Validation of a Portable eDNA Detection Kit for Invasive Carps
Abstract
:1. Introduction
2. Methods
2.1. LAMP Primer Design
2.2. LAMP Optimization
2.3. Limit of Detection
2.4. Mesocosm Validation
2.5. DNA Extractions
2.5.1. 500-mL Filter DNA Extraction
2.5.2. 50-mL Grab Sample DNA Extraction
2.6. LAMP Assay
2.7. Spike qPCR
2.8. Statistical Analysis
3. Results
3.1. Specificity Testing
3.2. Limit of Detection and Limit of Quantification
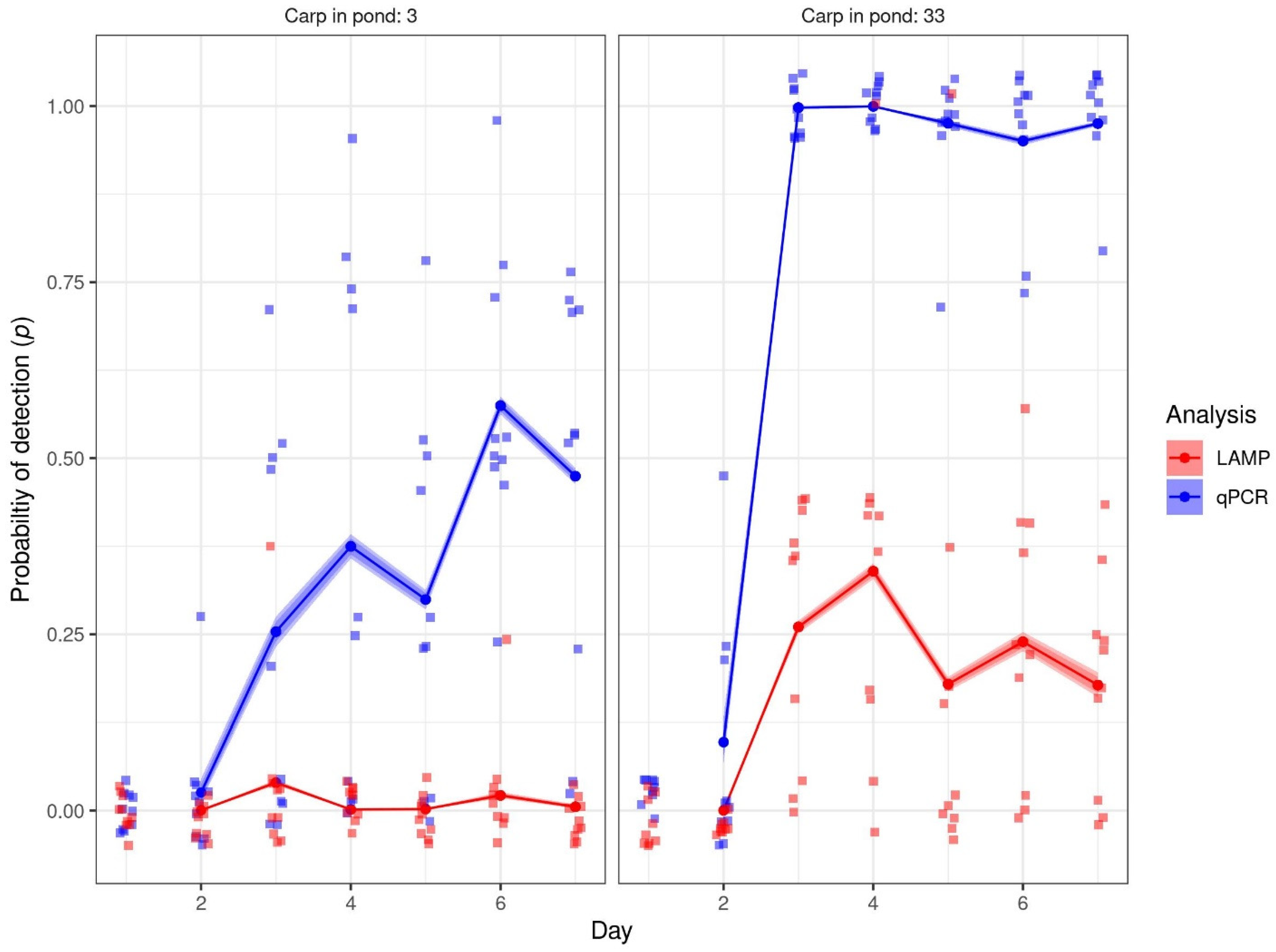
3.3. Mesocosm Validation
3.4. LAMP vs. qPCR
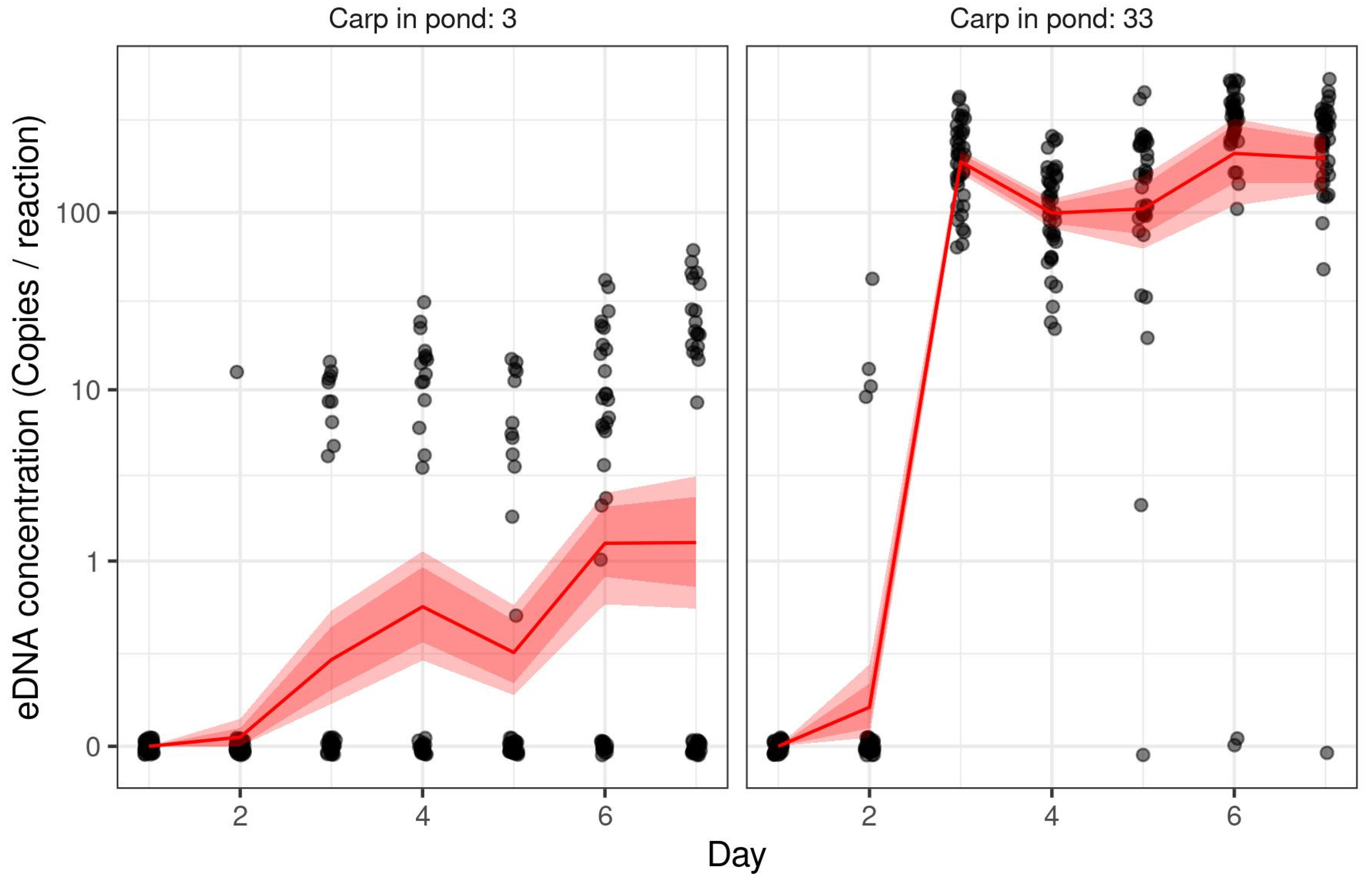
3.5. Accumulation of Grass Carp eDNA over Time
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pimentel, D.; Lach, L.; Zuniga, R.; Morrison, D. Environmental and economic costs of nonindigenous species in the United States. BioScience 2000, 50, 53–65. [Google Scholar] [CrossRef] [Green Version]
- Courtenay, W.R.; Robins, C.R. The grass carp enigma. BioScience 1972, 22, 210. [Google Scholar] [CrossRef]
- Burke, J.S.; Bayne, D.R.; Rea, H. Impact of silver and bighead carps on plankton communities of channel catfish ponds. Aquaculture 1986, 55, 59–68. [Google Scholar] [CrossRef]
- Lieberman, D.M. Use of silver carp (Hypophthalmichthys molotrix) and bighead carp (Aristichthys nobilis) for algae control in a small pond: Changes in water quality. J. Freshw. Ecol. 1996, 11, 391–397. [Google Scholar] [CrossRef]
- DiStefano, R.J.; Litvan, M.E.; Horner, P.T. The bait industry as a potential vector for alien crayfish introductions: Problem recognition by fisheries agencies and a Missouri evaluation. Fisheries 2009, 34, 586–597. [Google Scholar] [CrossRef]
- Drake, D.A.R.; Mandrak, N.E. Ecological risk of live bait fisheries: A new angle on selective fishing. Fisheries 2014, 39, 201–211. [Google Scholar] [CrossRef]
- Drake, D.A.R.; Mandrak, N.E. Bycatch, bait, anglers, and roads: Quantifying vector activity and propagule introduction risk across lake ecosystems. Ecol. Appl. 2014, 24, 877–894. [Google Scholar] [CrossRef]
- Mahon, A.R.; Nathan, L.R.; Jerde, C.L. Meta-genomic surveillance of invasive species in the bait trade. Conserv. Genet. Resour. 2014, 6, 563–567. [Google Scholar] [CrossRef]
- Strecker, A.L.; Campbell, P.M.; Olden, J.D. The aquarium trade as an invasion pathway in the Pacific Northwest. Fisheries 2011, 36, 74–85. [Google Scholar] [CrossRef] [Green Version]
- Patoka, J.; Magalhães, A.L.B.; Kouba, A.; Faulkes, Z.; Jerikho, R.; Vitule, J.R.S. Invasive aquatic pets: Failed policies increase risks of harmful invasions. Biodivers. Conser. 2018, 27, 3037–3046. [Google Scholar] [CrossRef]
- Schloesser, D.W.; Metcalfe-Smith, J.L.; Kovalak, W.P.; Longton, G.D.; Smithee, R.D. Extirpation of freshwater mussels (Bivalvia: Unionidae) following the invasion of dreissenid mussels in an interconnecting river of the Laurentian Great Lakes. Am. Midl. Nat. 2006, 155, 307–320. [Google Scholar] [CrossRef]
- Chakraborti, R.K.; Madon, S.; Kaur, J. Costs for controlling dreissenid mussels affecting drinking water infrastructure: Case studies. J. Am. Water Works Assoc. 2016, 108, E442–E453. [Google Scholar] [CrossRef]
- Prescott, T.H.; Claudi, R.; Prescott, K.L. Impact of dreissenid mussels on the infrastructure of dams and hydroelectric power plants. In Quagga and Zebra Mussels: Biology, Impacts, and Control; Nalepa, T.F., Schloesser, D.W., Eds.; CRC Press: Boca Raton, FL, USA, 2013; pp. 315–329. [Google Scholar]
- Department of Interior, U.S. Fish and Wildlife Service. Injurious Wildlife Species; Listing the Bighead Carp (Hypophthalmichthys nobilis) as Injurious Fish; Department of Interior, U.S. Fish and Wildlife Service: Fairfax, VA, USA, 2011; Volume 50, CFR Part 16, RIN 1018-AT49.
- Department of Interior, U.S. Fish and Wildlife Service. Injurious Wildlife Species; Silver Carp (Hypophthalmichthys molitrix) and Largescale Silver Carp (Hypophthalmichthys harmandi); Department of Interior, U.S. Fish and Wildlife Service: Fairfax, VA, USA, 2007; Volume 50, CFR Part 16, RIN 1018–AT29.
- Department of the Interior, U.S. Fish and Wildlife Service. Injurious Wildlife Species; Black Carp (Mylopharyngodon piceus); Department of Interior, U.S. Fish and Wildlife Service: Fairfax, VA, USA, 2007; Volume 50, CFR Part 16, RIN 1018-AG70.
- US Army Corps of Engineers. The Great Lakes and Mississippi River Interbasin Study—Brandon Road Final Integrated Feasibility Study and Environmental Impact Statement—Will County, Illinois; U.S. Army Corps of Engineers: Chicago, IL, USA, 2018; 516p. [Google Scholar]
- Mack, R.N. Invading Plants: Their Potential Contribution to Population Biology. In Studies on Plant Demography: A Festschrift for John L. Harper; White, J., Ed.; Academic Press: Cambridge, MA, USA, 1985; pp. 127–142. [Google Scholar]
- Kowarik, I. Time lags in biological invasions with regard to the success and failure of alien species. In Plant Invasions-General Aspects and Special Problems; Pysek, P., Prach, K., Rejmanek, M., Wade, M., Eds.; Academic Publishing: Amsterdam, The Netherlands, 1995; pp. 15–38. [Google Scholar]
- Sakai, A.K.; Allendorf, F.W.; Holt, J.S.; Lodge, D.M.; Molofsky, J.; With, K.A.; Baughman, S.; Cabin, R.J.; Cohen, J.E.; Ellstrand, N.C.; et al. The population biology of invasive species. Annu. Rev. Ecol. Syst. 2001, 32, 305–332. [Google Scholar] [CrossRef] [Green Version]
- Mehta, S.V.; Haight, R.G.; Homans, F.R.; Polasky, S.; Venette, R.C. Optimal detection and control strategies for invasive species management. Ecol. Econ. 2007, 61, 237–245. [Google Scholar] [CrossRef]
- Vander Zanden, M.J.; Hansen, G.J.A.; Higgins, S.N.; Kornis, M.S. A pound of prevention, plus a pound of cure: Early detection and eradication of invasive species in the Laurentian Great Lakes. J. Gt. Lakes Res. 2010, 36, 199–205. [Google Scholar] [CrossRef]
- Hayes, K.R.; Cannon, R.; Neil, K.; Inglis, G. Sensitivity and cost considerations for the detection and eradication of marine pests in ports. Mar. Pollut. Bull. 2005, 50, 823–834. [Google Scholar] [CrossRef]
- Jerde, C.L.; Mahon, A.R.; Chadderton, W.L.; Lodge, D.M. “Sight-unseen” detection of rare aquatic species using environmental DNA. Conserv. Lett. 2011, 4, 150–157. [Google Scholar] [CrossRef]
- Piaggio, A.J.; Engeman, R.M.; Hopken, M.W.; Humphrey, J.S.; Keacher, K.L.; Bruce, W.E.; Avery, M.L. Detecting an elusive invasive species: A diagnostic pcr to detect burmese python in Florida waters and an assessment of persistence of environmental DNA. Mol. Ecol. Resour. 2014, 14, 374–380. [Google Scholar] [CrossRef] [Green Version]
- Erickson, R.A.; Merkes, C.M.; Mize, E.L. Sampling designs for landscape-level eDNA monitoring programs. Integr. Environ. Assess. Manag. 2019, 15, 760–771. [Google Scholar] [CrossRef]
- Lor, Y.; Schreier, T.M.; Waller, D.L.; Merkes, C.M. Using environmental DNA (eDNA) to detect the endangered spectaclecase mussel (Margaritifera monodonta). Freshw. Sci. 2020, 39, 837–847. [Google Scholar] [CrossRef]
- Morisette, J.; Burgiel, S.; Brantley, K.; Daniel, W.M.; Darling, J.; Davis, J.; Franklin, T.; Gaddis, K.; Hunter, M.; Lance, R.; et al. Strategic considerations for invasive species managers in the utilization of environmental DNA (eDNA): Steps for incorporating this powerful surveillance tool. Manag. Biol. Invasions 2021, 12, 747–775. [Google Scholar] [CrossRef] [PubMed]
- Ficetola, G.F.; Miaud, C.; Pompanon, F.; Taberlet, P. Species detection using environmental DNA from water samples. Biol. Lett. 2008, 4, 423–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldberg, C.S.; Pilliod, D.S.; Arkle, R.S.; Waits, L.P. Molecular detection of vertebrates in stream water: A demonstration using Rocky Mountain tailed frogs and Idaho giant salamanders. PLoS ONE 2011, 6, e22746. [Google Scholar] [CrossRef] [PubMed]
- Guan, X.; Monroe, E.M.; Bockrath, K.D.; Mize, E.L.; Rees, C.B.; Lindsay, D.L.; Baerwaldt, K.L.; Nico, L.G.; Lance, R.F. Environmental DNA (eDNA) assays for invasive populations of black carp in North America. Trans. Am. Fish. Soc. 2019, 148, 1043–1055. [Google Scholar] [CrossRef] [Green Version]
- Hayer, C.-A.; Bayless, M.F.; George, A.; Thompson, N.; Richter, C.A.; Chapman, D.C. Use of environmental DNA to detect grass carp spawning events. Fishes 2020, 5, 27. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef] [Green Version]
- Kaneko, H.; Kawana, T.; Fukushima, E.; Suzutani, T. Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J. Biochem. Biophys. Methods 2007, 70, 499–501. [Google Scholar] [CrossRef]
- Francois, P.; Tangomo, M.; Hibbs, J.; Bonetti, E.-J.; Boehme, C.C.; Notomi, T.; Perkins, M.D.; Schrenzel, J. Robustness of a loop-mediated isothermal amplification reaction for diagnostic applications. FEMS Immunol. Med. Microbiol. 2011, 62, 41–48. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.; Wang, F.; Prinyawiwatkul, W.; Ge, B. Robustness of Salmonella loop-mediated isothermal amplification assays for food applications. J. Appl. Microbiol. 2014, 116, 81–88. [Google Scholar] [CrossRef]
- Kurosaki, Y.; Takada, A.; Ebihara, H.; Grolla, A.; Kamo, N.; Feldmann, H.; Kawaoka, Y.; Yasuda, J. Rapid and simple detection of Ebola virus by reverse transcription-loop-mediated isothermal amplification. J. Virol. Methods 2007, 141, 78–83. [Google Scholar] [CrossRef]
- Sirichaisinthop, J.; Buates, S.; Watanabe, R.; Han, E.-T.; Suktawonjaroenpon, W.; Krasaesub, S.; Takeo, S.; Tsuboi, T.; Sattabongkot, J. Evaluation of loop-mediated isothermal amplification (LAMP) for malaria diagnosis in a field setting. Am. J. Trop. Med. Hyg. 2011, 85, 594–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahn, S.J.; Baek, Y.H.; Lloren, K.K.S.; Choi, W.-S.; Jeong, J.H.; Antigua, K.J.C.; Kwon, H.; Park, S.-J.; Kim, E.-H.; Kim, Y.; et al. Rapid and simple colorimetric detection of multiple influenza viruses infecting humans using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. BMC Infect. Dis. 2019, 19, 676. [Google Scholar] [CrossRef] [Green Version]
- Park, G.-S.; Ku, K.; Baek, S.-H.; Kim, S.-J.; Kim, S.I.; Kim, B.-T.; Maeng, J.-S. Development of reverse transcription loop-mediated isothermal amplification assays targeting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). J. Mol. Diagn. 2020, 22, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.-I.; Park, N.-Y.; Cho, H.-S. Detection of canine parvovirus in fecal samples using loop-mediated isothermal amplification. J. Vet. Diagn. Investig. 2006, 18, 81–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Cui, S. Detection of porcine parvovirus by loop-mediated isothermal amplification. J. Virol. Methods 2009, 155, 122–125. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Zhang, Y.; Zhang, H.; Zhou, Y.; Cao, J.; Zhou, J. Comparison of loop-mediated isothermal amplification (LAMP) and real-time PCR method targeting a 529-bp repeat element for diagnosis of toxoplasmosis. Vet. Parasitol. 2012, 185, 296–300. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, J.A.; Boonham, N.; Dickinson, M. Development and evaluation of a one-hour DNA extraction and loop-mediated isothermal amplification assay for rapid detection of phytoplasmas. Plant Pathol. 2010, 59, 465–471. [Google Scholar] [CrossRef]
- Niu, J.H.; Jian, H.; Guo, Q.X.; Chen, C.L.; Wang, X.Y.; Liu, Q.; Guo, Y.D. Evaluation of loop-mediated isothermal amplification (LAMP) assays based on 5S rDNA-IGS2 regions for detecting Meloidogyne enterolobii. Plant Pathol. 2012, 61, 809–819. [Google Scholar] [CrossRef]
- Kyei-Poku, G.; Gauthier, D.; Quan, G. Development of a loop-mediated isothermal amplification assay as an early-warning tool for detecting emerald ash borer (Coleoptera: Buprestidae) incursions. J. Econ. Entomol. 2020, 113, 2480–2494. [Google Scholar] [CrossRef]
- Zhou, D.; Guo, J.; Xu, L.; Gao, S.; Lin, Q.; Wu, Q.; Wu, L.; Que, Y. Establishment and application of a loop-mediated isothermal amplification (LAMP) system for detection of Cry1Ac transgenic sugarcane. Sci. Rep. 2014, 4, 4912. [Google Scholar] [CrossRef] [Green Version]
- Abdulmawjood, A.; Grabowski, N.; Fohler, S.; Kittler, S.; Nagengast, H.; Klein, G. Development of loop-mediated isothermal amplification (LAMP) assay for rapid and sensitive identification of ostrich meat. PLoS ONE 2014, 9, e100717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aartse, A.; Scholtens, I.M.J.; van der A, H.J.G.; Boersma-Greve, M.M.; Prins, T.W.; van Ginkel, L.A.; Kok, E.J.; Bovee, T.F.H. Evaluation of a loop-mediated isothermal amplification (lamp) method for rapid on-site detection of horse meat. Food Control 2017, 81, 9–15. [Google Scholar] [CrossRef]
- Sul, S.; Kim, M.-J.; Kim, H.-Y. Development of a direct loop-mediated isothermal amplification (LAMP) assay for rapid and simple on-site detection of chicken in processed meat products. Food Control 2019, 98, 194–199. [Google Scholar] [CrossRef]
- Da Silva, S.J.R.; Pardee, K.; Pena, L. Loop-mediated isothermal amplification (LAMP) for the diagnosis of Zika virus: A review. Viruses 2020, 12, 19. [Google Scholar] [CrossRef] [Green Version]
- Nakajima, N.; Sakamoto, Y.; Goka, K. Rapid detection of the red fire ant Solenopsis invicta (Hymenoptera: Formicidae) by loop-mediated isothermal amplification. Appl. Entomol. Zool. 2019, 54, 319–322. [Google Scholar] [CrossRef] [Green Version]
- Centeno-Cuadros, A.; Abbasi, I.; Nathan, R. Sex determination in the wild: A field application of loop-mediated isothermal amplification successfully determines sex across three raptor species. Mol. Ecol. Resour. 2017, 17, 153–160. [Google Scholar] [CrossRef] [Green Version]
- Williams, M.R.; Stedtfeld, R.D.; Engle, C.; Salach, P.; Fakher, U.; Stedtfeld, T.; Dreelin, E.; Stevenson, R.J.; Latimore, J.; Hashsham, S.A. Isothermal amplification of environmental DNA (eDNA) for direct field-based monitoring and laboratory confirmation of Dreissena sp. PLoS ONE 2017, 12, e0186462. [Google Scholar] [CrossRef]
- Williams, K.E.; Huyvaert, K.P.; Piaggio, A.J. Clearing muddied waters: Capture of environmental DNA from turbid waters. PLoS ONE 2017, 12, e0179282. [Google Scholar] [CrossRef]
- Abbaszadegan, M.; Huber, M.; Gerba, C.; Pepper, L. Detection of enteroviruses in groundwater with the polymerase chain reaction. Appl. Environ. Microbiol. 1993, 59, 1318–1324. [Google Scholar] [CrossRef] [Green Version]
- Ijzerman, M.M.; Dahling, D.R.; Fout, G.S. A method to remove environmental inhibitors prior to the detection of waterborne enteric viruses by reverse transcription-polymerase chain reaction. J. Virol. Methods 1997, 63, 145–153. [Google Scholar] [CrossRef]
- Matheson, C.; Gurney, C.; Esau, N.; Lehto, R. Assessing PCR inhibition from humic substances. Open Enzym. Inhib. J. 2010, 3, 38. [Google Scholar] [CrossRef] [Green Version]
- Farrington, H.L.; Edwards, C.E.; Guan, X.; Carr, M.R.; Baerwaldt, K.; Lance, R.F. Mitochondrial genome sequencing and development of genetic markers for the detection of DNA of invasive bighead and silver carp (Hypophthalmichthys nobilis and H. molitrix) in environmental water samples from the United States. PLoS ONE 2015, 10, e0117803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erickson, R.A.; Merkes, C.M.; Jackson, C.A.; Goforth, R.R.; Amberg, J.J. Seasonal trends in eDNA detection and occupancy of bigheaded carps. J. Gt. Lakes Res. 2017, 43, 762–770. [Google Scholar] [CrossRef]
- Mahon, A.R.; Jerde, C.L.; Galaska, M.; Bergner, J.L.; Chadderton, W.L.; Lodge, D.M.; Hunter, M.E.; Nico, L.G. Validation of eDNA surveillance sensitivity for detection of Asian carps in controlled and field experiments. PLoS ONE 2013, 8, e58316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, M.; Belliveau, V.; Mandrak, N.E.; Gagné, N. Development of environmental DNA (eDNA) methods for detecting high-risk freshwater fishes in live trade in Canada. Biol. Invasions 2018, 20, 299–314. [Google Scholar] [CrossRef]
- Nathan, L.R.; Jerde, C.L.; Budny, M.L.; Mahon, A.R. The use of environmental DNA in invasive species surveillance of the Great Lakes commercial bait trade. Conserv. Biol. 2015, 29, 430–439. [Google Scholar] [CrossRef]
- Klymus, K.E.; Merkes, C.M.; Allison, M.J.; Goldberg, C.S.; Helbing, C.C.; Hunter, M.E.; Jackson, C.A.; Lance, R.F.; Mangan, A.M.; Monroe, E.M.; et al. Reporting the limits of detection and quantification for environmental DNA assays. Environ. DNA 2020, 2, 271–282. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Dorazio, R.M.; Erickson, R.A. Ednaoccupancy: An R package for multiscale occupancy modelling of environmental DNA data. Mol. Ecol. Resour. 2017, 18, 368–380. [Google Scholar] [CrossRef]
- Mize, E.L.; Erickson, R.A.; Merkes, C.M.; Berndt, N.; Bockrath, K.; Credico, J.; Grueneis, N.; Merry, J.; Mosel, K.; Tuttle-Lau, M.; et al. Refinement of eDNA as an early monitoring tool at the landscape-level: Study design considerations. Ecol. Appl. 2019, 29, e01951. [Google Scholar] [CrossRef]
- Stan Development Team. RStan: The R Interface to Stan, Version 2.21.2; 2020. Available online: http://mc-stan.org/ (accessed on 15 November 2022).
- Erickson, R.A.; Kageyama, S.A. Analysis of Grass Carp eDNA Data, U.S. Geological Survey Software Release; U.S. Geological Survey: Reston, VA, USA, 2022. [CrossRef]
- Kageyama, S.A.; Hoogland, M.R.; Tajjioui, T.; Schreier, T.M.; Erickson, R.A.; Merkes, C.M. Data Release for Validation of a Portable eDNA Detection Kit for Invasive Carps: U.S. Geological Survey Data Release; U.S. Geological Survey: Reston, VA, USA, 2022. [CrossRef]
- Wickham, H.; Navarro, D.; Pedersen, T.L. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Khan, M.; Wang, R.; Li, B.; Liu, P.; Weng, Q.; Chen, Q. Comparative evaluation of the LAMP assay and PCR-based assays for the rapid detection of Alternaria solani. Front. Microbiol. 2018, 9, 2089. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Lowe, S.B.; Gooding, J.J. Brief review of monitoring methods for loop-mediated isothermal amplification (LAMP). Biosens. Bioelectron. 2014, 61, 491–499. [Google Scholar] [CrossRef]
- Okiro, L.A.; Tancos, M.A.; Nyanjom, S.G.; Smart, C.D.; Parker, M.L. Comparative evaluation of LAMP, qPCR, conventional PCR, and ELISA to detect Ralstonia solanacearum in Kenyan potato fields. Plant Dis. 2019, 103, 959–965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anklam, K.; Kulow, M.; Yamazaki, W.; Döpfer, D. Development of real-time PCR and loop-mediated isothermal amplification (LAMP) assays for the differential detection of digital dermatitis associated treponemes. PLoS ONE 2017, 12, e0178349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahara, T.; Minamoto, T.; Yamanaka, H.; Doi, H.; Kawabata, Z. Estimation of fish biomass using environmental DNA. PLoS ONE 2012, 7, e35868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jane, S.F.; Wilcox, T.M.; McKelvey, K.S.; Young, M.K.; Schwartz, M.K.; Lowe, W.H.; Letcher, B.H.; Whiteley, A.R. Distance, flow and PCR inhibition: eDNA dynamics in two headwater streams. Mol. Ecol. Resour. 2015, 15, 216–227. [Google Scholar] [CrossRef]
- Klymus, K.E.; Richter, C.A.; Chapman, D.C.; Paukert, C. Quantification of eDNA shedding rates from invasive bighead carp Hypophthalmichthys nobilis and silver carp Hypophthalmichthys molitrix. Biol. Conserv. 2015, 183, 77–84. [Google Scholar] [CrossRef]
- Doi, H.; Uchii, K.; Takahara, T.; Matsuhashi, S.; Yamanaka, H.; Minamoto, T. Use of droplet digital PCR for estimation of fish abundance and biomass in environmental DNA surveys. PLoS ONE 2015, 10, e0122763. [Google Scholar] [CrossRef] [Green Version]
- Coulter, D.P.; Wang, P.; Coulter, A.A.; Van Susteren, G.E.; Eichmiller, J.J.; Garvey, J.E.; Sorensen, P.W. Nonlinear relationship between silver carp density and their eDNA concentration in a large river. PLoS ONE 2019, 14, e0218823. [Google Scholar] [CrossRef] [PubMed]
- Schultz, M.T.; Lance, R.F. Modeling the sensitivity of field surveys for detection of environmental DNA (eDNA). PLoS ONE 2015, 10, e0141503. [Google Scholar] [CrossRef] [Green Version]
- Sepulveda, A.J.; Schabacker, J.; Smith, S.; Al-Chokhachy, R.; Luikart, G.; Amish, S.J. Improved detection of rare, endangered and invasive trout in using a new large-volume sampling method for eDNA capture. Environ. DNA 2019, 1, 227–237. [Google Scholar] [CrossRef]
- Rees, H.C.; Maddison, B.C.; Middleditch, D.J.; Patmore, J.R.M.; Gough, K.C. REVIEW: The detection of aquatic animal species using environmental DNA—A review of eDNA as a survey tool in ecology. J. Appl. Ecol. 2014, 51, 1450–1459. [Google Scholar] [CrossRef]
- Eichmiller, J.J.; Bajer, P.G.; Sorensen, P.W. The relationship between the distribution of common carp and their environmental DNA in a small lake. PLoS ONE 2014, 9, e112611. [Google Scholar] [CrossRef] [Green Version]
- Maruyama, A.; Nakamura, K.; Yamanaka, H.; Kondoh, M.; Minamoto, T. The release rate of environmental DNA from juvenile and adult fish. PLoS ONE 2014, 9, e114639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maruyama, A.; Nakamura, K.; Yamanaka, H.; Kondoh, M.; Minamoto, T. Correction: The release rate of environmental DNA from juvenile and adult fish. PLoS ONE 2019, 14, e0212145. [Google Scholar] [CrossRef] [PubMed]
- Colle, D.E.; Shireman, J.V.; Rottmann, R.W. Food selection by grass carp fingerlings in a vegetated pond. Trans. Am. Fish. Soc. 1978, 107, 149–152. [Google Scholar] [CrossRef]
- Cui, Y.; Wootton, R.J. Bioenergetics of growth of a cyprinid, Phoxinus phoxinus: The effect of ration, temperature and body size on food consumption, faecal production and nitrogenous excretion. J. Fish Biol. 1988, 33, 431–443. [Google Scholar] [CrossRef]
- Bettoli, P.W.; Neill, W.H.; Kelsch, S.W. Temperature preference and heat resistance of grass carp, Ctenopharyngodon idella (Valenciennes), bighead carp, Hypophthalmichthys nobilis (Gray), and their F1 hybrid. J. Fish Biol. 1985, 27, 239–247. [Google Scholar] [CrossRef]
- Sepulveda, A.J.; Hutchins, P.R.; Massengill, R.L.; Dunker, K.J. Tradeoffs of a portable, field-based environmental DNA platform for detecting invasive northern pike (Esox lucius) in Alaska. Manag. Biol. Invasions 2018, 9, 253–258. [Google Scholar] [CrossRef]
- Thomas, A.C.; Tank, S.; Nguyen, P.L.; Ponce, J.; Sinnesael, M.; Goldberg, C.S. A system for rapid eDNA detection of aquatic invasive species. Environ. DNA 2020, 2, 261–270. [Google Scholar] [CrossRef] [Green Version]
- Enosawa, M.; Kageyama, S.; Sawai, K.; Watanabe, K.; Notomi, T.; Onoe, S.; Mori, Y.; Yokomizo, Y. Use of loop-mediated isothermal amplification of the IS900 sequence for rapid detection of cultured Mycobacterium avium subsp. paratuberculosis. J. Clin. Microbiol. 2003, 41, 4359–4365. [Google Scholar] [CrossRef] [Green Version]
- Kiddle, G.; Hardinge, P.; Buttigieg, N.; Gandelman, O.; Pereira, C.; McElgunn, C.J.; Rizzoli, M.; Jackson, R.; Appleton, N.; Moore, C.; et al. GMO detection using a bioluminescent real time reporter (BART) of loop mediated isothermal amplification (LAMP) suitable for field use. BMC Biotechnol. 2012, 12, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Litvak, M.K.; Mandrak, N.E. Ecology of freshwater baitfish use in Canada and the United States. Fisheries 1993, 18, 6–13. [Google Scholar] [CrossRef]
- Ludwig, H.R., Jr.; Leitch, J.A. Interbasin transfer of aquatic biota via anglers’ bait buckets. Fisheries 1996, 21, 14–18. [Google Scholar] [CrossRef]
- Kilian, J.V.; Klauda, R.J.; Widman, S.; Kashiwagi, M.; Bourquin, R.; Weglein, S.; Schuster, J. An assessment of a bait industry and angler behavior as a vector of invasive species. Biol. Invasions 2012, 14, 1469–1481. [Google Scholar] [CrossRef]
- Anderson, L.G.; White, P.C.L.; Stebbing, P.D.; Stentiford, G.D.; Dunn, A.M. Biosecurity and vector behaviour: Evaluating the potential threat posed by anglers and canoeists as pathways for the spread of invasive non-native species and pathogens. PLoS ONE 2014, 9, e92788. [Google Scholar] [CrossRef] [PubMed]
- Conover, G.; Simmonds, R.; Whalen, M. Management and Control Plan for Bighead, Black, Grass, and Silver Carps in the United States; Asian Carp Working Group, Aquatic Nuisance Species Task Force: Washington, DC, USA, 2007; 223p.
- Snyder, M.R.; Stepien, C.A.; Marshall, N.T.; Scheppler, H.B.; Black, C.L.; Czajkowski, K.P. Detecting aquatic invasive species in bait and pond stores with targeted environmental (e)DNA high-throughput sequencing metabarcode assays: Angler, retailer, and manager implications. Biol. Conserv. 2020, 245, 108430. [Google Scholar] [CrossRef]


| Assay Type | Oligo | Target Gene | Sequence (5′-3′) | Target Size (bp) |
|---|---|---|---|---|
| LAMP | AC1-F3 | 16S (invasive carp) | TTCCCCTAACARTATCAGGCT | 215 |
| LAMP | AC1-FIP | AGTGGTTTGTCCGATCTGGTCATGGAAGAAATTATGCTAAAATG | ||
| LAMP | AC1-FLP | CTTGGAGAAGAGCAGGTCT | ||
| LAMP | AC1-B3 | TAGCACTCCRGTGTGGGGT | ||
| LAMP | AC1-BIP | ATTAACGAACTCAACCCAAGAACGATTGTTTAATTGTGGGTTT | ||
| LAMP | AC1-BLP | GAGTAATGTRAAYAACAAAAAAACC | ||
| LAMP | 16S-F3 | 16S (bacteria) | AAGCCTGATGCAGCCATGC | 206 |
| LAMP | 16S-FIP | CGGGTAACGTCAATGAGCAAAGGGTATGAAGAAGGCCTTCGGG | ||
| LAMP | 16S-FLP | CCTTCCTCCCCGCTGAAAGTAC | ||
| LAMP | 16S-B3 | CGCCTGCGTGCGCTTTAC | ||
| LAMP | 16S-BIP | AAGCACCGGCTAACTCCGTGCCAGTAATTCCGATTAACGCTTGC | ||
| LAMP | 16S-BLP | AGCAGCCGCGGTAATACGGAG | ||
| PCR | ND2-F60 | ND2 | AATCAATACCTTAGCAATCATTCCA | 157 |
| PCR | ND2-R60 | TATTTATATCTCACTCTCCTGTAAT | ||
| Probe | ND2-probe | 56-FAM/AATAGCCCA/ZEN/ACACCACCACCCTC/3IABkFQ |
| Family | Species | Common Name | Result |
|---|---|---|---|
| Acipenseridae | Acipenser fulvescens | Lake sturgeon | − |
| Acipenseridae | Scaphirhynchus albus | Pallid sturgeon | − |
| Catostomidae | Ictiobus cyprinellus | Bigmouth buffalo | − |
| Centrarchidae | Lepomis macrochirus | Bluegill | − |
| Centrarchidae | Micropterus salmoides | Largemouth bass | − |
| Cichlidae | Oreochromis aureus × Oreochromis niloticus | Tilapia | − |
| Clupeidae | Dorosoma cepedianum | Gizzard shad | − |
| Cyprinidae | Ctenopharygodon idella | Grass carp | + |
| Cyprinidae | Cyprinella spiloptera | Spotfin shiner | − |
| Cyprinidae | Cyprinus carpio | Common carp | − |
| Cyprinidae | Hypophthalmichthys molitrix | Silver carp | + |
| Cyprinidae | Hypophthalmichthys nobilis | Bighead carp | + |
| Cyprinidae | Mylopharyngodon piceus | Black carp | + |
| Cyprinidae | Notemigonus crysoleucas | Golden shiner | − |
| Cyprinidae | Pimephales promelas | Fathead minnow | − |
| Ictaluridae | Ictalurus punctatus | Channel catfish | − |
| Percidae | Perca flavescens | Yellow perch | − |
| Percidae | Sander vitreus | Walleye | − |
| Petromyzontidae | Petromyzon marinus | Sea lamprey | − |
| Poeciliidae | Gambusia affinis | Mosquitofish | − |
| Polyodontidae | Polyodon spathula | Paddlefish | − |
| Salmonidae | Oncorhynchus mykiss | Rainbow trout | − |
| Salmonidae | Salvelinus fontinalis | Brook trout | − |
| Salmonidae | Salvelinus namaycush | Lake trout | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kageyama, S.A.; Hoogland, M.R.; Tajjioui, T.; Schreier, T.M.; Erickson, R.A.; Merkes, C.M. Validation of a Portable eDNA Detection Kit for Invasive Carps. Fishes 2022, 7, 363. https://doi.org/10.3390/fishes7060363
Kageyama SA, Hoogland MR, Tajjioui T, Schreier TM, Erickson RA, Merkes CM. Validation of a Portable eDNA Detection Kit for Invasive Carps. Fishes. 2022; 7(6):363. https://doi.org/10.3390/fishes7060363
Chicago/Turabian StyleKageyama, Stacie A., Matthew R. Hoogland, Tariq Tajjioui, Theresa M. Schreier, Richard A. Erickson, and Christopher M. Merkes. 2022. "Validation of a Portable eDNA Detection Kit for Invasive Carps" Fishes 7, no. 6: 363. https://doi.org/10.3390/fishes7060363






