Functional Characterization and Molecular Marker Development of the Proenkephalin as Biomarker of Food Addiction in Food Habit Domestication of Mandarin Fish (Siniperca chuatsi)
Abstract
:1. Introduction
2. Materials and Methods
2.1. PENK Gene Structure and Synteny Analysis
2.2. Experimental Fish and Experimental Approach
2.2.1. The Origin of Experimental Fish
2.2.2. The Experimental Approach of Intracerebroventricular Injection of Amino Acid
2.2.3. The Experimental Approach of Food Habit Domestication
2.2.4. The Experimental Approach of High-Carbohydrate Artificial Diet Feeding
2.2.5. The Preservation of Sample of Each Experimental Treatment
2.3. RNA Isolation and Reverse Transcription
2.4. Real-Time Quantitative PCR
2.5. Statistical Analysis
2.6. Association Analysis of penk Gene and Domestication Traits in Mandarin Fish
2.6.1. Genomic DNA Extraction and Primer Design
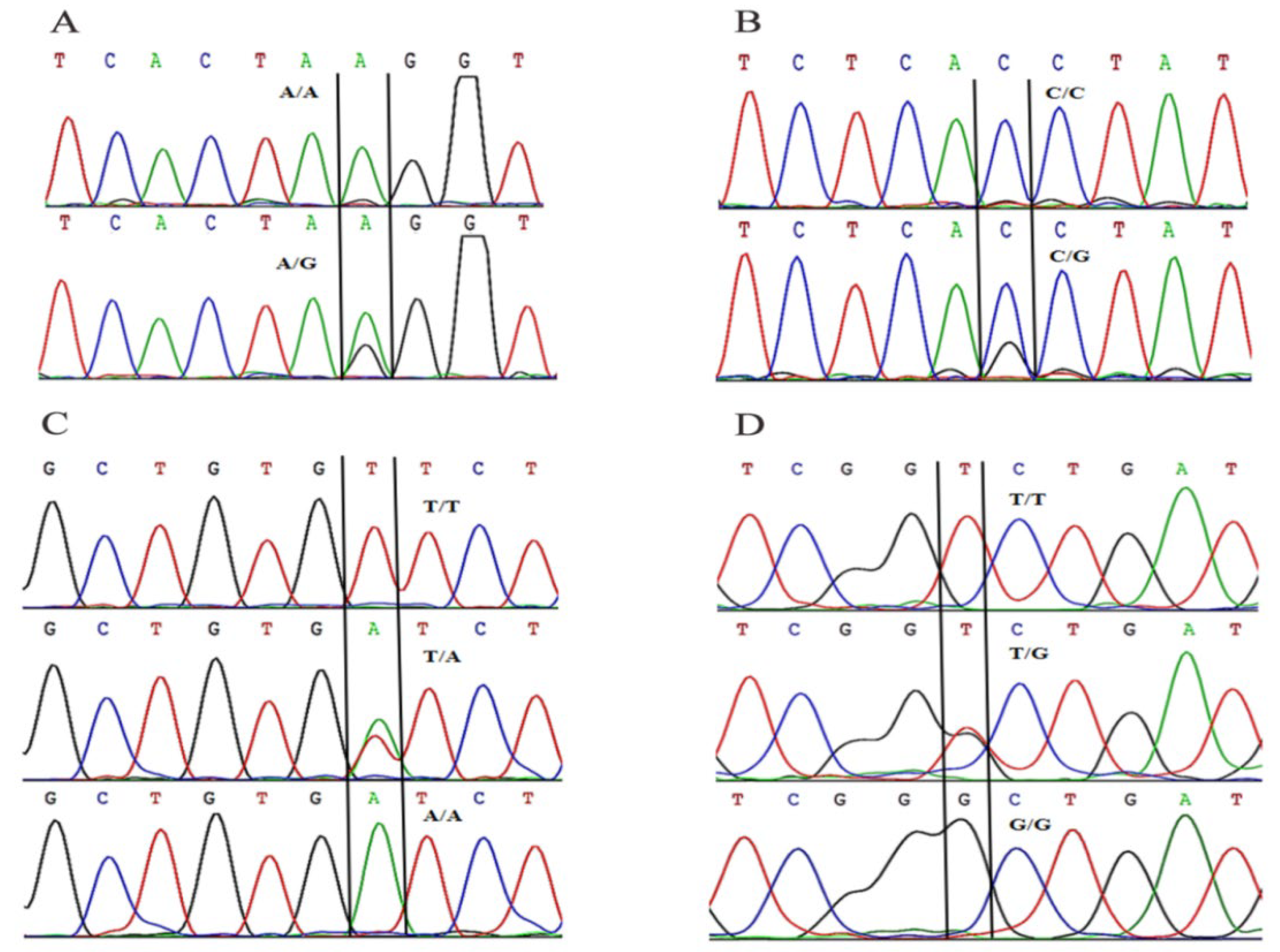
2.6.2. Screening and Typing of SNP Loci
2.6.3. Association Analysis of penk Gene and Domestication Trait in Mandarin Fish
3. Results
3.1. Bioinformatics Analysis of the PENK Gene
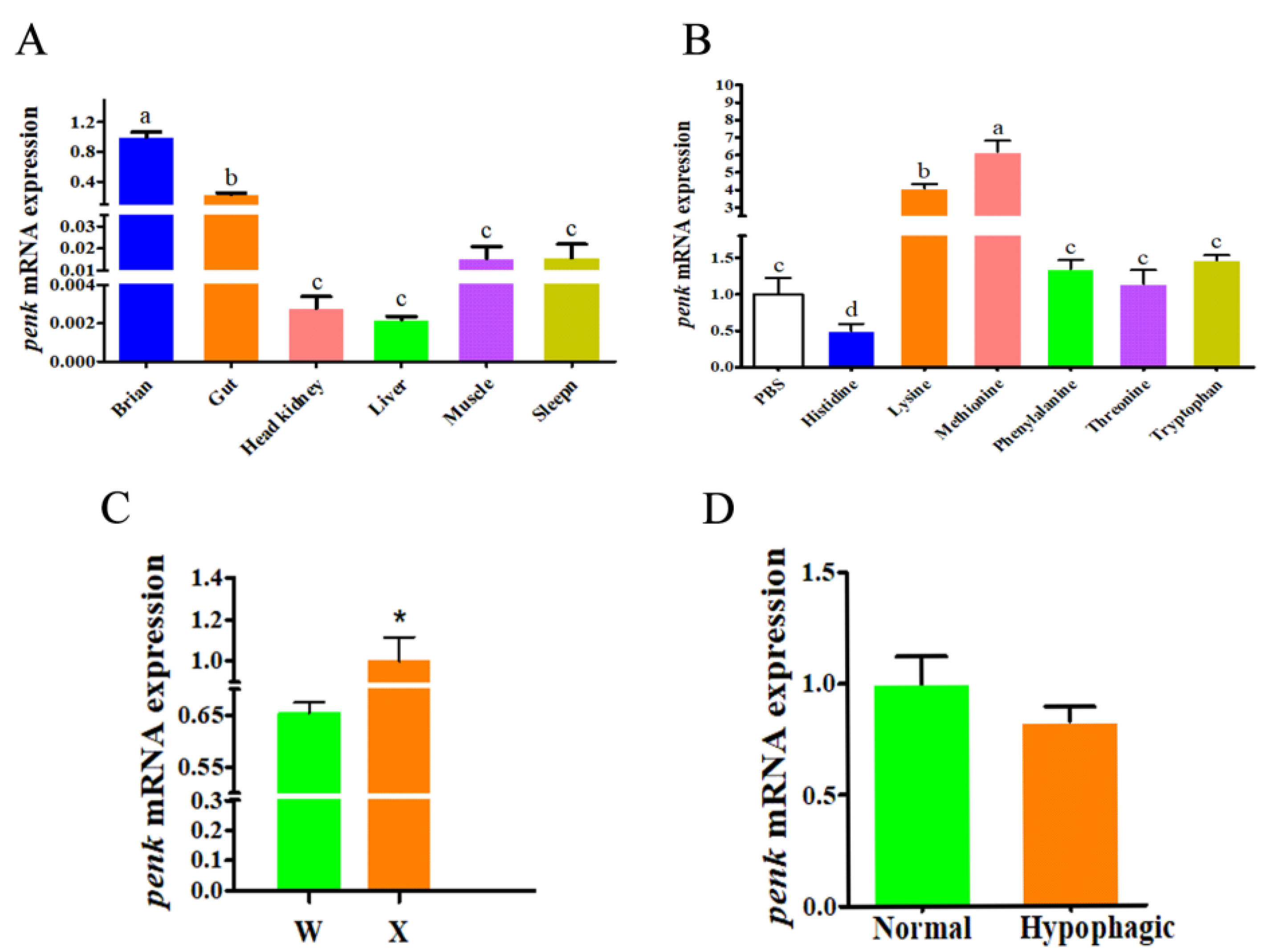
3.2. RT-PCR Analysis of the penk Gene in Mandarin Fish
3.3. Association Analysis of Mandarin Fish SNP Loci and Domestication Traits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Stern, A.S.; Lewis, R.V.; Kimura, S.; Rossier, J.; Gerber, L.D.; Brink, L.; Stein, S.; Udenfriend, S. Isolation of the opioid heptapeptide Met-enkephalin [Arg6,Phe7] from bovine adrenal medullary granules and striatum. Proc. Natl. Acad. Sci. USA 1979, 6, 6680–6683. [Google Scholar] [CrossRef] [Green Version]
- Kilpatrick, D.L.; Jones, B.N.; Kojima, K.; Udenfriend, S. Identification of the octapeptide [Met] enkephalin -Arg6-Gly7-Leu8 in extracts of bovine adrenal medulla. Biochem. Biophys. Res. Commun. 1981, 103, 698–705. [Google Scholar] [CrossRef]
- Kilpatrick, D.L.; Taniguchi, T.; Jones, B.N.; Stern, A.S.; Shively, J.E.; Hullihan, J.; Kimura, S.; Stein, S.; Udenfriend, S. A highly potent 3200-dalton adrenal opioid peptide that contains both a [Met] and [Leu] enkephalin sequence. Proc. Natl. Acad. Sci. USA 1981, 78, 3265–3268. [Google Scholar] [CrossRef] [Green Version]
- Nuñez, V.G.; Sarmiento, R.G.; Rodríguez, R.E. Characterization of zebrafish proenkephalin reveals novel opioid sequences. Brain Res. Mol. Brain Res. 2003, 114, 31–39. [Google Scholar] [CrossRef]
- Shao, Z.X.; Wu, H.Q. Research progress in enkephalin. Stroke Nerv. Disord 2013, 20, 58–60. [Google Scholar] [CrossRef]
- Eiden, L.E. The enkephalin-containing cell: Strategies for polypeptide synthesis and secretion throughout the neuroendocrine system. Cell Mol. Neurobiol. 1987, 4, 339–352. [Google Scholar] [CrossRef]
- Shan, Z.Z.; Dai, S.M.; Fang, F.; Su, D.F. Changes of central norepinephrine, beta-endorphin, LEU-enkehalin, peripheral arginine-vasopressin, and angiotensin II levels in acute and chronic phases sino-aortic denervati-on in rats. Cardiovas. Pharmacol. 2004, 43, 234–241. [Google Scholar] [CrossRef]
- Malendowicz, L.K.; Rebuffat, P.; Tortorella, C.; Nussdorfer, G.G.; Ziolkowska, A.; Hochol, A. Effects of met-enkephalin on cell proliferation in different models of adrenocortical-cell growth. Int. J. Mol. Med. 2005, 15, 841–845. [Google Scholar] [CrossRef]
- Haber, S.N.; Knutson, B. The reward circuit: Linking primate anatomy and human imaging. Neuropsychopharmacology 2010, 35, 4–26. [Google Scholar] [CrossRef] [Green Version]
- Juarez, B.; Han, M.H. Diversity of Dopaminergic Neural Circuits in Response to Drug Exposure. Neuropsychopharmacology 2016, 41, 2424–2446. [Google Scholar] [CrossRef] [Green Version]
- Vasiliu, O. Current Status of Evidence for a New Diagnosis: Food Addiction-A Literature Review. Front. Psychiatry 2022, 12, 824–936. [Google Scholar] [CrossRef]
- Vesnina, A.; Prosekov, A.; Kozlova, O.; Atuchin, V. Genes and Eating Preferences, Their Roles in Personalized Nutrition. Genes 2020, 11, 357. [Google Scholar] [CrossRef] [Green Version]
- Blasio, A.; Steardo, L.; Sabino, V.; Cottone, P. Opioid system in the medial prefrontal cortex mediates binge-like eating. Addict Biol. 2014, 19, 652–662. [Google Scholar] [CrossRef] [Green Version]
- Johnson, S.W.; North, R.A. Opioids excite dopamine neurons by hyperpolarization of local interneurons. J. Neurosci. 1992, 12, 483–488. [Google Scholar] [CrossRef] [Green Version]
- Comings, D.E.; Blake, H.; Dietz, G.; Gade-Andavolu, R.; Legro, R.S.; Saucier, G.; Johnson, P.; Verde, R.; MacMurray, J.P. The proenkephalin gene (PENK) and opioid dependence. Neuroreport 1999, 10, 1133–1135. [Google Scholar] [CrossRef]
- Heinsbroek, J.A.; Bobadilla, A.C.; Dereschewitz, E.; Assali, A.; Chalhoub, R.M.; Cowan, C.W.; Kalivas, P.W. Opposing Regulation of Cocaine Seeking by Glutamate and GABA Neurons in the Ventral Pallidum. Cell Rep. 2020, 30, 2018–2027. [Google Scholar] [CrossRef] [Green Version]
- Dores, R.M.; Sollars, C.; Lecaude, S.; Lee, J.; Danielson, P.; Alrubaian, J.; Lihrman, I.; Joss, J.M.; Vaudry, H. Cloning of prodynorphin cDNAs from the brain of Australian and African lungfish: Implications for the evolution of the prodynorphin gene. Neuroendocrinology 2004, 79, 185–196. [Google Scholar] [CrossRef]
- Li, C.Y.; Zhou, W.Z.; Zhang, P.W.; Johnson, C.; Wei, L.; Uhl, G.R. Meta-analysis and genome-wide interpretation of genetic susceptibility to drug addiction. BMC Genom. 2011, 12, 508. [Google Scholar] [CrossRef] [Green Version]
- Spanagel, R. Convergent functional genomics in addiction research-a translational approach to study candidate genes and gene networks. Silico Pharmacol. 2013, 1, 18. [Google Scholar] [CrossRef] [Green Version]
- Liang, X.F.; Liu, J.K.; Huang, B.Y. The role of sense organs in the feeding behaviour of Chinese perch. J. Fish Biol. 1998, 52, 1058–1067. [Google Scholar] [CrossRef]
- Chen, K.; Zhang, Z.; Li, J.; Xie, S.; Shi, L.J.; He, H.M.; Liang, X.F.; Zhu, Q.S.; He, S. Different regulation of branched-chain amino acid on food intake by TOR signaling in Chinese perch (Siniperca chuatsi). Aquaculture 2020, 530, 735–792. [Google Scholar] [CrossRef]
- De Pedro, N.; Pinillos, M.L.; Valenciano, A.I.; Alonso-Bedate, M.; Delgado, M.J. Inhibitory effect of serotonin on feeding behavior in goldfish: Involvement of CRF. Peptides 1998, 19, 505–511. [Google Scholar] [CrossRef]
- Moeller, S.J.; Beebe-Wang, N.; Schneider, K.E.; Konova, A.B.; Parvaz, M.A.; Alia-Klein, N.; Hurd, Y.L.; Goldstein, R.Z. Effects of an opioid (proenkephalin) polymorphism on neural response to errors in health and cocaine use disorder. Behav. Brain Res. 2015, 293, 18–26. [Google Scholar] [CrossRef] [Green Version]
- Liang, X.F.; Zheng, W.Y.; Wang, Y.L. Visual characteristics of mandarin fish (Siniperca chuatsi) in relation to its feeding habit: I Photo-sensitivity and spectral sensitivity of electroretinogram. Acta Hydrobiol. Sin. 1994, 18, 247–253. [Google Scholar] [CrossRef]
- Zhu, Q.S.; He, S.; Liang, X.F.; Xu, J.; Zhang, Y.P. Effect of six essential amino acids on the regulation of warped mouth mandarin feeding. Aquat. Sci. Technol. Intell. 2020, 47, 154–161. [Google Scholar] [CrossRef]
- He, S.; Liang, X.F.; Sun, J.; Li, L.; Yu, Y.; Huang, W.; Qu, C.M.; Cao, L.; Bai, X.L.; Tao, Y.X. Insights into food preference in hybrid F1 of Siniperca chuatsi (♀) × Siniperca scherzeri (♂) mandarin fish through transcriptome analysis. BMC Genom. 2013, 14, 601. [Google Scholar] [CrossRef] [Green Version]
- He, S.; You, J.J.; Liang, X.F.; Zhang, Z.L.; Zhang, Y.P. Transcriptome sequencing and metabolome analysis of food habits domestication from live prey fish to artificial diets in mandarin fish (Siniperca chuatsi). BMC Genom. 2021, 22, 129. [Google Scholar] [CrossRef]
- Liang, X.F.; Oku, H.; Ogata, H.Y.; Liu, J.; He, X. Weaning Chinese perch (Siniperca chuatsi) basilewsky onto artificial diets based upon its specific sensory modality in feeding. Aquac. Res. 2001, 32, 76–82. [Google Scholar] [CrossRef] [Green Version]
- You, J.J.; Ren, P.; He, S.; Liang, X.F.; Xiao, Q.Q.; Zhang, Y.P. Histone Methylation of H3K4 Involved in the Anorexia of Carnivorous Mandarin Fish (Siniperca chuatsi) After Feeding on a Carbohydrate-Rich Diet. Front. Endocrinol. 2020, 11, 323. [Google Scholar] [CrossRef]
- Vandesompele, J.; Preter, K.D.; Pattyn, F.; Poppe, B.; Roy, N.V.; Paepe, A.D. Accuratenormalization of real-time quantitative RT-PCR data by geometric averagingof multiple internal control genes. Genome Biol. 2018, 3, research0034.1. [Google Scholar] [CrossRef] [Green Version]
- Liang, H.; He, S.; Liang, X.F.; Lu, H.L.; Chen, K. Feeding habit transition induced bysocial learning through CAMKII signaling in chinese perch (Siniperca chuatsi). Aquaculture 2020, 533, 736211. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Bostein, D.; White, R.L.; Skolnick, M.; DAVIS, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar]
- Hazra, A.; Gogtay, N. Biostatistics Series Module 6: Correlation and Linear Regression. Indian J. Dermatol. 2016, 61, 593–601. [Google Scholar] [CrossRef]
- Hook, V.; Funkelstein, L.; Lu, D.; Bark, S.; Wegrzyn, J.; Hwang, S.R. Proteases for processing proneuropeptides into peptide neurotransmitters and hormones. Annu. Rev. Pharmacol. Toxicol. 2008, 48, 393–423. [Google Scholar] [CrossRef] [Green Version]
- Dores, R.M.; McDonald, L.K.; Goldsmith, A.; Deviche, P.; Rubin, D.A. The phylogeny of enkephalins: Speculations on the origins of opioid precursors. Cell. Physiol. Biochem. 1993, 3, 231–244. [Google Scholar] [CrossRef]
- Bojnik, E.; Magyar, A.; Tóth, G.; Bajusz, S.; Borsodi, A.; Benyhe, S. Binding studies of novel, non-mammalian enkephalins, structures predicted from frog and lungfish brain cDNA sequences. Neuroscience 2009, 158, 867–874. [Google Scholar] [CrossRef]
- Bojnik, E.; Babos, F.; Magyar, A.; Borsodi, A.; Benyhe, S. Bioinformatic and biochemical studies on the phylogenetic variability of proenkephalin-derived octapeptides. Neuroscience 2010, 165, 542–552. [Google Scholar] [CrossRef]
- Bojnik, E.; Boynik, E.; Corbani, M.; Babos, F.; Magyar, A.; Borsodi, A.; Benyhe, S. Phylogenetic diversity and functional efficacy of the C-terminally expressed heptapeptide unit in the opioid precursor polypeptide proenkephalin A. Neuroscience 2011, 178, 56–67. [Google Scholar] [CrossRef]
- Hughes, J.; Smith, T.W.; Kosterlitz, H.W.; Fothergill, L.A.; Morgan, B.A.; Morris, H.R. Identification of two related pentapeptides from the brain with potent opiate agonist activity. Nature 1975, 258, 577–580. [Google Scholar] [CrossRef]
- Hughes, J.; Kosterlitz, H.W.; Smith, T.W. The distribution of methionine-enkephalin and leucine-enkephalin in the brain and peripheral tissues. Br. J. Pharmacol. 1977, 120, 639–647. [Google Scholar] [CrossRef] [Green Version]
- Bojnik, E.; Kleczkowska, P.; de Velasco, E.M.F.; Corbani, M.; Babos, F.; Lipkowski, A.W.; Magyar, A.; Benyhe, S. Bioactivity studies on atypical natural opioid hexapeptides processed from proenkephalin (PENK) precursor polypeptides. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2014, 174, 29–35. [Google Scholar] [CrossRef]
- Bodnar, R.J. Endogenous opiates and behavior: 2016. Peptides 2018, 101, 167–212. [Google Scholar] [CrossRef] [PubMed]
- Kah, O.; Dufour, S. Conserved and divergent features of reproductive neu-roendocrinology in teleost fishes. In Hormones and Reproduction of Vertebrates; Norris, D.O., Lopez, K.H., Eds.; Academic Press: London, UK, 2011; Volume 1, pp. 15–42. [Google Scholar]
- Chalmers, J.; Arnolda, L.; Kapoor, V.; Llewellyn-Smith, I.; Minson, J.; Pilowsky, P. Amino acid neurotransmitters in the central control of blood pressure and in experimental hypertension. J. Hypertens. Suppl. 1992, 10, S27–S37. [Google Scholar] [CrossRef]
- Holaday, J.W. Cardiovascular effects of endogenous opiate systems. Annu. Rev. Pharmacol. Toxicol. 1983, 23, 541–594. [Google Scholar] [CrossRef]
- Helfman, G.; Collette, B.; Facey, D. The Diversity of Fishes. In Blackwell Science; Fricke, H., Ed.; Oxford Press: Malden, MA, USA, 1998; pp. 454–455. [Google Scholar] [CrossRef]
- Demski, L.S. In a Fish’s Mind’s eye: The visual pallium of Teleosts. In Sensory Processing in Aquatic Environments; Collin, S.P., Marshall, N.J., Eds.; Springer: New York, NY, USA, 2003; pp. 404–419. [Google Scholar]
- Salas, C.; Broglio, C.; Durán, E.; Gómez, A.; Ocaña, F.M.; Jiménez-Moya, F.; Rodríguez, F. Neuropsychology of learning and memory in teleost fish. Zebrafish 2006, 3, 157–171. [Google Scholar] [CrossRef]
- Heeley, N.; Blouet, C. Central amino acid sensing in the control of feeding behavior. Front. Endocrinol. 2016, 7, 148. [Google Scholar] [CrossRef] [Green Version]
- Laeger, T.; Reed, S.D.; Henagan, T.M.; Fernandez, D.H.; Taghavi, M.; Addington, A.; Münzberg, H.; Martin, R.J.; Hutson, S.M.; Morrison, C.D. Leucine acts in the brain to suppress food intake but does not function as a physiological signal of low dietary protein. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2014, 307, R310–R320. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Bailo, B.; Toguri, C.; Eny, K.M.; El-Sohemy, A. Genetic variation in taste and its influence on food selection. OMICS 2009, 1, 69–80. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.A. The adaptive significance of behavioural ontogeny in some. Cent. Fishes 1985, 13, 25–34. [Google Scholar] [CrossRef]
- Warburton, K. Learning of foraging skills by fish. Fish Fishes 2003, 4, 203–215. [Google Scholar] [CrossRef]
- Gendall, K.A.; Joyce, P.R.; Abbott, R.M. The effects of meal composition on subsequent craving and binge eating. Addict. Behav. 1999, 24, 305–315. [Google Scholar] [CrossRef]
- Lennerz, B.; Lennerz, J.K. Food Addiction, High-Glycemic-Index Carbohydrates, and Obesity. Clin. Chem. 2018, 64, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Hemre, G.I.; Mommsen, T.P.; Krogdahl, A. Carbohydrates in fish nutrition: Effects on growth, glucose metabolism and hepatic enzymes. Aquac. Nutr. 2002, 8, 175–194. [Google Scholar] [CrossRef]
- Polakof, S.; Míguez, J.M.; Soengas, J.L. Dietary carbohydrates induce changes in glucosensing capacity and food intake of rainbow trout. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008, 295, R478–R489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeomans, M.R.; Gray, R.W. Opioid peptides and the control of human ingestive behaviour. Neurosci. Biobehav. Rev. 2002, 26, 713–728. [Google Scholar] [CrossRef]
- Park, S.G.; Hannenhalli, S.; Choi, S.S. Conservation in fifirst introns is positively associated with the number of exons within genes and the presence of regulatory epigenetic signals. BMC Genom. 2014, 15, 526. [Google Scholar] [CrossRef] [Green Version]
- Jo, S.S.; Choi, S.S.; Hurst, L. Analysis of the functional relevance of epigenetic chromatin marks in the first intron associated with specifific gene expression patterns. Genome Biol. Evol. 2019, 11, 786–797. [Google Scholar] [CrossRef]
- Chorev, M.; Bekker, A.J.; Goldberger, J.; Carmel, L. Identifification of introns harboring functional sequence elements through positional conservation. Sci. Rep. 2017, 7, 4201. [Google Scholar] [CrossRef] [Green Version]
- Hou, J.; Lu, D.; Mason, A.S.; Li, B.; Xiao, M.; An, S.; Fu, D. Non-coding RNAs and transposable elements in plant genomes: Emergence, regulatory mechanisms and roles in plant development and stress responses. Planta 2019, 250, 23–40. [Google Scholar] [CrossRef]
- Huang, M.T.F.; Gorman, C.M. Intervening sequences increase efficiency of RNA 3′ processing and accumulation of cytoplasmic RNA. Nucleic Acids Res. 1990, 18, 937–947. [Google Scholar] [CrossRef] [PubMed]
- CaUis, J.; Fromm, M.; Walbot, V. lntrons increase gene expression in cultured maize cells. Genes Dev. 1987, 1, 1183–1200. [Google Scholar] [CrossRef] [Green Version]
- McElroy, D.; Zhang, W.; Cao, J.; Wu, R. Isolation of an efficient actin promoter for use in rice transformation. Plant Cell 1990, 2, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Maas, C.; Laufs, J.; Grant, S.; Korfhage, C.; Werr, W. The combination of a novel stimulatory element in the first exon of the maize Shrunken-I gene with the following intron 1 enhances reporter gene expression up to 1000- fold. Plant Mol. Biol. 1991, 16, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Collis, P.; Antoniou, M.; Grosveld, F. Definition of the minimal requirements within the human /3-globin gene and the dominant control region for high level expression. EMBO J. 1990, 9, 233–240. [Google Scholar] [CrossRef]
- Kang, J.H.; Lee, S.J.; Park, S.R.; Ryu, H.Y. DNA polymorphism in the growth hormone gene and its association with weight in olive flounder Paralichthys olivaceus. Fish. Sci. 2002, 68, 494–498. [Google Scholar] [CrossRef]

| Species | EMBL/GenBank Databases |
|---|---|
| Mandarin fish (Siniperca chuatsi) | PENK: SC_LG18_22463 |
| Human (Homo sapiens) | PENK: NP_001129162.1 |
| Mouse (Mus musculus) | PENK: NP_001335138.1 |
| Zebrafish (Danio rerio) | PENKA: NP_956377.2 |
| Zebrafish (Danio rerio) | PENKB: NP_878303.1 |
| European seabass (Dicentrarchus labrax) | PENK: DLA_LG10_000290 |
| Chicken (Gallus gallus) | PENK: XP_040520680.1 |
| Japanese medaka (Oryzias latipes) | PENK: XP_011484397.1 |
| African clawed frog (Xenopus laevis) | PENK: XP_018079238.1 |
| Torafugu (Takifugu rubripes) | PENK: XP_011614671.1 |
| Spotted green pufferfish (Tetraodon nigroviridis) | PENK: ALD51518.1 |
| Blackstrip livebearer (Poeciliopsis prolifica) | PENK: JAO05419.1 |
| Nile tilapia (Oreochromis niloticus) | PENK: XP_003456767.1 |
| Grass carp (Ctenopharyngodon idellus) | PENK: ROL48303.1 |
| Japanese medaka (Oryzias latipes) | PENK: XP_011484397.1 |
| Gene Name | Primer | (5′–3′) Primer Sequence | Tm/°C |
|---|---|---|---|
| penk | F | ACAGCAGTCTGGTTTTCCCT | 58 |
| R | ATGTTGCGTCCTCTTCGTCA | ||
| rpl13 | F | TATCCCCCCACCCTATGACA | 58 |
| R | ACGCCCAAGGAGAGCGAACT | ||
| penk-1 | F | ATGGCTGCCCCTGCACA | 61.2 |
| R | AGGGGACAACAAATACCAGATATGGAC | ||
| penk-2 | F | CTTTCCATCTTTACACAATTATTACTTGTTGTATGCT | 59.5 |
| R | GTGAGGCCTCTCTCTCTCT | ||
| penk-3 | F | AATTCATCACGGCACAAAATTAATATGAAC | 55.7 |
| R | CTAGTCCATGAACCCACCATACC |
| Locus | Group | Ne | Ho | He | PIC | p-Value (HWE) |
|---|---|---|---|---|---|---|
| penk-A | X = 25 | 0.5000 | 1.0000 | 0.5102 | 0.3750 | 0.0000 * |
| W = 30 | 0.4978 | 0.9333 | 0.5062 | 0.3740 | 0.0000 * | |
| penk-B | X = 25 | 0.4968 | 0.9200 | 0.5069 | 0.3730 | 0.0001 * |
| W = 30 | 0.4800 | 0.8000 | 0.4881 | 0.3650 | 0.0013 * | |
| penk-C | X = 25 | 0.1128 | 0.1200 | 0.1151 | 0.1060 | 0.9503 |
| W = 30 | 0.3578 | 0.3333 | 0.3638 | 0.2980 | 0.9276 | |
| penk-D | X = 25 | 0.1800 | 0.2000 | 0.1837 | 0.1640 | 0.3817 |
| W = 30 | 0.2994 | 0.2333 | 0.3045 | 0.2600 | 0.4950 | |
| Mean | 0.3656 | 0.5675 | 0.3723 | 0.2894 | 0.3445 |
| Locus | Genotype | Genotypic Frequency | Pearson χ2 | Pearson Correlation Coefficient (r) | |
|---|---|---|---|---|---|
| X (n = 25) | W (n = 30) | ||||
| penk-A A/G | AA | 0.00 (0) | 0.07 (2) | 1.730 | −0.177 |
| AG | 1.00 (25) | 0.93 (28) | 1.730 | ||
| penk-B C/G | CC | 0.08 (2) | 0.20 (6) | 1.580 | −0.169 |
| CG | 0.92 (23) | 0.80 (24) | 1.580 | ||
| penk-C T/A | TT | 0.88 (22) | 0.60 (18) | 5.390 * | 0.323 |
| TA | 0.12 (3) | 0.33 (10) | 3.438 | ||
| AA | 0.00 (0) | 0.07 (2) | 1.730 | ||
| penk-D T/G | TT | 0.80 (20) | 0.70 (21) | 0.719 | 0.157 |
| TG | 0.20 (5) | 0.23 (7) | 0.089 | ||
| GG | 0.00 (0) | 0.07 (2) | 1.730 | ||
| Transcription Factor | Score | Relative Score | Strand | Start | End | Predicted Sequence |
|---|---|---|---|---|---|---|
| ZNF85 | 18.63 | 0.95 | − | 1646 | 1661 | AAAGAGATTACAACAG |
| NeuroD2 | 15.17 | 0.96 | − | 1632 | 1646 | GTCAACAGATGGTTT |
| TAL1::TCF3 | 15.16 | 0.98 | + | 1632 | 1643 | AAACCATCTGTT |
| NeuroD1 | 13.94 | 0.95 | − | 1632 | 1644 | CAACAGATGGTTT |
| Atoh1 | 13.92 | 0.98 | − | 1633 | 1643 | AACAGATGGTT |
| NeuroG2 | 13.89 | 0.97 | − | 1633 | 1645 | TCAACAGATGGTT |
| Ptf1a | 13.53 | 0.96 | − | 1632 | 1644 | CAACAGATGGTTT |
| Atoh1 | 13.33 | 0.99 | − | 1634 | 1641 | CAGATGGT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Miao, Y.; Liang, X.; He, S. Functional Characterization and Molecular Marker Development of the Proenkephalin as Biomarker of Food Addiction in Food Habit Domestication of Mandarin Fish (Siniperca chuatsi). Fishes 2022, 7, 118. https://doi.org/10.3390/fishes7030118
Li Y, Miao Y, Liang X, He S. Functional Characterization and Molecular Marker Development of the Proenkephalin as Biomarker of Food Addiction in Food Habit Domestication of Mandarin Fish (Siniperca chuatsi). Fishes. 2022; 7(3):118. https://doi.org/10.3390/fishes7030118
Chicago/Turabian StyleLi, Yingcen, Yunliang Miao, Xufang Liang, and Shan He. 2022. "Functional Characterization and Molecular Marker Development of the Proenkephalin as Biomarker of Food Addiction in Food Habit Domestication of Mandarin Fish (Siniperca chuatsi)" Fishes 7, no. 3: 118. https://doi.org/10.3390/fishes7030118





