Inexpensive, Accurate, and Stable Method to Quantitate Blood Alanine Aminotransferase (ALT) Levels
Abstract
:1. Introduction
2. Experimental Design
2.1. Materials
- Tris Base (#BP152-1, Fisher Scientific, Waltham, MA, USA);
- Alpha-ketoglutarate (#75890-25G, Sigma Aldrich, St. Louis, MO, USA);
- L-alanine (#A7627-100G, Sigma Aldrich, St. Louis, MO, USA);
- Ethylenediaminetetraacetic acid (EDTA) (#E177-500ML, VWR, Radnor, PA, USA);
- LDH (#350-20-1, Lee Bio, Maryland Heights, MO, USA);
- NADH (#16078, Cayman Chemical, Ann Arbor, MI, USA);
- Phosphate-buffered saline (PBS) (#BP66150, Fisher Scientific, Waltham, MA, USA);
- Validate GC3 ALT Linearity Test Kit (#1300sd, LGC Maine Standards/LGC Clinical Diagnostics, Inc., Cumberland Foreside, ME, USA).
2.2. Equipment
- 96-well flat-bottom plates (#07-200-656, Corning, Corning, NY, USA);
- 15 mL or 50 mL Falcon tube (#1495949B or #1495949A, Fisher Scientific, Waltham, MA, USA);
- Whatman Puradisc 25 mm 0.2 µM polyethersulfone (PES) membrane filter (#6780-2502, Cytiva, Marlborough, MA, USA);
- Disposable reagent reservoirs (#89108-008, Axygen Scientific, Union City, CA, USA);
- VersaMaxTM Absorbance Microplate Reader (#VERSAMAX, Molecular Devices, San Jose, CA, USA);
- SoftMax Pro GxP Software version 5.4 (#SMP54-GXP-10UL, Molecular Devices, San Jose, CA, USA).
2.3. Methods for ALT Reagent Formulations, Mouse Studies, Triglyceride Experiments and Statistics
2.3.1. Various ALT Reagent Formulations
2.3.2. Blood Samples
2.3.3. Triglyceride Experiments
2.3.4. Statistics
3. Procedure
3.1. Preparation of Plasma Sample
- Obtain blood from rodents, humans or other organisms. We recommend adding 5 µL of EDTA per 500 µL blood to prevent coagulation.
- Spin blood at 10,000× g for 15 min at 4 degrees Celsius.
- Transfer clear supernatant (=plasma) to a new sterile Eppendorf tube and store at −80 degrees Celsius or use immediately for the ALT assay.
3.2. Preparation of ALT Reagent
- Preparation of 100 mL ALT reagent–with 97 millimolar (mM) Tris Base (stored at room temperature), 13 mM alpha-ketoglutarate (stored at 4 degrees Celsius), 440 mM L-alanine (stored at room temperature), 5 mM EDTA (stored at room temperature), 3200 units/L LDH (stored at −20 degrees Celsius), and 1.5 mM NADH (stored at −20 degrees Celsius):
- Add 1.175 g Tris Base to a glass beaker.
- Fill up to ~50 mL with sterile water.
- Start stirring with a magnet stir bar in the beaker.
- Add 190.0 mg alpha-ketoglutarate.
- Add 3.92 g L-alanine.
- Add 1 mL of 0.5 M solution of the preservative EDTA.
- Dissolve 1000 units of LDH in a vial with 1 mL PBS, and add 320 µL of LDH solution (=320 units) to ALT reagent (store remaining LDH in vial at −20 degrees Celsius).
- Continue stirring with the magnet stir bar, until all particles are dissolved in the ALT reagent solution.
- Adjust pH to 7.80 at room temperature.
- Fill up to final volume of 100 mL with sterile water, transfer this prepared ALT reagent to a sterile bottle, and
PAUSE STEP store at 4 degrees Celsius protected from a light source (e.g., wrap bottle with aluminum foil) until use.
CRITICAL STEP Prior to use, obtain the required volume and add the same amount of NADH in mg as the required volume in mL (e.g., add 10 mg NADH to 10 mL prepared ALT reagent) and vortex for 15–30 s (NADH is stable in the ALT reagent only for a short period of time, i.e., approximately a couple of weeks, hence the recommendation of adding NADH to the ALT reagent right before use). If no ALT values above 250 U/L are expected, one can consider adding a lower amount of NADH, i.e., 5 mg NADH to 10 mL prepared ALT reagent.
CRITICAL STEP Filter the required volume of ALT reagent using Whatman Puradisc 25 mm 0.2 µM PES membrane filter to remove possible remaining small particles from the solution.
CRITICAL STEP Warm up the required filtered volume in a 15 mL or 50 mL Falcon tube for 5 min in a water bath maintained at 37 degrees Celsius immediately prior to pipetting to induce the kinetic reaction required for the assay to work appropriately.
3.3. Preparation of 96-Well Plate and ALT Measurement
- 15.
- Add 10 µL of ALT standards ranging from 0 (=water) to 1000 units/L (Validate GC3 ALT Linearity Test Kit) and plasma samples in duplicates or triplicates into 96-well flat-bottom plates.
- 16.
- Add 90 µL of filtered, warmed up ALT reagent to each sample using a multi-pipet and a disposable reagent reservoir. It is good practice to prepare 1 mL in excess of the calculated required volume to ensure rapid pipetting with the multi-pipet. Shake plate slightly after completion of pipetting. Remove (large) bubbles by bursting them with a small needle.
- 17.
- Read plate without lid in VersaMax Microplate Reader with SoftMax Pro Software, or similar microplate reader and software.
CRITICAL STEP Use the kinetic enzyme assay function with 24 reads at 340 nm every 13 s for a total of ~5 min. The wavelength of 340 nm is chosen to quantitate NADH, as the reduced form (NADH) can absorb light at 340 nm, whereas the oxidized from (NAD+) does not [17]. The more enzyme ALT is present, the more pyruvate is produced, which then in the subsequent reaction is metabolized with NADH to L-lactate and NAD+ in a reaction catalyzed by LDH. The decrease of NADH over time is detected by the kinetic assay at 340 nm and correlates with the activity of ALT [18]. The maximum reaction velocity Vmax is displayed in milliunits/min.
4. Expected Results and Discussion
4.1. The Stability of the Standard Curve Depends on the NADH Concentration in the ALT Reagents
4.2. The ALT Reagent with a High NADH Content Shows Similarly Excellent Performance as a Commercial ALT and Longer Stability of the Results
4.3. Anticoagulation with EDTA or Heparin Results in Lower ALT Levels Compared to No Anticoagulation
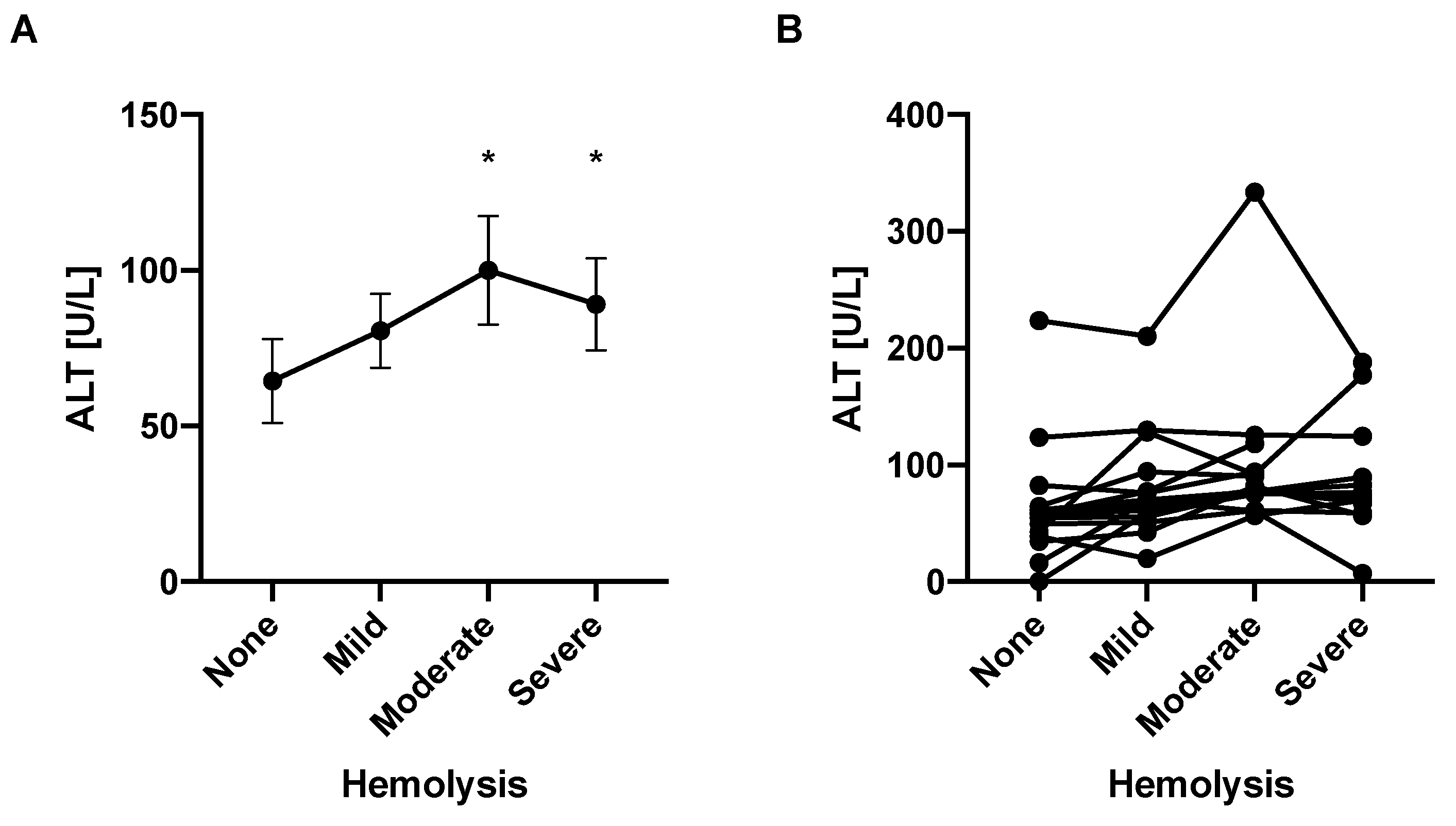
4.4. Hemolysis Is Associated with Elevated ALT Levels
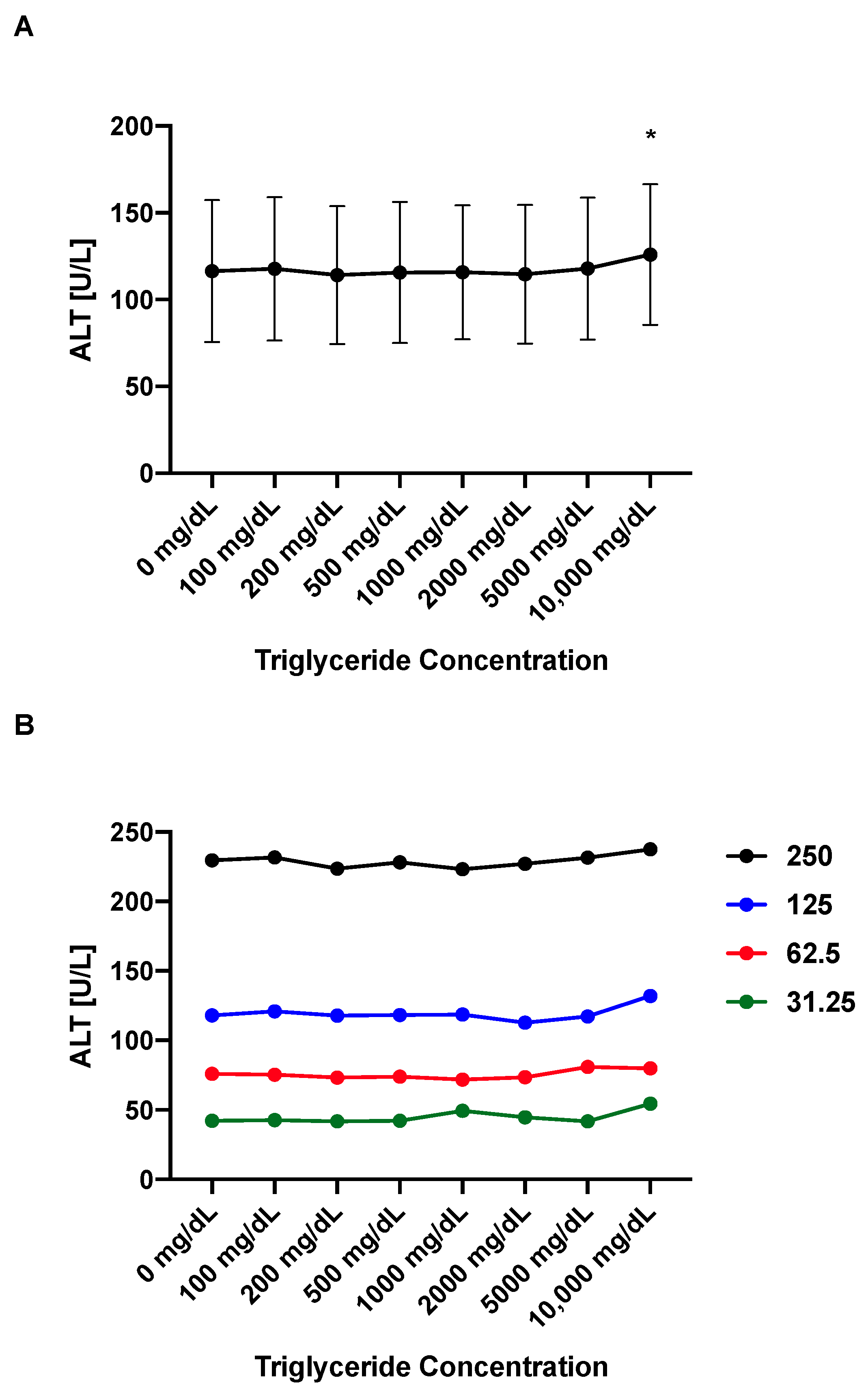
4.5. ALT Values Remain Stable over a Broad Range of Triglyceride Concentrations
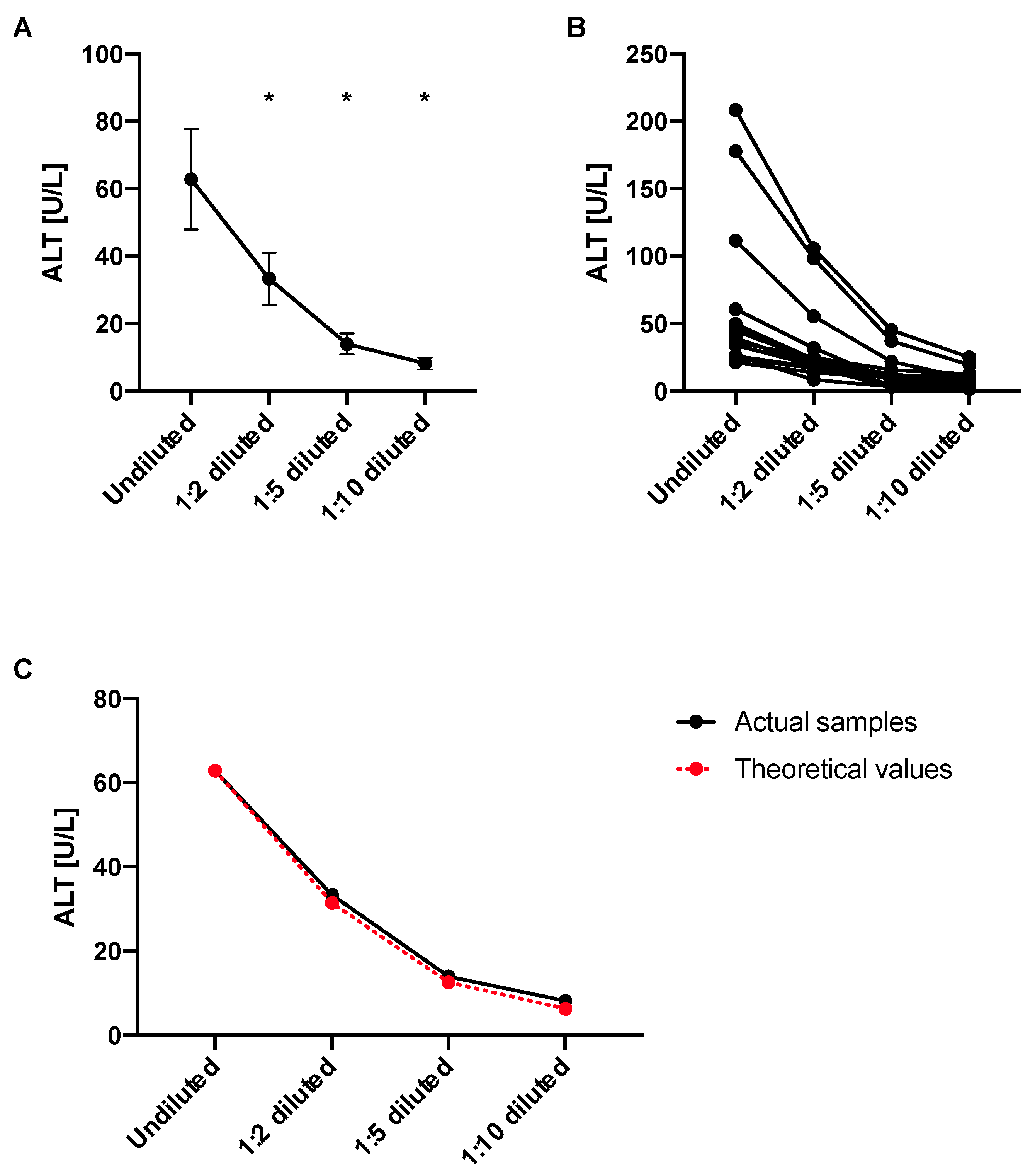
4.6. ALT Levels Decrease in Diluted Samples Proportionally to the Dilution Factor
4.7. The Presented ALT Assay Is Less Expensive Than Commercial ALT Kits
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hartmann, P.; Lang, S.; Zeng, S.; Duan, Y.; Zhang, X.; Wang, Y.; Bondareva, M.; Kruglov, A.; Fouts, D.E.; Stärkel, P.; et al. Dynamic Changes of the Fungal Microbiome in Alcohol Use Disorder. Front. Physiol. 2021, 12, 699253. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, P.; Chen, P.; Wang, H.J.; Wang, L.; McCole, D.F.; Brandl, K.; Stärkel, P.; Belzer, C.; Hellerbrand, C.; Tsukamoto, H.; et al. Deficiency of intestinal mucin-2 ameliorates experimental alcoholic liver disease in mice. Hepatology 2013, 58, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, P.; Hochrath, K.; Horvath, A.; Chen, P.; Seebauer, C.T.; Llorente, C.; Wang, L.; Alnouti, Y.; Fouts, D.E.; Stärkel, P.; et al. Modulation of the intestinal bile acid/farnesoid X receptor/fibroblast growth factor 15 axis improves alcoholic liver disease in mice. Hepatology 2018, 67, 2150–2166. [Google Scholar] [CrossRef]
- Brandl, K.; Hartmann, P.; Jih, L.J.; Pizzo, D.P.; Argemi, J.; Ventura-Cots, M.; Coulter, S.; Liddle, C.; Ling, L.; Rossi, S.J.; et al. Dysregulation of serum bile acids and FGF19 in alcoholic hepatitis. J. Hepatol. 2018, 69, 396–405. [Google Scholar] [CrossRef]
- Demir, M.; Lang, S.; Hartmann, P.; Duan, Y.; Martin, A.; Miyamoto, Y.; Bondareva, M.; Zhang, X.; Wang, Y.; Kasper, P.; et al. The fecal mycobiome in non-alcoholic fatty liver disease. J. Hepatol. 2022, 76, 788–799. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, P.; Duan, Y.; Miyamoto, Y.; Demir, M.; Lang, S.; Hasa, E.; Stern, P.; Yamashita, D.; Conrad, M.; Eckmann, L.; et al. Colesevelam ameliorates non-alcoholic steatohepatitis and obesity in mice. Hepatol. Int. 2022, 16, 359–370. [Google Scholar] [CrossRef]
- Wang, L.; Hartmann, P.; Haimerl, M.; Bathena, S.P.; Sjöwall, C.; Almer, S.; Alnouti, Y.; Hofmann, A.F.; Schnabl, B. Nod2 deficiency protects mice from cholestatic liver disease by increasing renal excretion of bile acids. J. Hepatol. 2014, 60, 1259–1267. [Google Scholar] [CrossRef]
- Ginès, P.; Castera, L.; Lammert, F.; Graupera, I.; Serra-Burriel, M.; Allen, A.M.; Wong, V.W.; Hartmann, P.; Thiele, M.; Caballeria, L.; et al. Population screening for liver fibrosis: Toward early diagnosis and intervention for chronic liver diseases. Hepatology 2022, 75, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.A.; Kim, S.; Park, M.; Park, H.J.; Kim, J.H.; Park, S.; Hwang, J.R.; Kim, Y.C.; Jun Kim, Y.; Cho, Y.; et al. Ciclopirox inhibits Hepatitis B Virus secretion by blocking capsid assembly. Nat. Commun. 2019, 10, 2184. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Tan, H.Y.; Li, S.; Wang, N.; Feng, Y. Glutamic-Pyruvic Transaminase 1 Facilitates Alternative Fuels for Hepatocellular Carcinoma Growth-A Small Molecule Inhibitor, Berberine. Cancers 2020, 12, 1854. [Google Scholar] [CrossRef] [PubMed]
- WROBLEWSKI, F.; LADUE, J.S. Serum glutamic pyruvic transaminase in cardiac with hepatic disease. Proc. Soc. Exp. Biol. Med. 1956, 91, 569–571. [Google Scholar] [CrossRef] [PubMed]
- Hsueh, C.J.; Wang, J.H.; Dai, L.; Liu, C.C. Determination of alanine aminotransferase with an electrochemical nano ir-C biosensor for the screening of liver diseases. Biosensors 2011, 1, 107–117. [Google Scholar] [CrossRef]
- Hwang, I.; Lee, E.J.; Park, H.; Moon, D.; Kim, H.S. Retinol from hepatic stellate cells via STRA6 induces lipogenesis on hepatocytes during fibrosis. Cell Biosci. 2021, 11, 3. [Google Scholar] [CrossRef]
- Pandurangan, M.; Kim, D.H. ZnO nanoparticles augment ALT, AST, ALP and LDH expressions in C2C12 cells. Saudi J. Biol. Sci. 2015, 22, 679–684. [Google Scholar] [CrossRef]
- Beydoun, M.A.; Beydoun, H.A.; MacIver, P.H.; Hossain, S.; Canas, J.A.; Evans, M.K.; Zonderman, A.B. Biochemical and Hematological Correlates of Elevated Homocysteine in National Surveys and a Longitudinal Study of Urban Adults. Nutrients 2020, 12, 950. [Google Scholar] [CrossRef]
- Per 2001–2002 National Health and Nutrition Examination Survey (NHANES) Data Documentation, Codebook, and Frequencies—Standard Biochemistry Profile (L40_B). Available online: https://wwwn.cdc.gov/nchs/nhanes/2001-2002/L40_B.htm (accessed on 4 October 2022).
- Mayevsky, A.; Rogatsky, G.G. Mitochondrial function in vivo evaluated by NADH fluorescence: From animal models to human studies. Am. J. Physiol. Cell Physiol. 2007, 292, C615–C640. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.F.; Taylor, R.H. Comparison of two-point (AutoAnalyzer II) with kinetic methods for transaminase assay. J. Clin. Pathol. 1973, 26, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Tholen, D.; Kallner, A.; Kennedy, J.; Krouwer, J.; Meier, K. Evaluation of Precision Performance of Quantitative Measurement Methods; Approved Guideline; CLSI EP5-A2; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2004. [Google Scholar]
- Mohri, M.; Allhyari, L.; Sardari, K. Effects of Common Anticoagulants on Routine Plasma Biochemistry of Horse and Comparison with Serum. J. Equine Vet. Sci. 2007, 27, 313–316. [Google Scholar] [CrossRef]
- Mohri, M.; Rezapoor, H. Effects of heparin, citrate, and EDTA on plasma biochemistry of sheep: Comparison with serum. Res. Vet. Sci. 2009, 86, 111–114. [Google Scholar] [CrossRef]
- Koseoglu, M.; Hur, A.; Atay, A.; Cuhadar, S. Effects of hemolysis interferences on routine biochemistry parameters. Biochem. Med. 2011, 21, 79–85. [Google Scholar] [CrossRef]
- Brady, J.; O’Leary, N. Interference due to haemolysis in routine photometric analysis—A survey. Ann. Clin. Biochem. 1998, 35 Pt 1, 128–134. [Google Scholar] [CrossRef]
- Garay-García, K.; Gaete, P.V.; Mendivil, C.O. Severe hypertriglyceridemia secondary to splice-site and missense variants in LMF1 in three patients from Ecuador. J. Clin. Lipidol. 2022, 16, 277–280. [Google Scholar] [CrossRef] [PubMed]
- Please see Package Insert of Commercial ALT Kit Used in Figure 2, ALT Kinetic Assay, #A524-150, Teco Diagnostics. Available online: https://www.tecodiagnostics.com/_files/ugd/be31e8_bfe352a786304fb6af5731de685bf2d4.pdf (accessed on 4 October 2022).



| 0.12 mM NADH | 0.75 mM NADH | 1.5 mM NADH | |
|---|---|---|---|
| First Run | |||
| 0 to 1000 | −0.219 | 0.996 | 0.998 |
| 7.8125 to 1000 | −0.489 | 0.997 | 0.998 |
| 0 to 500 | −0.138 | 0.987 | 0.992 |
| Second Run | |||
| 0 to 1000 | −0.427 | 0.274 | 0.998 |
| 7.8125 to 1000 | −0.405 | 0.188 | 0.998 |
| 0 to 500 | −0.438 | 1.000 | 0.999 |
| Third Run | |||
| 0 to 1000 | −0.112 | −0.054 | 0.997 |
| 7.8125 to 1000 | −0.024 | −0.166 | 0.997 |
| 0 to 500 | −0.148 | 0.253 | 0.989 |
| 1.5 mM NADH | Commercial Kit | |
|---|---|---|
| First Run | ||
| 0 to 1000 | 0.999 | 0.999 |
| 7.8125 to 1000 | 1.000 | 1.000 |
| 0 to 500 | 0.996 | 0.998 |
| Second Run | ||
| 0 to 1000 | 0.993 | 0.276 |
| 7.8125 to 1000 | 0.999 | 0.185 |
| 0 to 500 | 0.972 | 0.979 |
| Third Run | ||
| 0 to 1000 | 0.998 | −0.085 |
| 7.8125 to 1000 | 0.999 | −0.212 |
| 0 to 500 | 0.996 | 0.240 |
| 1 Month | 2 Months | 5 Months | |
|---|---|---|---|
| First Run | |||
| 0 to 1000 | 0.993 | 0.980 | 0.990 |
| 7.8125 to 1000 | 0.998 | 0.997 | 0.991 |
| 0 to 500 | 0.970 | 0.918 | 0.969 |
| Second Run | |||
| 0 to 1000 | 0.999 | 0.998 | 0.999 |
| 7.8125 to 1000 | 1.000 | 0.998 | 1.000 |
| 0 to 500 | 0.997 | 0.991 | 0.996 |
| Third Run | |||
| 0 to 1000 | 0.999 | 0.997 | 0.997 |
| 7.8125 to 1000 | 1.000 | 0.999 | 0.997 |
| 0 to 500 | 0.997 | 0.992 | 0.989 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hartmann, P.; Schnabl, B. Inexpensive, Accurate, and Stable Method to Quantitate Blood Alanine Aminotransferase (ALT) Levels. Methods Protoc. 2022, 5, 81. https://doi.org/10.3390/mps5050081
Hartmann P, Schnabl B. Inexpensive, Accurate, and Stable Method to Quantitate Blood Alanine Aminotransferase (ALT) Levels. Methods and Protocols. 2022; 5(5):81. https://doi.org/10.3390/mps5050081
Chicago/Turabian StyleHartmann, Phillipp, and Bernd Schnabl. 2022. "Inexpensive, Accurate, and Stable Method to Quantitate Blood Alanine Aminotransferase (ALT) Levels" Methods and Protocols 5, no. 5: 81. https://doi.org/10.3390/mps5050081







