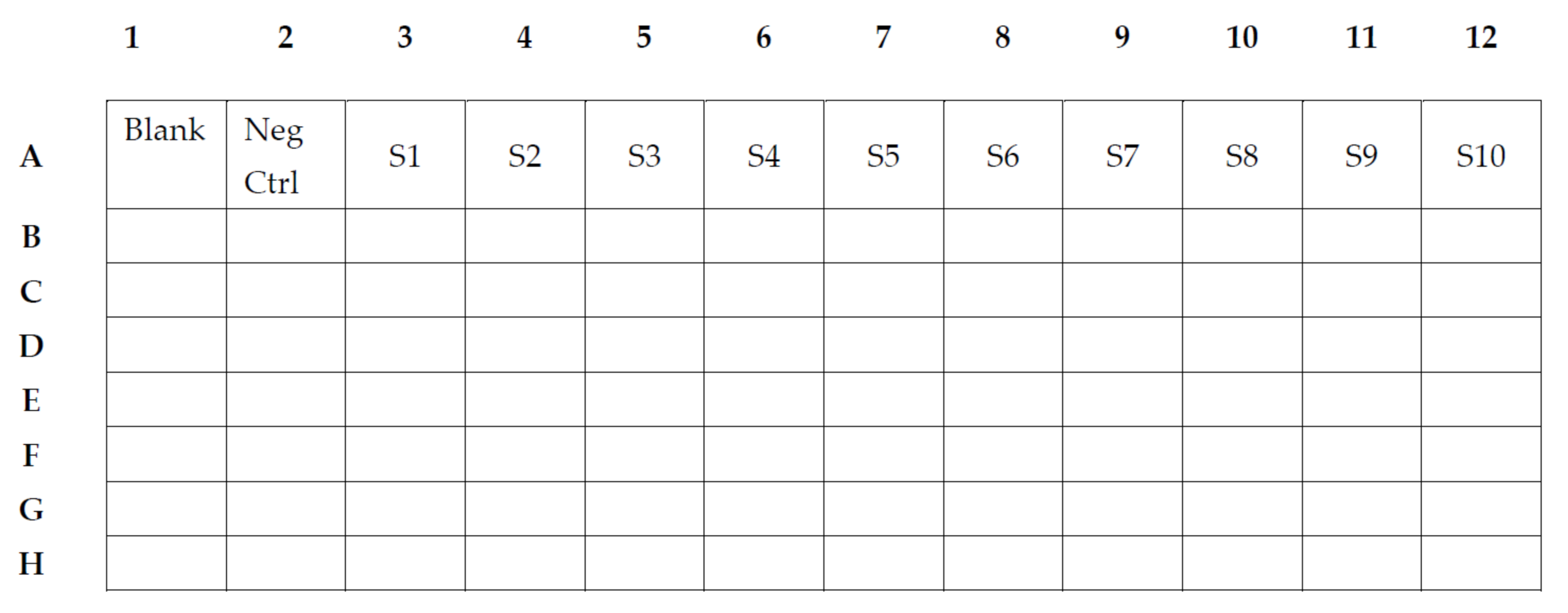
- Samples and standard curves are pre-diluted in a 96-well round-bottom plate (dilution plate).
- In the 96-well round-bottom dilution plate, we use one well of blank, consisting of 100 µL PBT buffer (Figure 1).
- One well of negative control, consisting of 99 µL PBT and 1 µL of the negative plasma or 90 µL PBT and 10 µL of the negative saliva (Figure 2).
- In wells A3–A12, we add 75 µL PBT to each well, except in well A3, in which a total of 147 µL PBT and 3 µL of the positive plasma pool or 135 µL PBT and 15 µL of the positive saliva pool will be added to achieve a 1:50 dilution or 1:10 dilution, respectively (S1). Two-fold serial dilution of the standard is achieved by transferring 75 µL after thorough mixing from A3 to A4, and consecutively from each well to the next thereafter until A12. This will provide a dilution series of the positive pool from 1:50 to 1:25,600 (Figure 2(S1–S10)) and 1:10 to 1:5120 for a saliva pool.
- Individual samples to be analyzed are pre-diluted in PBT (99 µL for plasma samples and 80 µL for saliva samples) in the remaining wells of the 96-well round-bottom dilution plate. Plasma (1 µL/well) or saliva (20 µL/well) is added to yield a 1:100 dilution and 1:5 dilution, respectively.
- Magnetic beads are prepared and added to a 96-well flat-bottom plate (assay plate). The magnetic bead mix is prepared by vortexing the individual coupled bead regions for 30 s each, before adding 0.1 µL/well of each (total amount is based on the number of wells used) to a total of 50 µL PBT/well in a master bead mix. A volume of 50 µL PBT, containing 0.1 µL of each of the coupled beads, are added to each well of the 96-well flat-bottom assay plate.
- After the addition of the bead mix to all wells, 50 µL of the plasma or saliva dilutions and control dilutions are added to each well on the black flat-bottom assay plate. This mix of coupled beads and plasma/saliva samples is incubated for 30 min (IgG and IgM) or 2 h (IgA) at room temperature on a shaker in the dark, followed by washing the plate 3× with 100 µL PBT, using a magnetic plate washer or a hand magnet.
- After the washings, add 100 µL of a 1:100 dilution of PE-conjugated Donkey F(ab’)2 anti-human IgG, IgM or IgA to the plate, mix well and incubate for 15 min at room temperature on a shaker in the dark.
- After a further washing of the plate, add 80 µL PBT to each well, mix well and keep on a shaker until plate is inserted and read on a compatible Luminex instrument.
- At least 15 beads of each region/antigen are then acquired and analyzed.
4. Expected Results
5. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Long, Q.X.; Liu, B.Z.; Deng, H.J.; Wu, G.C.; Deng, K.; Chen, Y.K.; Liao, P.; Qiu, J.F.; Lin, Y.; Cai, X.F.; et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020, 26, 845–848. [Google Scholar] [CrossRef]
- Iyer, A.S.; Jones, F.K.; Nodoushani, A.; Kelly, M.; Becker, M.; Slater, D.; Mills, R.; Teng, E.; Kamruzzaman, M.; Garcia-Beltran, W.F.; et al. Persistence and decay of human antibody responses to the receptor binding domain of SARS-CoV-2 spike protein in COVID-19 patients. Sci. Immunol. 2020, 5, eabe0367. [Google Scholar] [CrossRef] [PubMed]
- Choe, P.G.; Kim, K.H.; Kang, C.K.; Suh, H.J.; Kang, E.; Lee, S.Y.; Kim, N.J.; Yi, J.; Park, W.B.; Oh, M.D. Antibody Responses 8 Months after Asymptomatic or Mild SARS-CoV-2 Infection. Emerg. Infect. Dis. 2021, 27, 928–931. [Google Scholar] [CrossRef] [PubMed]
- Dan, J.M.; Mateus, J.; Kato, Y.; Hastie, K.M.; Yu, E.D.; Faliti, C.E.; Grifoni, A.; Ramirez, S.I.; Haupt, S.; Frazier, A.; et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 2021, 371, eabf4063. [Google Scholar] [CrossRef] [PubMed]
- Sabino, E.C.; Buss, L.F.; Carvalho, M.P.S.; Prete, C.A., Jr.; Crispim, M.A.E.; Fraiji, N.A.; Pereira, R.H.M.; Parag, K.V.; da Silva Peixoto, P.; Kraemer, M.U.G.; et al. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet 2021, 397, 452–455. [Google Scholar] [CrossRef]
- Dugas, M.; Grote-Westrick, T.; Merle, U.; Fontenay, M.; Kremer, A.E.; Hanses, F.; Vollenberg, R.; Lorentzen, E.; Tiwari-Heckler, S.; Duchemin, J.; et al. Lack of antibodies against seasonal coronavirus OC43 nucleocapsid protein identifies patients at risk of critical COVID-19. J. Clin. Virol. 2021, 139, 104847. [Google Scholar] [CrossRef]
- Dugas, M.; Grote-Westrick, T.; Vollenberg, R.; Lorentzen, E.; Brix, T.; Schmidt, H.; Tepasse, P.R.; Kuhn, J. Less severe course of COVID-19 is associated with elevated levels of antibodies against seasonal human coronaviruses OC43 and HKU1 (HCoV OC43, HCoV HKU1). Int. J. Infect. Dis. 2021, 105, 304–306. [Google Scholar] [CrossRef] [PubMed]
- Sterlin, D.; Mathian, A.; Miyara, M.; Mohr, A.; Anna, F.; Claer, L.; Quentric, P.; Fadlallah, J.; Devilliers, H.; Ghillani, P.; et al. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci. Transl. Med. 2021, 13, eabd2223. [Google Scholar] [CrossRef]
- Mazhari, R.; Brewster, J.; Fong, R.; Bourke, C.; Liu, Z.S.J.; Takashima, E.; Tsuboi, T.; Tham, W.H.; Harbers, M.; Chitnis, C.; et al. A comparison of non-magnetic and magnetic beads for measuring IgG antibodies against Plasmodium vivax antigens in a multiplexed bead-based assay using Luminex technology (Bio-Plex 200 or MAGPIX). PLoS ONE 2020, 15, e0238010. [Google Scholar] [CrossRef]
- Dobano, C.; Vidal, M.; Santano, R.; Jimenez, A.; Chi, J.; Barrios, D.; Ruiz-Olalla, G.; Rodrigo Melero, N.; Carolis, C.; Parras, D.; et al. Highly Sensitive and Specific Multiplex Antibody Assays To Quantify Immunoglobulins M, A, and G against SARS-CoV-2 Antigens. J. Clin. Microbiol. 2021, 59, e01731-20. [Google Scholar] [CrossRef]
- Filomena, A.; Pessler, F.; Akmatov, M.K.; Krause, G.; Duffy, D.; Gartner, B.; Gerhard, M.; Albert, M.L.; Joos, T.O.; Schneiderhan-Marra, N. Development of a Bead-Based Multiplex Assay for the Analysis of the Serological Response against the Six Pathogens HAV, HBV, HCV, CMV, T. gondii, and H. pylori. High-Throughput 2017, 6, 14. [Google Scholar] [CrossRef] [Green Version]
- Franca, C.T.; He, W.Q.; Gruszczyk, J.; Lim, N.T.; Lin, E.; Kiniboro, B.; Siba, P.M.; Tham, W.H.; Mueller, I. Plasmodium vivax Reticulocyte Binding Proteins Are Key Targets of Naturally Acquired Immunity in Young Papua New Guinean Children. PLoS Negl. Trop. Dis. 2016, 10, e0005014. [Google Scholar] [CrossRef] [Green Version]
- Franca, C.T.; Hostetler, J.B.; Sharma, S.; White, M.T.; Lin, E.; Kiniboro, B.; Waltmann, A.; Darcy, A.W.; Li Wai Suen, C.S.; Siba, P.; et al. An Antibody Screen of a Plasmodium vivax Antigen Library Identifies Novel Merozoite Proteins Associated with Clinical Protection. PLoS Negl. Trop. Dis. 2016, 10, e0004639. [Google Scholar] [CrossRef] [Green Version]
- Liechti, T.; Kadelka, C.; Ebner, H.; Friedrich, N.; Kouyos, R.D.; Gunthard, H.F.; Trkola, A. Development of a high-throughput bead based assay system to measure HIV-1 specific immune signatures in clinical samples. J. Immunol. Methods 2018, 454, 48–58. [Google Scholar] [CrossRef]
- Longley, R.J.; White, M.T.; Takashima, E.; Brewster, J.; Morita, M.; Harbers, M.; Obadia, T.; Robinson, L.J.; Matsuura, F.; Liu, Z.S.J.; et al. Development and validation of serological markers for detecting recent Plasmodium vivax infection. Nat. Med. 2020, 26, 741–749. [Google Scholar] [CrossRef] [PubMed]
- Lopez, E.; Haycroft, E.R.; Adair, A.; Mordant, F.L.; O’Neill, M.T.; Pymm, P.; Redmond, S.J.; Lee, W.S.; Gherardin, N.A.; Wheatley, A.K.; et al. Simultaneous evaluation of antibodies that inhibit SARS-CoV-2 variants via multiplex assay. JCI Insight 2021, 6, e150012. [Google Scholar] [CrossRef] [PubMed]
- Rosado, J.; Pelleau, S.; Cockram, C.; Merkling, S.H.; Nekkab, N.; Demeret, C.; Meola, A.; Kerneis, S.; Terrier, B.; Fafi-Kremer, S.; et al. Multiplex assays for the identification of serological signatures of SARS-CoV-2 infection: An antibody-based diagnostic and machine learning study. Lancet Microbe 2021, 2, e60–e69. [Google Scholar] [CrossRef]
- Selva, K.J.; van de Sandt, C.E.; Lemke, M.M.; Lee, C.Y.; Shoffner, S.K.; Chua, B.Y.; Davis, S.K.; Nguyen, T.H.O.; Rowntree, L.C.; Hensen, L.; et al. Systems serology detects functionally distinct coronavirus antibody features in children and elderly. Nat. Commun. 2021, 12, 2037. [Google Scholar] [CrossRef]
- Wu, L.; Hall, T.; Ssewanyana, I.; Oulton, T.; Patterson, C.; Vasileva, H.; Singh, S.; Affara, M.; Mwesigwa, J.; Correa, S.; et al. Optimisation and standardisation of a multiplex immunoassay of diverse Plasmodium falciparum antigens to assess changes in malaria transmission using sero-epidemiology. Wellcome Open Res. 2019, 4, 26. [Google Scholar] [CrossRef] [PubMed]
- Coppock, M.B.; Stratis-Cullum, D.N. A universal method for the functionalization of dyed magnetic microspheres with peptides. Methods 2019, 158, 12–16. [Google Scholar] [CrossRef]
- Reuter, G.; Gabius, H.J. Eukaryotic glycosylation: Whim of nature or multipurpose tool? Cell Mol. Life Sci. 1999, 55, 368–422. [Google Scholar] [CrossRef]
- Clausen, H.; Wandall, H.H.; Steentoft, C.; Stanley, P.; Schnaar, R.L. Glycosylation Engineering. In Essentials of Glycobiology, 3rd ed.; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Darvill, A.G., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2015; pp. 713–728. [Google Scholar]
- Caulley, L.; Corsten, M.; Eapen, L.; Whelan, J.; Angel, J.B.; Antonation, K.; Bastien, N.; Poliquin, G.; Johnson-Obaseki, S. Salivary Detection of COVID-19. Ann. Intern. Med. 2021, 174, 131–133. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.; Rashid, F.A.; Sabri, F.; Jamil, N.N.; Zain, R.; Hashim, R.; Amran, F.; Kok, H.T.; Samad, M.A.A.; Ahmad, N. Comparing Nasopharyngeal Swab and Early Morning Saliva for the Identification of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Clin. Infect. Dis. 2021, 72, e352–e356. [Google Scholar] [CrossRef]
- Senok, A.; Alsuwaidi, H.; Atrah, Y.; Al Ayedi, O.; Al Zahid, J.; Han, A.; Al Marzooqi, A.; Al Heialy, S.; Altrabulsi, B.; AbdelWareth, L.; et al. Saliva as an Alternative Specimen for Molecular COVID-19 Testing in Community Settings and Population-Based Screening. Infect. Drug Resist. 2020, 13, 3393–3399. [Google Scholar] [CrossRef]

| Pathogen | Antigen Region | Expression System | Supplier | Catalogue Number | Description |
|---|---|---|---|---|---|
| SARS-CoV-2 | S1 | HEK293 | The Native Antigen Company, Oxford, UK | REC31806 | SARS-CoV-2-Spike Glycoprotein (S1), recombinant antigen AA 1–674 |
| SARS-CoV-2 | S2 | HEK293 | The Native Antigen Company, Oxford, UK | REC31807 | SARS-CoV-2-Spike Glycoprotein (S2) recombinant antigen AA 685-1211 |
| SARS-CoV-2 | NP | E. coli | ProSpec Protein Specialists, Rehovot, Israel | sars-013 | SARS-CoV-2 Nucleocapsid protein, full length AA 1-419 |
| SARS-CoV-2 | NP | E. coli | MP Biomedicals, Santa Ana, CA, USA | 8720501 | SARS-CoV-2 Nucleocapsid protein, full length AA 1-431 |
| SARS-CoV-2 | NP | HEK293 | AcroBiosystems, Newark, DE, USA | NUN-C5227 | SARS-CoV-2 Nucleocapsid protein, full length AA 1-419 |
| SARS-CoV-2 | S-RBD | HEK293 | Wai-Hong Tham (WEHI) *2 | - | SARS-CoV-2-Receptor Binding Domain (RBD) of S1 |
| SARS-CoV-2 | Spike (Trimer) | HEK293 | Adam Wheatley (PDI) *3 | - | SARS-CoV-2 Full length Spike trimer |
| SARS-CoV-2 | Plpro *1 | E. coli | David Komander (WEHI) *2 | - | SARS-CoV-2-Papain-like protease |
| SARS-CoV-2 | Main protease | E. coli | Peter Czabotar (WEHI) *2 | - | SARS-CoV-2-Main protease |
| SARS | S1 | HEK293 | The Native Antigen Company, Oxford, UK | REC31809 | SARS-CoV-1 S1 subunit |
| SARS | S1-RBD | HEK | Wai-Hong Tham (WEHI) *2 | - | |
| MERS | S1 | E. coli | ProSpec Protein Specialists, Rehovot, Israel | sars-002 | Recombinant S1 subunit AA 56-295 |
| Seasonal Coronaviruses | |||||
| HCoV-229E | NP | E. coli | The Native Antigen Company, Oxford, UK | REC31758 | Recombinant NP, 359 AA |
| HCoV-NL63 | NP | E. coli | ProSpec Protein Specialists, Rehovot, Israel | sars-003 | Recombinant c-terminal NP, contains 130 AA |
| HCoV-NL63 | S1 | HEK293 | Sino Biological, Beijing, China | 40600-V08H | Recombinant S1 subunit, AA 13–756 |
| HCoV-229E | S1 | HEK293 | Sino Biological, Beijing, China | 40601-V08H | Recombinant S1 subunit, AA 19-717 |
| HCoV-HKU1 | S1 | HEK293 | Sino Biological, Beijing, China | 40602-V08H | Recombinant S1 subunit, AA 13-756 |
| HCoV-HKU1 | Spike | HEK293 | Adam Wheatley (PDI) *3 | - | |
| HCoV-OC43 | S1 + S2 + ECD | Bac-Ins cells | Sino Biological, Beijing, China | 40607-V08B | Recombinant Spike protein (S1 + S2 + ECD), AA 1-1304 |
| Influenza Viruses | |||||
| IVA Michigan H1N1-NA-45/2015 | Neuraminidase | HEK293 | Sino Biological, Beijing, China | 40568-V07H | Influenza A Neuraminidase, AA 36-469 |
| IVA Switzerland H3N2-2013 | Hemagglutinin | HEK293 | Sino Biological, Beijing, China | 40497-V08H1 | Influenza A HA1, AA 1-345 |
| IVB Victoria 02/1987 | Hemagglutinin | HEK293 | Sino Biological, Beijing, China | 40163-V08H | Influenza B HA1, AA 1-362 |
| IVB Phuket 3073/2013 | Hemagglutinin | HEK293 | Sino Biological, Beijing, China | 40498-V08H1 | Influenza B HA1, AA 1-361 |
| IVA California H1N1-HA | Hemagglutinin | HEK293 | Adam Wheatley (PDI) *3 | - | Influenza A HA1 |
| Tetanus Toxoid | Sigma-Aldrich, St. Louis, MO, USA | 582231-25UG | Clostridium tetani (formaldehyde inactivation) |
| Pathogen | Antigen Region | Protein Range Tested (µg) | Optimized Trial Protein Concentration (µg Ag/6.25 × 105 Beads) | Protein Bulk Coupling (µg Ag/2.5 × 106 Beads) | Saturated Protein Concentration-Bulk Coupling (µg Ag/2.5 × 106 Beads) |
|---|---|---|---|---|---|
| SARS-CoV-2 | S1 | 0.1–10 | 5 | 10 | 40 |
| SARS-CoV-2 | S2 | 0.1–10 | 5 | 10 | 40 |
| SARS-CoV-2 | NP | 0.1–5 | 1 | 2 | |
| SARS-CoV-2 | NP | 0.1–5 | |||
| SARS-CoV-2 | NP | 0.1–5 | 0.5 | 1 | 10 |
| SARS-CoV-2 | S-RBD | 0.2–5 | 2.5 | 5 | 20 |
| SARS-CoV-2 | Spike (Trimer) | 0.1–5 | 2.5 | 5 | 20 |
| SARS-CoV-2 | Plpro *1 | 1–5 | - | ||
| SARS-CoV-2 | Main protease | 1–5 | - | ||
| SARS | S1 | 0.5–5 | - | ||
| SARS | S1-RBD | 0.5–5 | - | ||
| MERS | S1 | 0.5–5 | - | ||
| Seasonal Coronaviruses | |||||
| HCoV-229E | NP | 0.2–5 | - | ||
| HCoV-NL63 | NP | 0.5–5 | 5 | 10 | 40 |
| HCoV-NL63 | S1 | 1–5 | - | ||
| HCoV-229E | S1 | 0.2–5 | 1 | 2 | 12 |
| HCoV-HKU1 | S1 | 0.5–5 | - | ||
| HCoV-HKU1 | Spike | 0.2–5 | 1 | 2 | 12 |
| HCoV-OC43 | S1 + S2 + ECD | 0.1–5 | 0.25 | 0.5 | 4 |
| Influenza Viruses | |||||
| IVA Michigan H1N1-NA-45/2015 | Neuraminidase | 0.5–5 | - | ||
| IVA Switzerland H3N2-2013 | Hemagglutinin | 0.2–5 | 1 | 2 | |
| IVB Victoria 02/1987 | Hemagglutinin | 0.2–5 | 1 | 2 | |
| IVB Phuket 3073/2013 | Hemagglutinin | 0.2–5 | 1 | 2 | |
| IVA California H1N1-HA | Hemagglutinin | 0.2–5 | 0.4 | 0.8 | 6 |
| Tetanus Toxoid | 0.5–5 | 1 | 2 | 8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mazhari, R.; Ruybal-Pesántez, S.; Angrisano, F.; Kiernan-Walker, N.; Hyslop, S.; Longley, R.J.; Bourke, C.; Chen, C.; Williamson, D.A.; Robinson, L.J.; et al. SARS-CoV-2 Multi-Antigen Serology Assay. Methods Protoc. 2021, 4, 72. https://doi.org/10.3390/mps4040072
Mazhari R, Ruybal-Pesántez S, Angrisano F, Kiernan-Walker N, Hyslop S, Longley RJ, Bourke C, Chen C, Williamson DA, Robinson LJ, et al. SARS-CoV-2 Multi-Antigen Serology Assay. Methods and Protocols. 2021; 4(4):72. https://doi.org/10.3390/mps4040072
Chicago/Turabian StyleMazhari, Ramin, Shazia Ruybal-Pesántez, Fiona Angrisano, Nicholas Kiernan-Walker, Stephanie Hyslop, Rhea J. Longley, Caitlin Bourke, Catherine Chen, Deborah A. Williamson, Leanne J. Robinson, and et al. 2021. "SARS-CoV-2 Multi-Antigen Serology Assay" Methods and Protocols 4, no. 4: 72. https://doi.org/10.3390/mps4040072