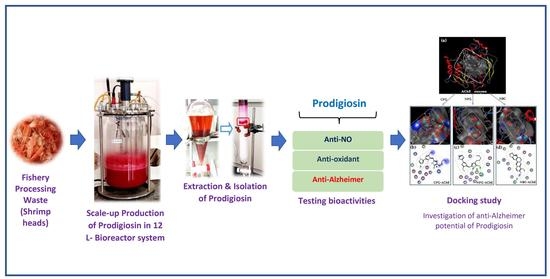
Bioproduction of Prodigiosin from Fishery Processing Waste Shrimp Heads and Evaluation of Its Potential Bioactivities
Abstract
:1. Introduction
2. Results and Discussion
2.1. Establishment of Production Process of PG in Small Erlenmeyer Flask Scale
2.1.1. Biosynthesis of PG by Different Bacterial Strains of Serratia marcescens
2.1.2. Effect of Free Protein and Salt Compositions Added into Culture Medium on PG Production by S. marcescens CC17
2.1.3. The Effect of Fermentation Parameters on PG Productivity by CC17 Strain
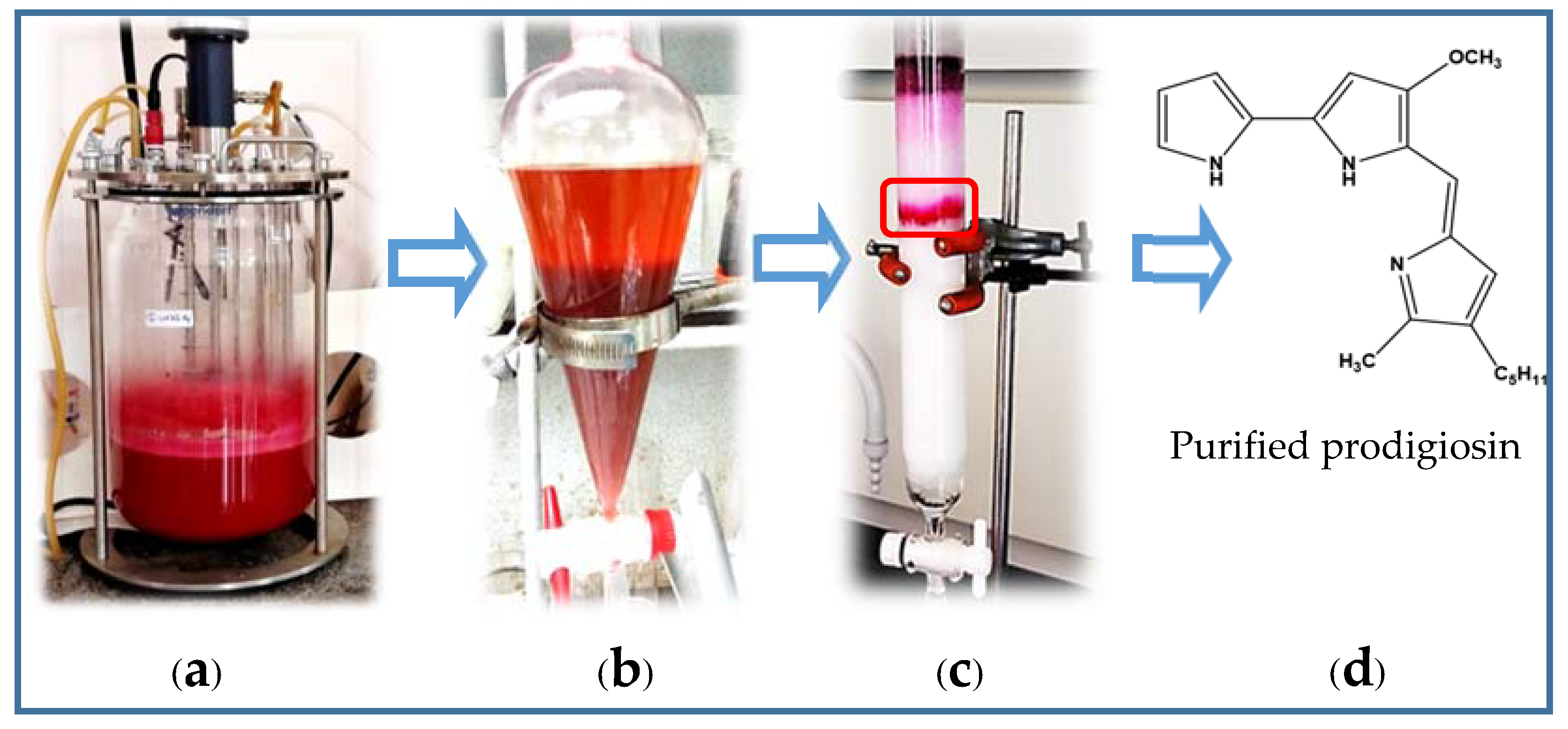
2.2. Scale-Up of PG Production to a 12 L-Bioreactor System
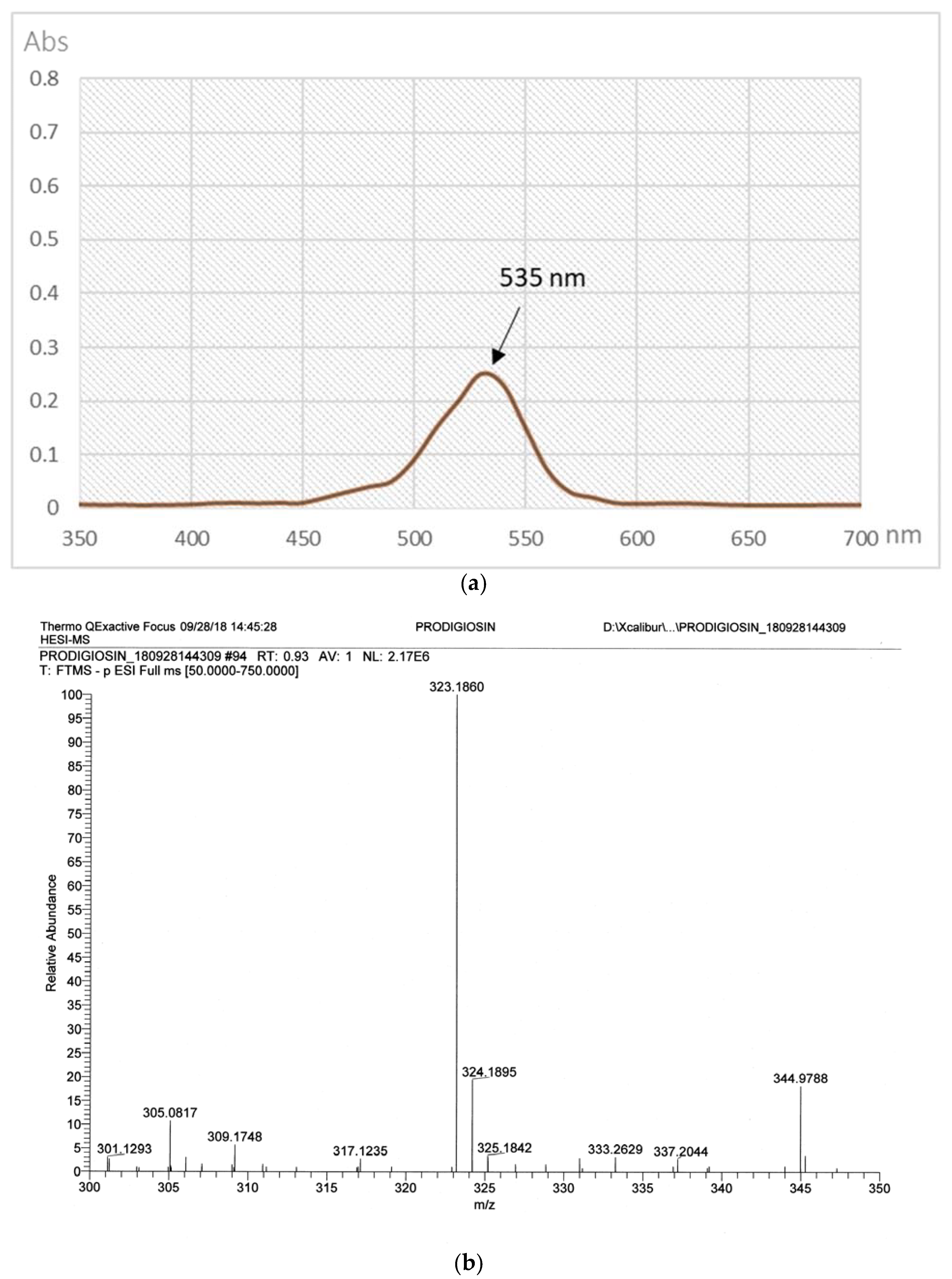
2.3. The Purification of PG in Culture Broth
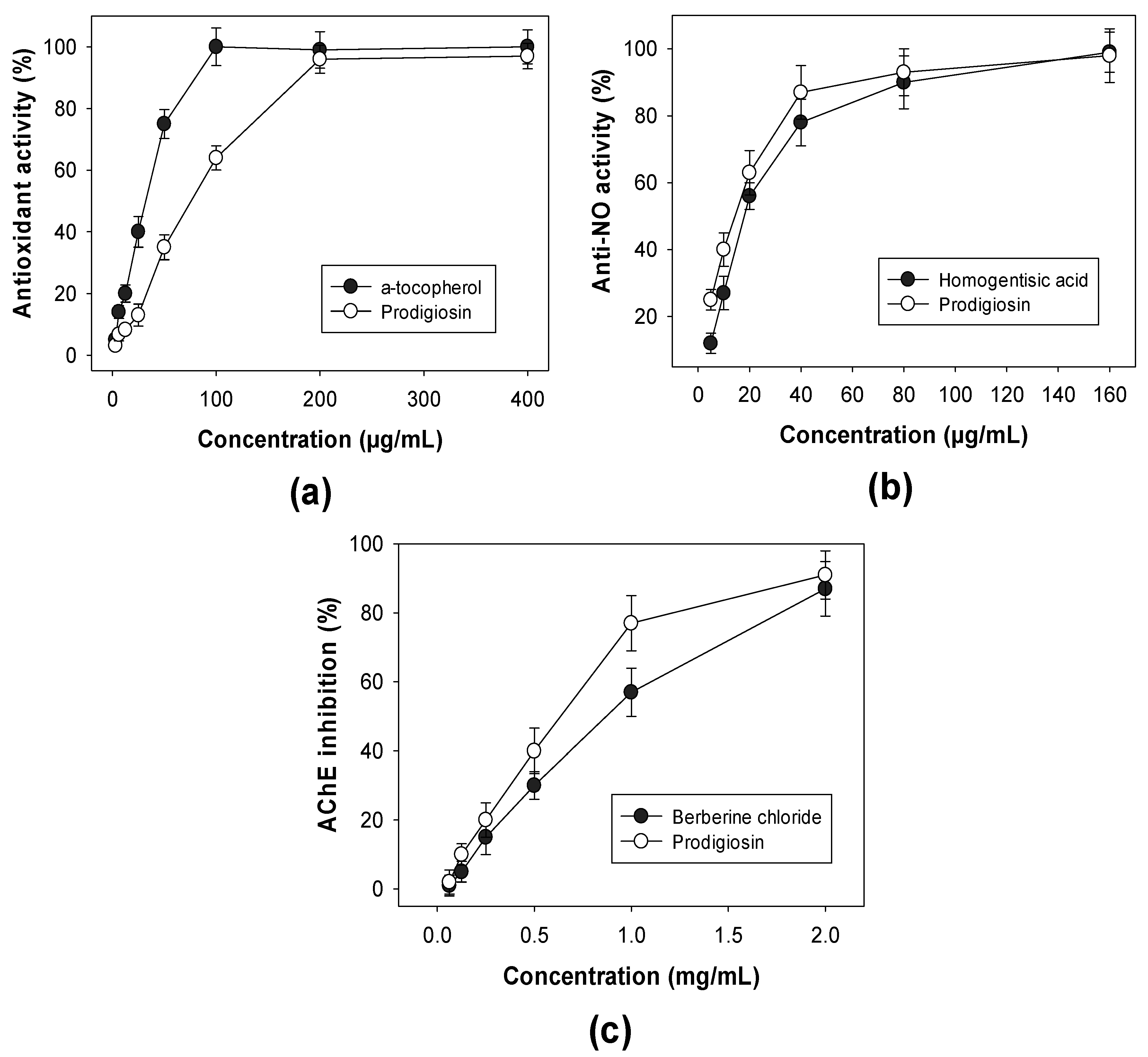
2.4. Detection of Biological Activities of Purified PG
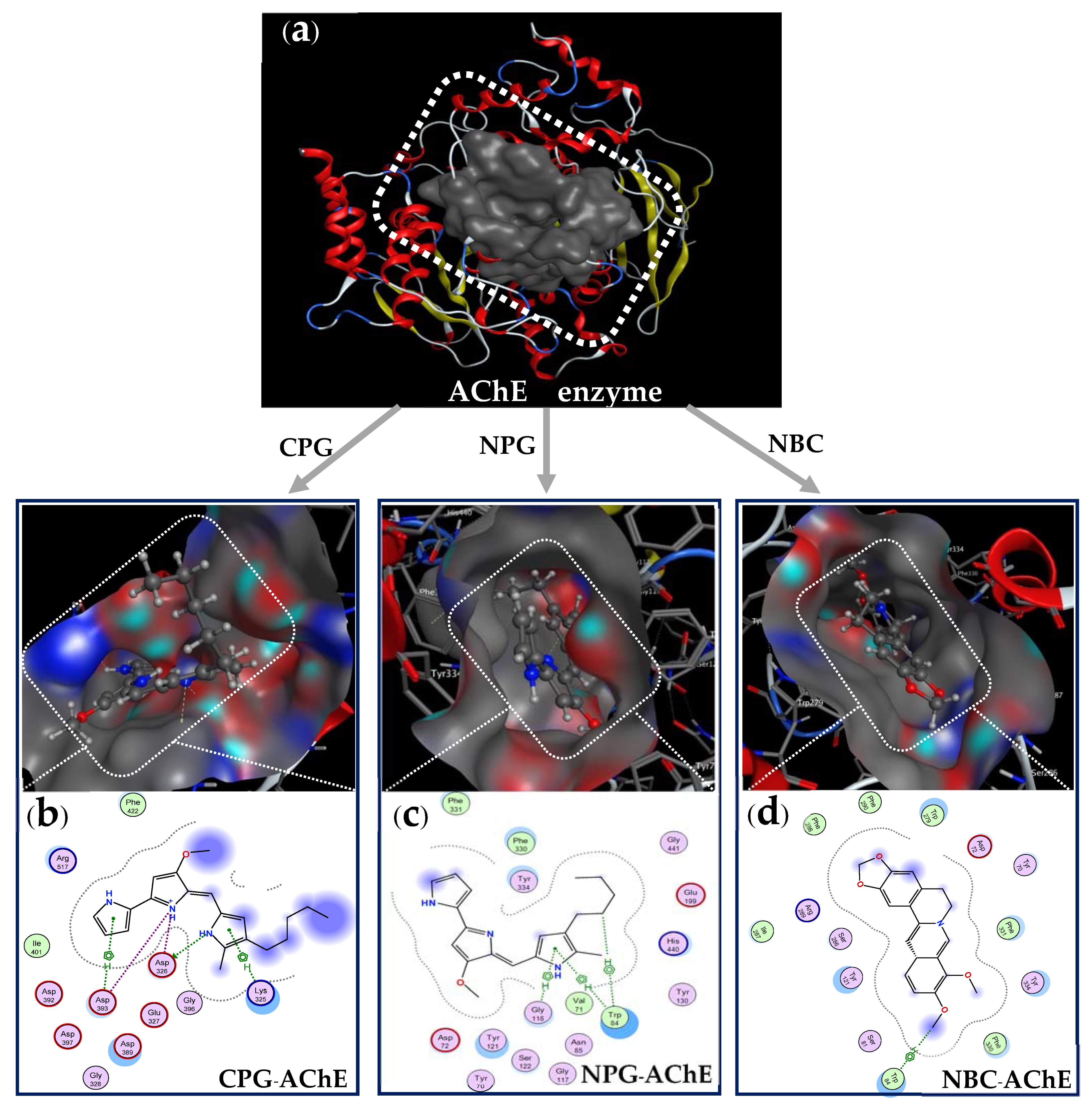
2.5. Docking Study
3. Materials and Methods
3.1. Materials
3.2. Methods
3.2.1. Experiments of Fermentation for PG Biosynthesis in Flask and 12 L-Bioreactor
Experiments of PG Biosynthesized by Different PG Producing Strains
The Effect of Added Free Protein in Culture Medium on PG Yield
The Effect of Phosphate Salts on PG Production
The Effects of Sulfate Salts on PG Production
The Effects of Fermentation Parameters on PG Productivity Produced by CC17 Strain
The Experiments to Scale-Up PG Biosynthesis to 12 L-Bioreactor System
3.2.2. Experiments of Qualification and Purification of PG
3.2.3. High-Performance Liquid Chromatography (HPLC) Analysis of PG
3.2.4. Biological Activity Assays
3.2.5. Docking Study Protocol
- Preparation of AChE enzyme before docking performance: The structure data of AChE (DOI:10.2210/pdb1GQR/pdb) was obtained from Worldwide Protein Data Bank. The protein and its 3D protonation were prepared by using the functionality of MOE QuickPrep (Figure 7d) based on the positions of ligand within 4.5 Å and the presence of important amino acids. All of the water molecules were removed before the recreation of enzymic action zones. The active zone (binding site, Figure 8a) of the ligands on the target protein was determined using the site finder function in MOE.
- Preparation of ligands(inhibitor compounds) before docking performance: The structures of the inhibitor compounds (ligands, Figure 7a,b) were prepared using ChemBioOffice 2018 software. The optimized structures of the ligands prepared using the MOE system with optimization parameters of Force field MMFF94x; R-Field 1: 80; cutoff, Rigid water molecules, space group p1, cell size 10, 10, 10; cell shape 90, 90, 90; and gradient 0.01 RMS kcal mol−1 A−2 [56,69].
- Docking performance between ligands and protein enzyme: The docking simulation calculations were performed on the ligands of cation-PG, neutral-PG, and neutral-BC and towards the target enzyme protein (AChE) using MOE-2015.10 software. The out-put included amino acid compositions in the binding site on AChE, docking score (DS), Root Mean Square Deviation (RMSD), interaction types (bonds), amino acids interacted with by the ligands as well as the distances of the linkages.
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Caruso, G.; Floris, R.; Serangeli, C.; Di Paola, L. Fishery wastes as a yet undiscovered treasure from the sea: Biomolecules sources, extraction methods and valorization. Mar. Drugs 2020, 18, 622. [Google Scholar] [CrossRef]
- Wang, C.H.; Doan, C.T.; Nguyen, V.B.; Nguyen, A.D.; Wang, S.L. Reclamation of fishery processing waste: A mini-review. Molecules 2019, 24, 2234. [Google Scholar] [CrossRef] [Green Version]
- Pradit, S.; Noppradit, P.; Goh, B.P.; Sornplang, K.; Ong, M.C.; Towatana, P. Occurrence of microplastics and trace metals in fish and shrimp from Songkhla Lake, Thailand during the Covid-19 pandemic. Appl. Ecol. Environ. Res. 2021, 19, 1085–1106. [Google Scholar] [CrossRef]
- Hue, H.T.T.; Pradit, S.; Lim, A.; Goncalo, C.; Nitiratsuwan, T. Shrimp and fish catch landing trends in Songkhla Lagoon, Thailand during 2003–2016. Appl. Ecol. Environ. Res. 2018, 16, 3061–3078. [Google Scholar] [CrossRef]
- Venugopal, V. Valorization of seafood processing discards: Bioconversion and biorefinery approaches. Front. Sustain. Food Syst. 2021, 5, 611835. [Google Scholar] [CrossRef]
- Nguyen, V.B.; Wang, S.L. Production of potent antidiabetic compounds from shrimp head powder via Paenibacillus conversion. Process. Biochem. 2019, 76, 18–24. [Google Scholar] [CrossRef]
- Kandra, P.; Challa, M.M.; Kalangi, P.J.H. Efficient use of shrimp waste: Present and future trends. Appl. Microbiol. Biotechnol. 2012, 93, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Shahidi, F.; Brown, J.A. Carotenoid pigments in seafood and aquaculture. CRC Cri. Rev. Food Sci. 1998, 38, 1–67. [Google Scholar] [CrossRef] [PubMed]
- Gildberg, A.; Stenberg, E. A new process for advanced utilization of shrimp waste. Process. Biochem. 2001, 36, 809–812. [Google Scholar] [CrossRef]
- Guo, N.; Sun, J.; Zhang, Z.; Mao, X. Recovery of chitin and protein from shrimp head waste by endogenous enzyme autolysis and fermentation. J. Ocean. Univ. China 2019, 18, 719–726. [Google Scholar] [CrossRef]
- Doan, C.T.; Tran, T.N.; Wang, C.-L.; Wang, S.-L. Microbial conversion of shrimp heads to proteases and chitin as an effective dye adsorbent. Polymers 2020, 12, 2228. [Google Scholar] [CrossRef]
- Doan, C.T.; Tran, T.N.; Nguyen, V.B.; Nguyen, A.D.; Wang, S.-L. Production of a thermostable chitosanase from shrimp heads via Paenibacillus mucilaginosus TKU032 Conversion and its application in the preparation of bioactive chitosan oligosaccharides. Mar. Drugs 2019, 17, 217. [Google Scholar] [CrossRef] [Green Version]
- Tran, T.N.; Doan, C.T.; Nguyen, M.T.; Nguyen, V.B.; Vo, T.P.K.; Nguyen, A.D.; Wang, S.-L. An exochitinase with n-acetyl-β-glucosaminidase-like activity from shrimp head conversion by Streptomyces speibonae and Its application in hydrolyzing β-chitin powder to produce N-acetyl-d-glucosamine. Polymers 2019, 11, 1600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nargis, A.; Ahmed, K.; Ahmed, G.; Hossain, M.; Rahman, M. Nutritional value and use of shrimp head waste as fish meal. Bangladesh J. Sci Ind Res. 2007, 41, 63–66. [Google Scholar] [CrossRef]
- Chatragadda, R.; Dufossé, L. Ecological and biotechnological aspects of pigmented microbes: A way forward in development of food and pharmaceutical grade pigments. Microorganisms 2021, 9, 637. [Google Scholar] [CrossRef]
- Ramesh, C.; Vinithkumar, N.V.; Kirubagaran, R.; Venil, C.K.; Dufossé, L. Multifaceted applications of microbial pigments: Current knowledge, challenges and future directions for public health implications. Microorganisms 2019, 7, 186. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.L.; Nguyen, V.B.; Doan, C.T.; Tran, T.N.; Nguyen, M.T.; Nguyen, A.D. Production and potential applications of bioconversion of chitin and protein-containing fishery byproducts into prodigiosin: A review. Molecules 2020, 25, 2744. [Google Scholar] [CrossRef]
- Ramesh, C.; Vinithkumar, N.V.; Kirubagaran, R.; Venil, C.K.; Dufossé, L. Applications of prodigiosin extracted from marine red pigmented bacteria zooshikella sp. and actinomycete Streptomyces sp. Microorganisms 2020, 8, 556. [Google Scholar] [CrossRef]
- Furstner, A. Chemistry and biology of roseophilin and the prodigiosin alkaloids: A survey of the last 2500 years. Chem. Int. Ed. Engl. 2003, 42, 3582–3603. [Google Scholar] [CrossRef] [PubMed]
- Cerdeno, A.M.; Bibb, M.J.; Challis, G.L. Analysis of the prodiginine biosynthesis gene cluster of Streptomyces coelicolor A3(2): New mechanisms for chain initiation and termination in modular multienzymes. Chem. Biol. 2001, 8, 817–829. [Google Scholar] [CrossRef] [Green Version]
- Liang, T.W.; Chen, S.Y.; Chen, Y.C.; Chen, Y.C.; Yen, Y.H.; Wang, S.L. Enhancement of prodigiosin production by Serratia marcescens TKU011 and its insecticidal activity relative to food colourants. J. Food Sci. 2013, 78, 1743–1751. [Google Scholar] [CrossRef] [PubMed]
- de Casullo, A.H.W.; Fukushima, K.; Campos, T.G.M. Prodigiosin production by Serratia marcescens UCP 1549 using renewable-resources as a low-cost substrate. Molecules 2010, 15, 6931–6940. [Google Scholar] [CrossRef]
- Samrot, A.V.; Chandana, K.; Senthilkumar, P.; Narendra, K.G. Optimization of prodigiosin production by Serratia marcescens SU-10 and evaluation of its bioactivity. Int. Res. J. Biotechnol. 2011, 2, 128–133. [Google Scholar]
- Patricia, H.V.; Irene, M.A.; Melissa, R.D.; José Manuel, R.D.; Donato, L.M.; Francisco Guadalupe, A.A.; Juan Francisco, V.C. Photoelectric evaluation of dye-sensitized solar cells based on prodigiosin pigment derived from Serratia marcescens 11E. Dye. Pigment. 2020, 177, 108278–108287. [Google Scholar]
- Nguyen, T.H.; Wang, S.L.; Nguyen, D.N.; Nguyen, A.D.; Nguyen, T.H.; Doan, M.D.; Ngo, V.A.; Doan, C.T.; Kuo, Y.H.; Nguyen, V.-B. Bioprocessing of marine chitinous wastes for the production of bioactive prodigiosin. Molecules 2021, 26, 3138. [Google Scholar] [CrossRef] [PubMed]
- Gulani, C.; Bhattacharya, S.; Das, A. Assessment of process parameters influencing the enhanced production of prodigiosin from Serratia marcescens and evaluation of its antimicrobial, antioxidant and dyeing potential. Malays J. Microbiol. 2012, 8, 116–122. [Google Scholar]
- Wei, Y.H.; Yu, W.J.; Chen, W.C. Enhanced undecylprodigiosin production from Serratia marcescens SS-1 by medium formulation and amino-acid supplementation. J. Biosci. Bioeng. 2005, 100, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Haddix, P.L.; Werner, T.F. Spectrophotometric assay of gene expression: Serratia marcescens pigmentation. Bioscene 2000, 26, 3–13. [Google Scholar]
- Wang, X.; Tao, J.; Wei, D.; Shen, Y.; Tong, W. Development of an adsorption procedure for the direct separation and purification of prodigiosin from culture broth. Biotechnol. Appl. Biochem. 2004, 40, 277–280. [Google Scholar]
- Montaner, B.; Navarro, S.; Piqué, M.; Vilaseca, M.; Martinell, M.; Giralt, E.; Gil, J.; Perez-Tomas, P. Prodigiosin from the supernatant of Serratia marcescens induces apoptosis in haematopoietic cancer cell lines. Br. J. Pharmacol. 2000, 131, 585–593. [Google Scholar] [CrossRef] [Green Version]
- Solé, M.; Rius, N.; Francia, A.; Lorén, J.G. The effect of pH on prodigiosin production by non-proliferating cells of Serratia marcescens. Lett. Appl. Microbiol. 1994, 19, 341–344. [Google Scholar] [CrossRef]
- Lin, C.; Jia, X.; Fang, Y.; Chen, L.; Zhang, H.; Lin, R.; Chen, J. Enhanced production of prodigiosin by Serratia marcescens FZSF02 in the form of pigment pellets. Electron. J. Biotechnol. 2019, 40, 58–64. [Google Scholar] [CrossRef]
- Wei, Y.H.; Chen, W.C. Enhanced production of prodigiosin-like pigment from Serratia marcescens SMdeltaR by medium improvement and oil-supplementation strategies. J. Biosci. Bioeng. 2005, 99, 616–622. [Google Scholar] [CrossRef]
- Chang, C.C.; Chen, W.C.; Ho, S.F.; Wu, H.S.; Wei, Y.H. Development of natural anti-tumor drugs by microorganisms. J. Biosci. Bioeng. 2011, 111, 501–511. [Google Scholar] [CrossRef]
- Giri, A.V.; Anandkumar, N.; Muthukumaran, G.; Pennathur, G. A novel medium for the enhanced cell growth and production of prodigiosin from Serratia marcescens isolated from soil. BMC Microbiol. 2004, 4, 11–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elkenawy, N.M.; Yassin, A.S.; Elhifnawy, H.N.; Amin, M.A. Optimization of prodigiosin production by Serratia marcescens using crude glycerol and enhancing production using gamma radiation. Biotechnol. Rep. 2017, 14, 47–53. [Google Scholar] [CrossRef]
- Nguyen, V.B.; Nguyen, A.D.; Wang, S.L. Utilization of fishery processing by-product squid pens for α-glucosidase inhibitors production by Paenibacillus sp. Mar. Drugs 2017, 15, 274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, V.B.; Wang, S.L. Reclamation of marine chitinous materials for the production of α-glucosidase inhibitors via microbial conversion. Mar. Drugs 2017, 15, 350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, V.B.; Nguyen, T.H.; Doan, C.T.; Tran, T.N.; Nguyen, A.D.; Kuo, Y.H.; Wang, S.L. Production and bioactivity-guided isolation of antioxidants with α-glucosidase inhibitory and anti-NO properties from marine chitinous materials. Molecules 2018, 23, 1124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, C.H.; Nguyen, V.B.; Nguyen, A.D.; Wang, S.L. Conversion of shrimp heads to α-glucosidase inhibitors via co-culture of Bacillus mycoides TKU040 and Rhizobium sp. TKU041. Res. Chem. Intermed. 2017, 44, 4597–4607. [Google Scholar] [CrossRef]
- Wang, S.L.; Su, Y.C.; Nguyen, V.B.; Nguyen, A.D. Reclamation of shrimp heads for the production of α-glucosidase inhibitors by Staphylococcus sp. TKU043. Res. Chem. Intermed. 2018, 44, 4929–4937. [Google Scholar] [CrossRef]
- Nguyen, V.B.; Nguyen, D.N.; Nguyen, A.D.; Ngo, V.A.; Ton, T.Q.; Doan, C.T.; Pham, T.P.; Tran, T.P.H.; Wang, S.L. Utilization of crab waste for cost-effective bioproduction of prodigiosin. Mar. Drugs 2020, 18, 523. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.B.; Nguyen, D.N.; Wang, S.L. Microbial reclamation of chitin and protein-containing marine by-products for the production of prodigiosin and the evaluation of its bioactivities. Polymers 2020, 12, 1328. [Google Scholar] [CrossRef]
- Nguyen, V.B.; Chen, S.P.; Nguyen, T.H.; Nguyen, M.T.; Tran, T.T.T.; Doan, C.T.; Tran, T.N.; Nguyen, A.D.; Kuo, Y.H.; Wang, S.L. Novel efficient bioprocessing of marine chitins into active anticancer prodigiosin. Mar. Drugs 2020, 18, 15. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, V.B.; Wang, S.L.; Nguyen, T.H.; Nguyen, T.H.; Trinh, T.H.T.; Nong, T.T.; Nguyen, T.U.; Nguyen, V.N.; Nguyen, A.D. Reclamation of rhizobacteria newly isolated from black pepper plant roots as potential biocontrol agents of root-knot nematodes. Res. Chem. Intermed. 2019, 45, 5293–5307. [Google Scholar] [CrossRef]
- Chávez-Castilla, L.R.; Aguilar, O. An integrated process for the in-situ recovery of prodigiosin using micellar ATPS from a culture of Serratia marcescens. J. Chem. Technol. Biotechnol. 2016, 91, 2896–2903. [Google Scholar]
- Tao, J.; Wang, X.; Shen, Y.; Wei, D. Strategy for the improvement of prodigiosin production by a Serratia marcescens mutant through fed-batch fermentation. World. J. Microbiol. Biotechnol. 2005, 21, 969–972. [Google Scholar] [CrossRef]
- Sura, J.M.; Khalid, J.K. A kinetic model for prodigiosin production by Serratia marcescens as a bio-colorant in bioreactor. AIP Conf. Proc. 2020, 2213, 020027. [Google Scholar]
- Chidambaram, K.V.; Zainul, A.Z.; Wan, A.A. Optimization of culture conditions for flexirubin production by Chryseobacterium artocarpi CECT 8497 using response surface methodology. Acta Biochim. Pol. 2014, 62, 185–190. [Google Scholar]
- Song, M.J.; Bae, J.D.; Lee, D.S.; Kim, C.H.; Kim, J.S.; Kim, S.W.; Hong, S.I. Purification and characterization of prodigiosin produced by integrated bioreactor from Serratia sp. KH-95. J. Biosci. Bioeng. 2006, 101, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Lobo, V.; Patil, A.; Phatak, A.; Chandra, N. Free radicals, antioxidants and functional foods: Impact on human health. Pharmacogn. Rev. 2010, 4, 118–126. [Google Scholar] [CrossRef] [Green Version]
- Muthukumar, A.; Pradeep, P.; Thigale, I.; Mohanasrinivasan, V.; Jemimah, N.S.; Devi, C.S. Exploring the bioactive potential of Serratia marcescens VITAPI (Acc: 1933637) isolatedfrom soil. Front. Biol. 2016, 11, 476–480. [Google Scholar] [CrossRef]
- Chung, M.J.; Park, J.K.; Park, Y.I. Anti-inflammatory effects of low-molecular weight chitosan oligosaccharides in IgE-antigen complex-stimulated RBL-2H3 cells and asthma model mice. Int. Immunopharmcol. 2012, 12, 453–459. [Google Scholar] [CrossRef]
- Santos, T.C.; Gomes, T.M.; Pinto, B.A.S.; Camara, A.L.; Paes, A.M.A. Naturally occurring acetylcholinesterase inhibitors and their potential use for Alzheimer’s disease therapy. Front. Pharmacol. 2018, 9, 1192. [Google Scholar] [CrossRef] [Green Version]
- Gligorić, E.; Igić, R.; Suvajdžić, L.; Grujić-Letić, N. Species of the Genus Salix, L.: Biochemical screening and molecular docking approach to potential acetylcholinesterase inhibitors. Appl. Sci. 2019, 9, 1842. [Google Scholar] [CrossRef] [Green Version]
- Chandra, B.T.M.; Rajesh, S.S.; Bhaskar, B.V.; Devi, S.; Rammohan, A.; Sivaraman, T.; Rajendra, W. Molecular docking, molecular dynamics simulation, biological evaluation and 2D QSAR analysis of flavonoids from Syzygium alternifolium as potent anti-Helicobacter pylori agents. RSC Adv. 2017, 7, 18277–18292. [Google Scholar] [CrossRef] [Green Version]
- Tran, T.P.T.; Thanh, Q.B.; Phan, T.Q.; Nguyen, C.B.; Tran, V.L.; Tran, V.C.; Nguyen, L.C.; Nguyen, V.T.; Tran, V.S.; Nguyen, T.A.N. Isolation, semi-synthesis, docking-based prediction, and bioassay-based activity of Dolichandrone spathacea iridoids: New catalpol derivatives as glucosidase inhibitors. RSC Adv. 2021, 11, 11959–11975. [Google Scholar]
- Ding, Y.; Fang, Y.; Moreno, J.; Ramanujam, J.; Jarrell, M.; Brylinski, M. Assessing the similarity of ligand binding conformations with the Contact Mode Score. Comput. Biol. Chem. 2016, 64, 403–413. [Google Scholar]
- Krishna, P.S.; Vani, K.; Prasad, M.R.; Samatha, B.; Bindu, N.S.; Charya, M.A.; Reddy, S.P. In-silico molecular docking analysis of prodigiosin and cycloprodigiosin as COX-2 inhibitors. Springer Plus. 2013, 2, 172. [Google Scholar] [CrossRef] [Green Version]
- El-Batal, A.; El-Hendawy, H.; Faraag, A. In silico and in vitro cytotoxic effect of prodigiosin-conjugated silver nanoparticles on liver cancer cells (HepG2). BioTechnologia 2017, 98, 225–243. [Google Scholar] [CrossRef]
- Boopathi, B.; Rajaiah, A.; Dharmaraj, R.D. Exploration of the optimized parameters for bioactive prodigiosin mass production and its biomedical applications in vitro as well as in silico. Biocatal. Agric. Biotechnol. 2019, 22, 101385. [Google Scholar]
- Mangesh, V.S.; Samadhan, R.W.; Nidhi, B.; Prafulla, B.C.; Tejashri, B.H.; Yogesh, S.S. Isolation and virtual screening of antimicrobial prodigiosin pigment from oxalotrophic Serratia marcescens OX_R strain. J. Appl. Pharm. Sci. 2016, 6, 52–58. [Google Scholar]
- Campàs, C.; Dalmau, M.; Montaner, B.; Barragán, M.; Bellosillo, B.; Colomer, D.; Pons, G.; Pérez-Tomás, R.; Gil, J. Prodigiosin induces apoptosis of B and T cells from B-cell chronic lymphocytic leukemia. Leukemia 2003, 17, 746–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.L.; Wang, C.Y.; Yen, Y.H.; Liang, T.W.; Chen, S.Y.; Chen, C.H. Enhanced production of insecticidal prodigiosin from Serratia marcescens TKU011 in media containing squid pen. Process. Biochem. 2012, 47, 1684–1690. [Google Scholar] [CrossRef]
- Nguyen, V.B.; Ton, T.Q.; Nguyen, D.N.; Nguyen, T.T.; Ngu, T.N.; Nguyen, T.H.; Doan, C.T.; Tran, T.N.; Nguyen, M.T.; Ho, N.D. Reclamation of beneficial bioactivities of herbal antioxidant condensed tannin extracted from Euonymus laxiflorus. Res. Chem. Intermed. 2020, 46, 4751–4766. [Google Scholar] [CrossRef]
- Nguyen, V.B.; Wang, S.L.; Nguyen, A.D.; Lin, Z.H.; Doan, C.T.; Tran, T.N.; Huang, H.T.; Kuo, Y.H. Bioactivity-guided purification of novel herbal antioxidant and anti-NO compounds from Euonymus laxiflorus Champ. Molecules 2019, 24, 120. [Google Scholar] [CrossRef] [Green Version]
- Tarasova, O.; Poroikov, V.; Veselovsky, A. Molecular docking studies of HIV-1 resistance to reverse transcriptase inhibitors: Mini-review. Molecules 2018, 23, 1233. [Google Scholar] [CrossRef] [Green Version]
- Labute, P. A widely applicable set of descriptors. J. Mol. Graph. Model. 2000, 18, 464–477. [Google Scholar] [CrossRef]
- Bui, T.P.T.; Le, T.H.; Tran, T.A.M.; Nguyen, T.T.H.; Huynh, T.P.L.; Nguyen, T.T.T.; Nguyen, T.T.; Tran, T.V.A.; Nguyen, T.X.D.; Phan, T.Q.; et al. Screening for Streptococcus pyogenes antibacterial and Candida albicans antifungal bioactivities of organic compounds in natural essential oils of Piper betle L., Cleistocalyx operculatus L. and Ageratum conyzoides L. Chem. Pap. 2021, 75, 1507–1519. [Google Scholar]




| No. | PG Producing-Strain | PG Yield Detected in Fermented Medium (mg L−1) |
|---|---|---|
| 01 | S. marcescens TKU011 | 2.43 ± 0.078 |
| 02 | S. marcescens TNU01 | 2.5746 ± 0.142 |
| 03 | S. marcescens TNU02 | 2.711 ± 0.136 |
| 04 | S. marcescens CC17 | 3.862 ± 0.145 |
| Control | PG not detected |
| Compound Form (Inhibitor) | Symbol (Inhibitor-Enzyme) | DS (kcal/mol) | RMSD (Å) | Number of Interactions (Bonds) | Amino Acids Interacting with the Ligand (Inhibitor Compound) |
|---|---|---|---|---|---|
| Cation-PG | CPG–AchE | −12.3 | 1.35 | 6 bonds (1 H-donor, 3 ionic, and 2 pi-H) | Asp 326 (3.2 Å), Asp 326 (3.89 Å), Asp 393 (2.75 Å), Asp 393 (3.87 Å), Lys 325 (4.13 Å), Asp 393 (3.62 Å) |
| Neutral-PG | NPG–AchE | −11.1 | 1.75 | 3 bonds (1 H-pi, and 2 pi-H) | Trp 84 (3.94 Å), Trp 84 (4.51 Å), Gly 118 (3.85 Å) |
| Neutral-BC | NBC–AchE | −10.8 | 1.93 | 1 bond (1 H-pi) | Trp 84 (4.62 Å) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, V.B.; Wang, S.-L.; Nguyen, A.D.; Phan, T.Q.; Techato, K.; Pradit, S. Bioproduction of Prodigiosin from Fishery Processing Waste Shrimp Heads and Evaluation of Its Potential Bioactivities. Fishes 2021, 6, 30. https://doi.org/10.3390/fishes6030030
Nguyen VB, Wang S-L, Nguyen AD, Phan TQ, Techato K, Pradit S. Bioproduction of Prodigiosin from Fishery Processing Waste Shrimp Heads and Evaluation of Its Potential Bioactivities. Fishes. 2021; 6(3):30. https://doi.org/10.3390/fishes6030030
Chicago/Turabian StyleNguyen, Van Bon, San-Lang Wang, Anh Dzung Nguyen, Tu Quy Phan, Kuaanan Techato, and Siriporn Pradit. 2021. "Bioproduction of Prodigiosin from Fishery Processing Waste Shrimp Heads and Evaluation of Its Potential Bioactivities" Fishes 6, no. 3: 30. https://doi.org/10.3390/fishes6030030