A Novel C-Type Lectin and Its Potential Role in Feeding and Feed Selection in Ruditapes philippinarum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Clams and Microalgae
2.2. Cloning of Rpcl cDNA
2.3. Sequence Analysis of Rpcl
2.4. qRT-PCR
2.5. Recombinant Rpcl Protein Expression
2.6. Agglutination Activity of Rpcl
2.7. Expression of Rpcl in the Lip and Gill of R. philippinarum after Feeding Microalgae
2.8. Statistical Methods
3. Results
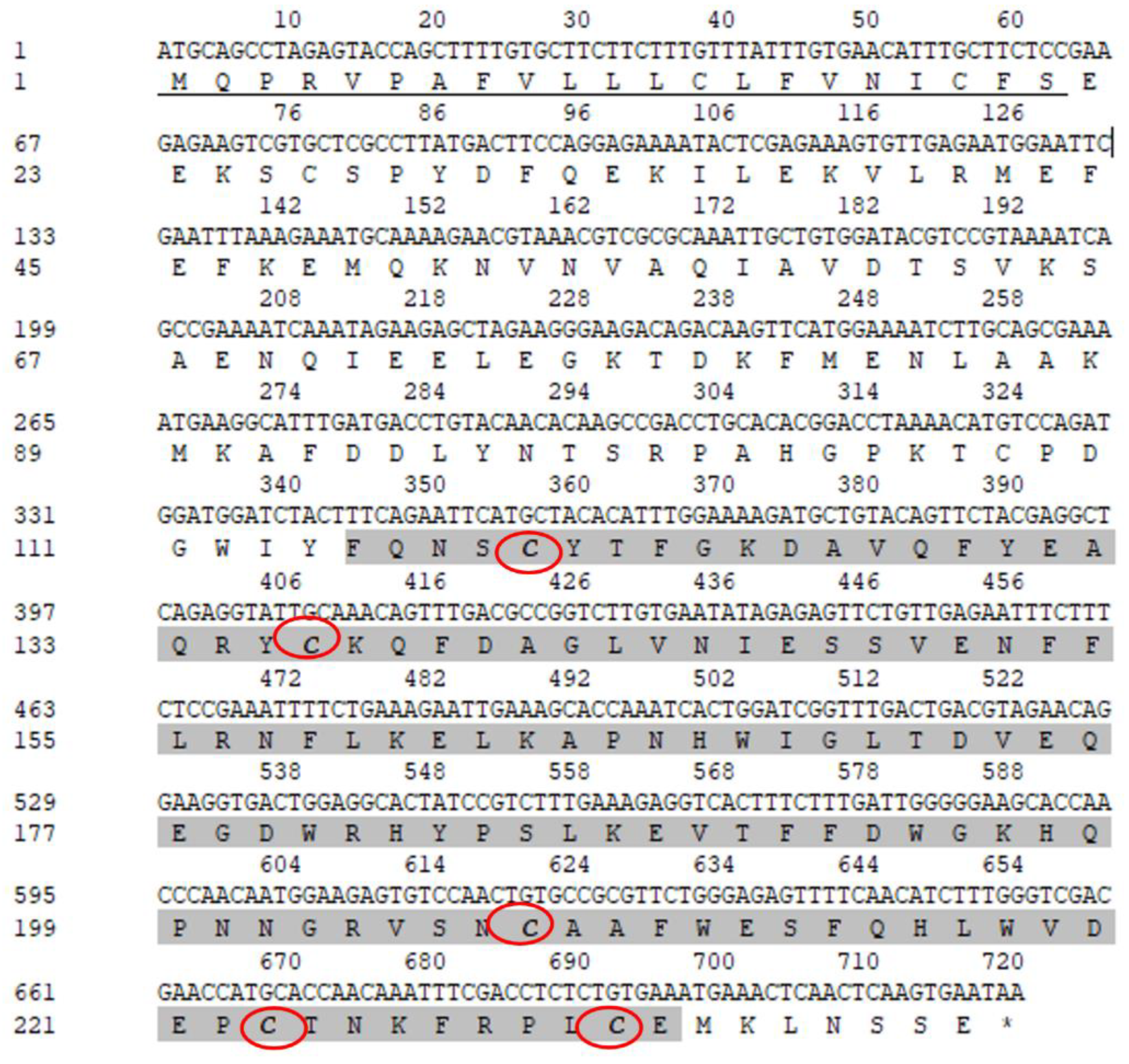
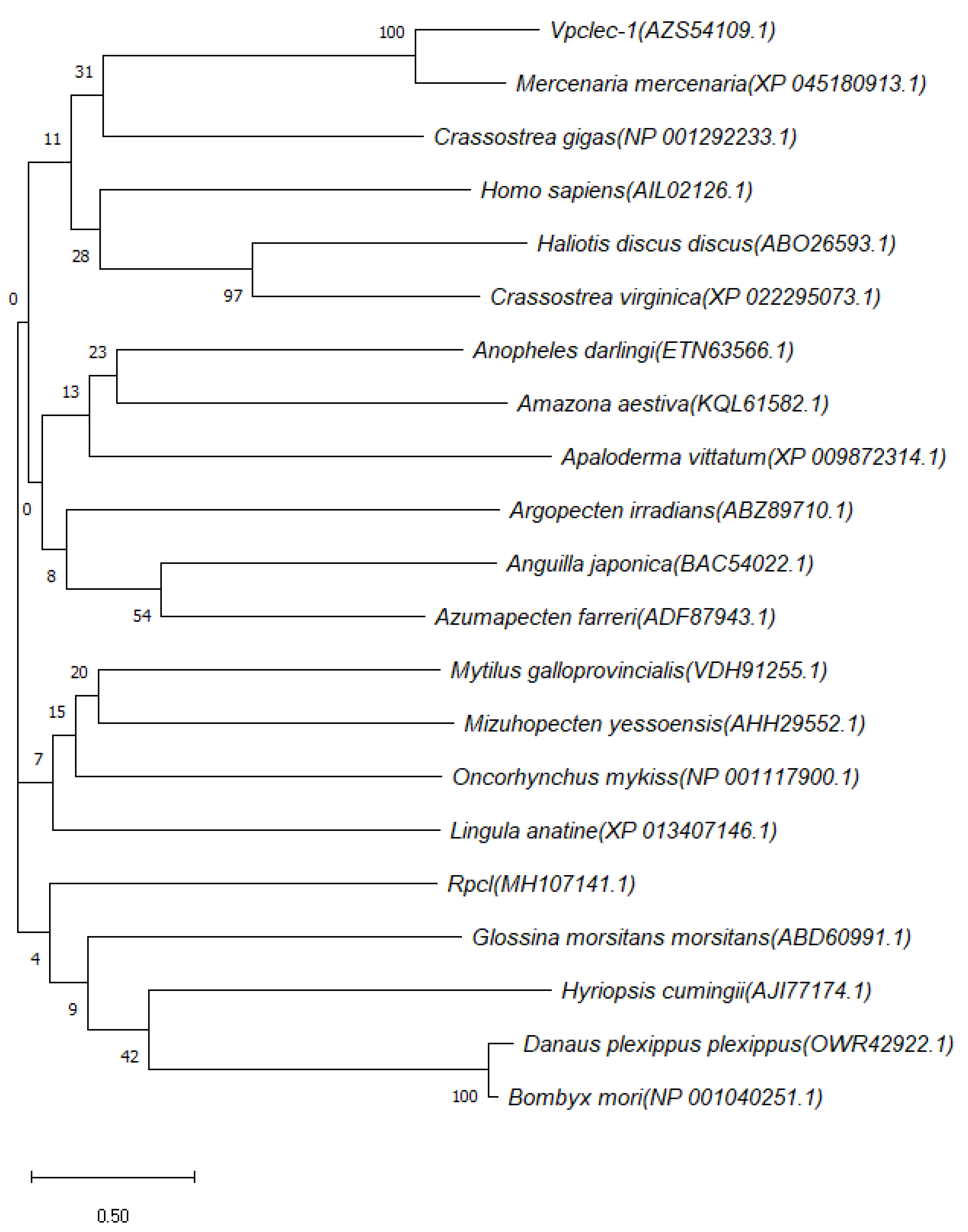
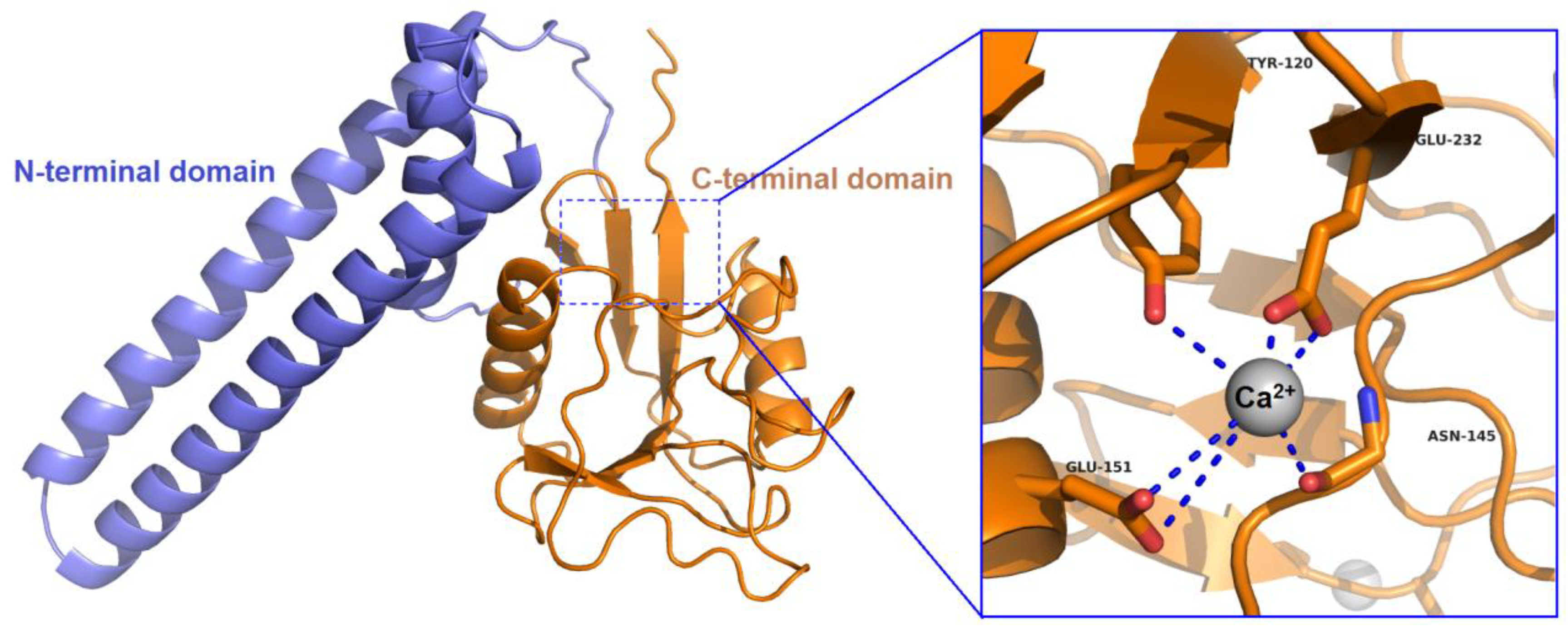
3.1. Molecular Characterization of Rpcl
3.2. Tissue Distribution of Rpcl
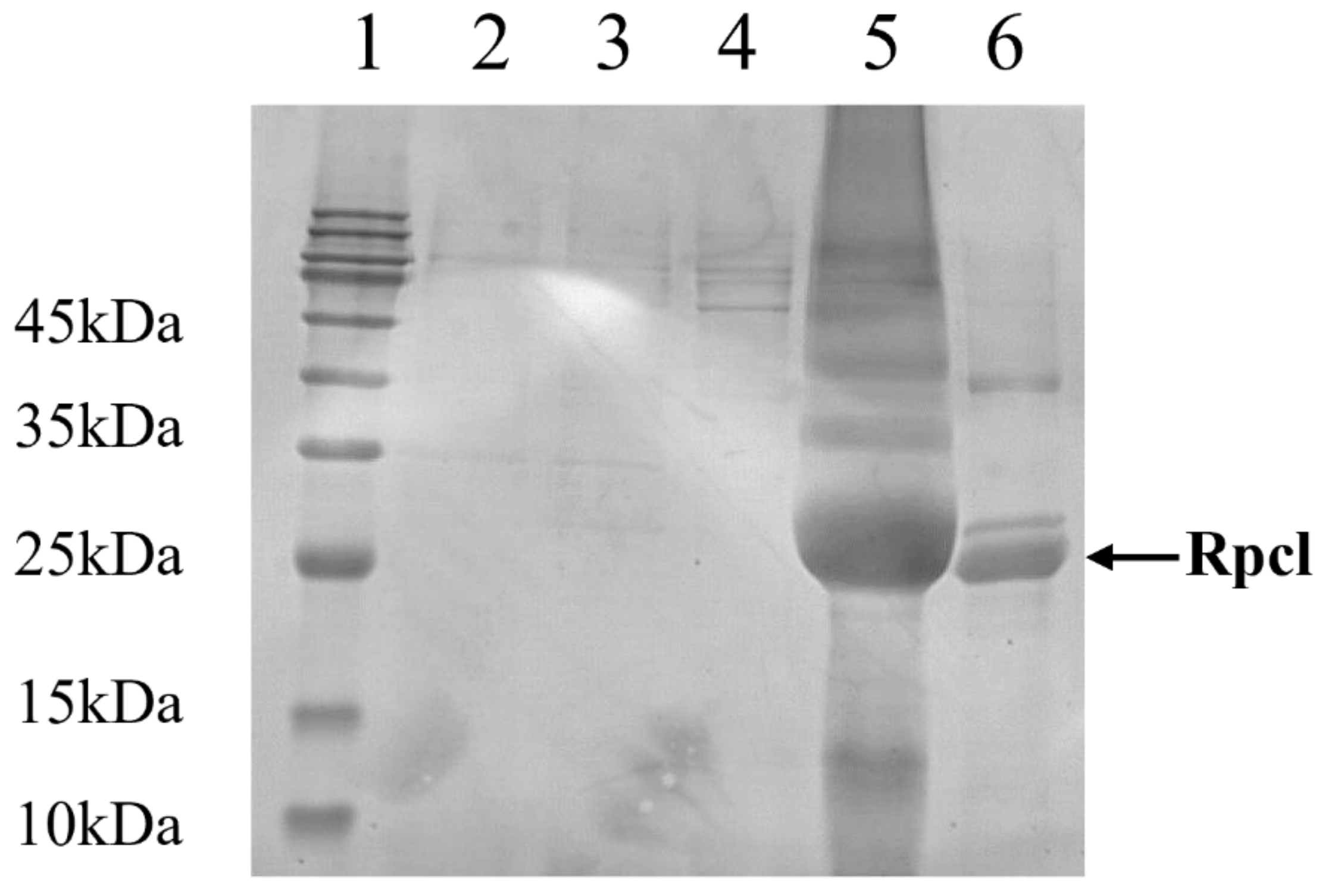
3.3. Heterologous Expression of Rpcl
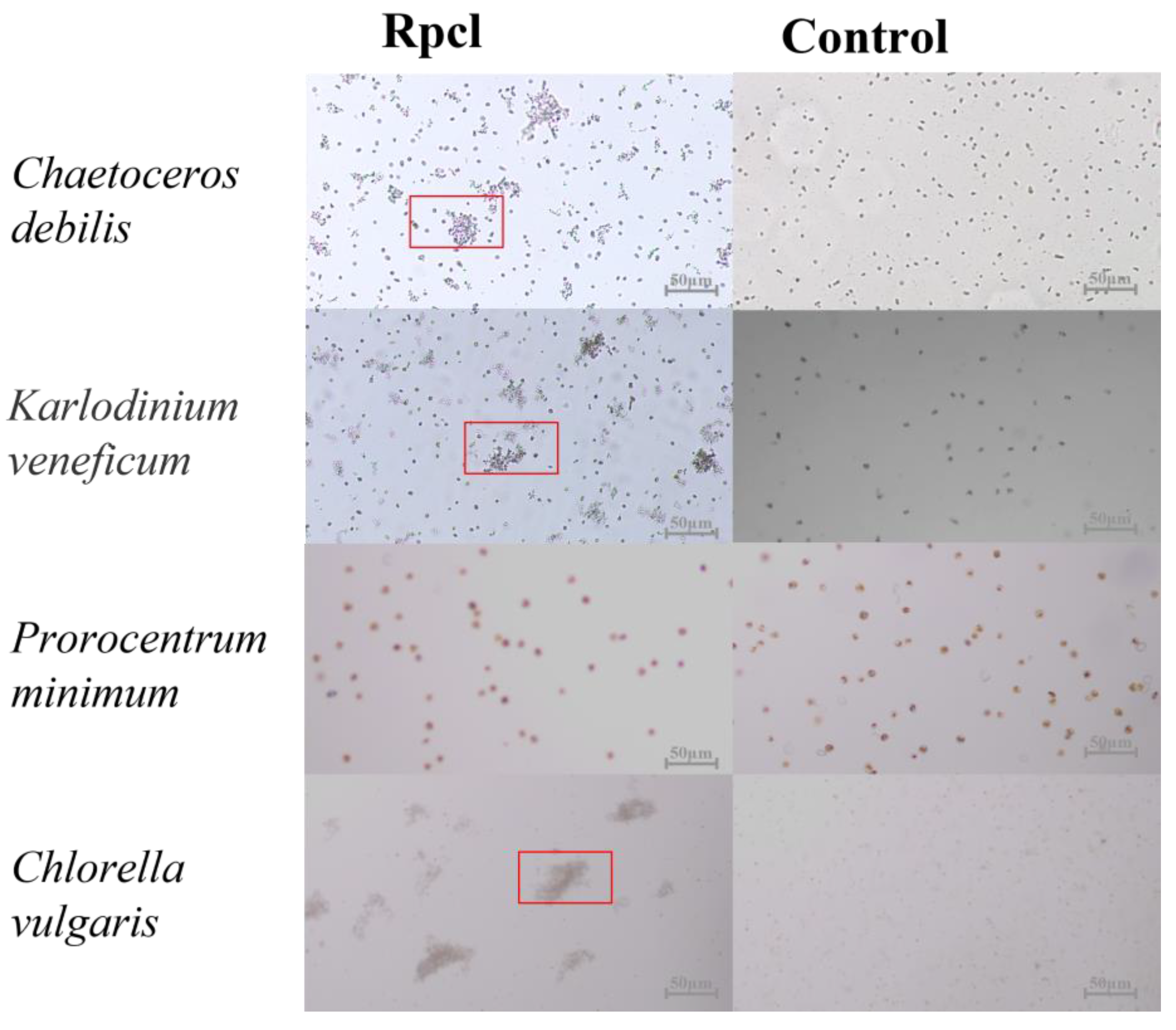
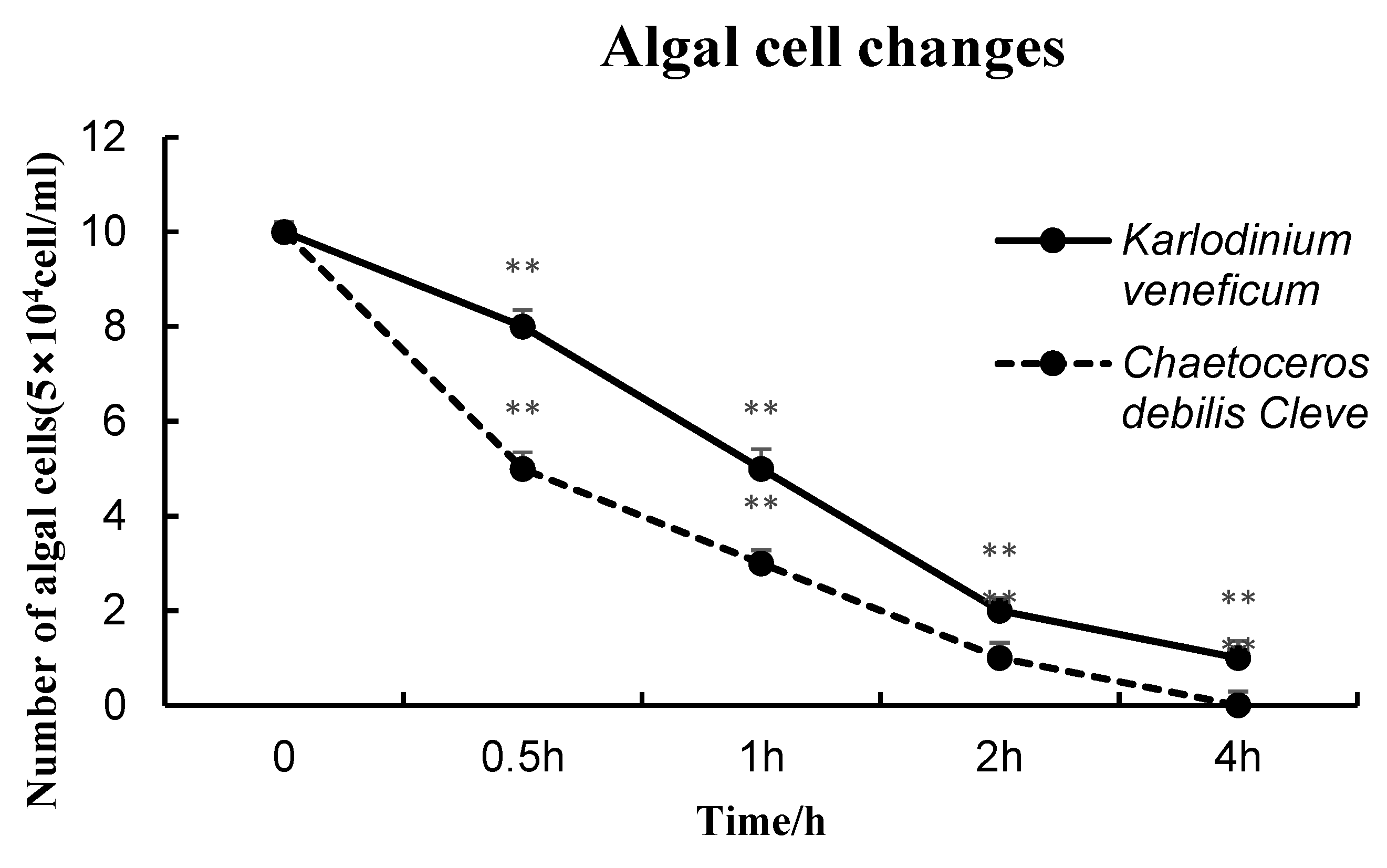
3.4. Microalgae Agglutinating Activity of Rpcl
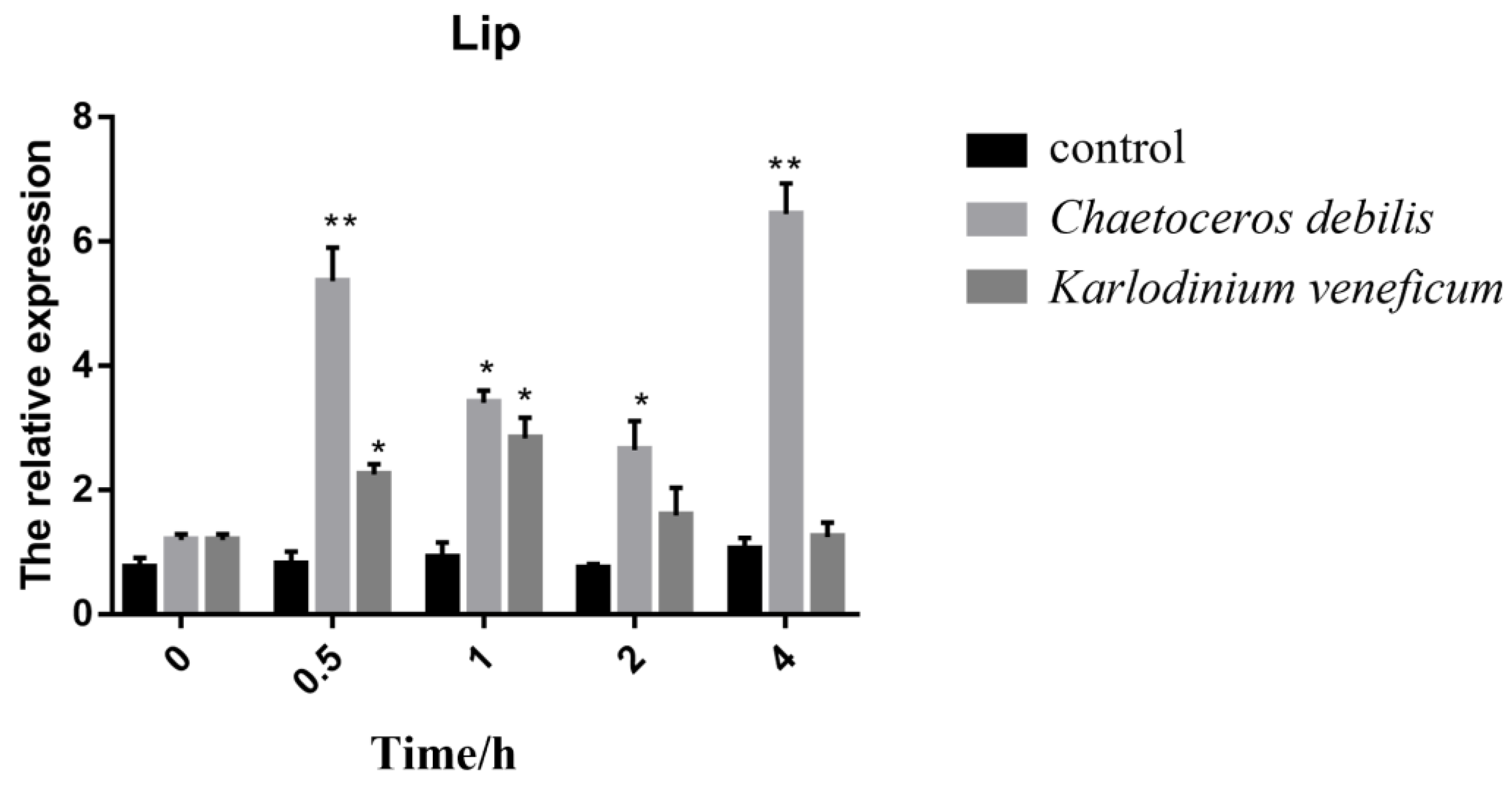
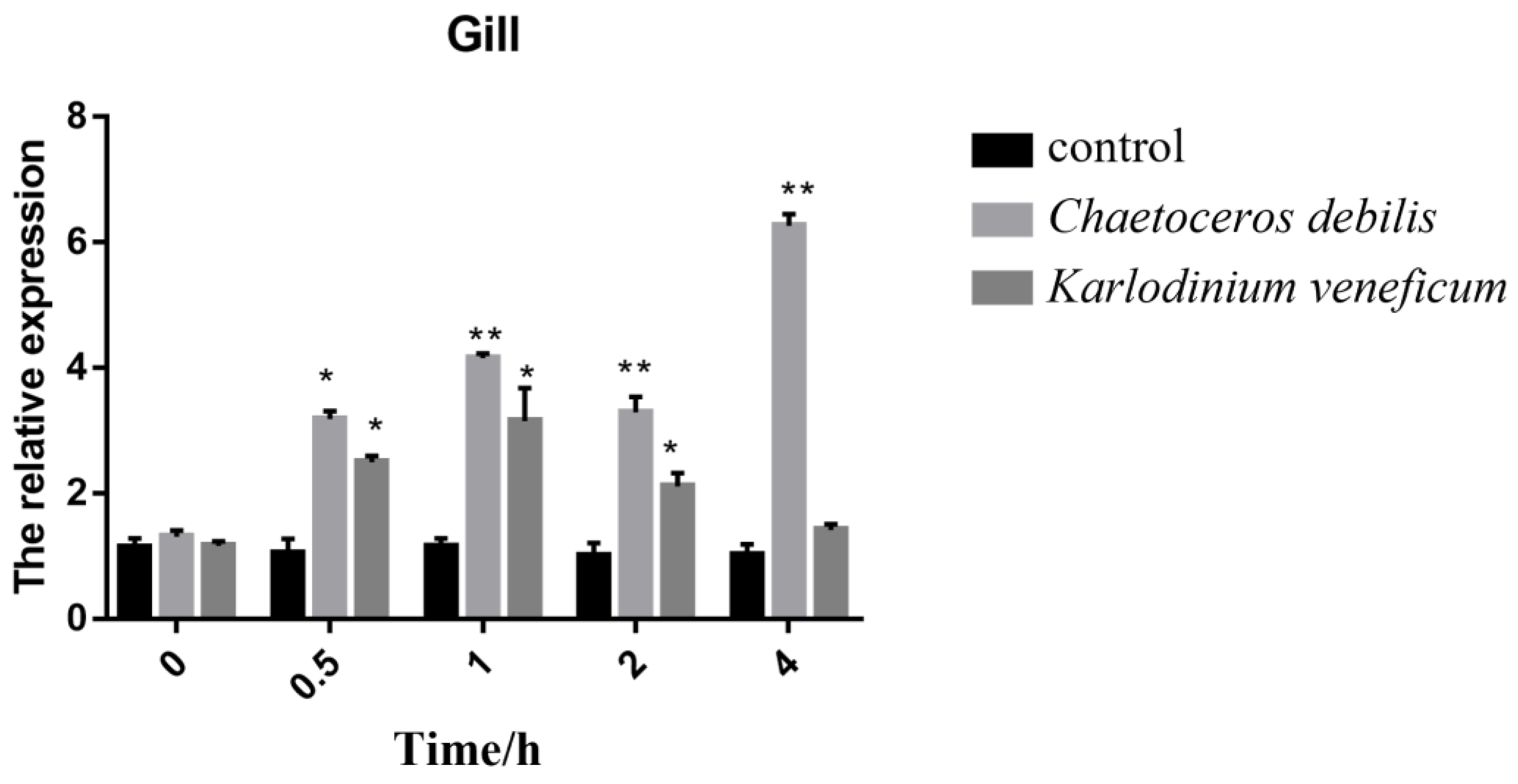
3.5. Expression of Rpcl in Feeding Experiments
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Bureau of Fisheries, Ministry of Agriculture and Rural Affairs, National Fisheries Technology Extension Center, China Society of Fisheries. China Fishery Statistical Yearbook 2021; China Agriculture Press: Beijing, China, 2021.
- Zuo, S.; Jiang, K.; Li, D.; Yan, X.; Nie, H. Transcriptomic analysis of Manila clam Ruditapes philippinarum under lipopolysaccharide challenge provides molecular insights into immune response. Fish Shellfish. Immunol. 2020, 106, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Mckindsey, C.W.; Landry, T.; O’Beirn, F.X.; Davies, I.M. Bivalve aquaculture and exotic species: A review of ecological considerations and management issues. J. Shellfish. Res. 2007, 26, 281–294. [Google Scholar] [CrossRef]
- Vasta, G.; Feng, C.; Bianchet, M.; Bachvaroff, T.; Tasumi, S. Structural, functional, and evolutionary aspects of galectins in aquatic mollusks: From a sweet tooth to the Trojan horse. Fish Shellfish. Immunol. 2015, 46, 94–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varki, A. Biological roles of glycans. Glycobiology 2017, 27, 3–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zelensky, A.N.; Gready, J.E. The C-type lectin-like domain superfamily. FEBS J. 2005, 272, 6179–6217. [Google Scholar] [CrossRef]
- Drickamer, K. Two distinct classes of carbohydrate-recognition domeins in animal’s lectins. J. Biol. Chem. 1988, 263, 9557–9560. [Google Scholar] [CrossRef]
- Weis, W.I.; Taylor, M.E.; Drickamer, K. The C-type lectin superfamily in the immune system. Immunol. Rev. 1998, 163, 19–34. [Google Scholar] [CrossRef]
- Kikuno, R.; Sato, A.; Mayer, W.E.; Shintani, S.; Aoki, T.; Klein, J. Clustering of C-type lectin natural killer receptor-like loci in the bony fish Oreochromis niloticus. Scand. J. Immunol. 2004, 59, 133–142. [Google Scholar] [CrossRef] [Green Version]
- Hurtado, C.; Granja, A.G.; Bustos, M.J.; Nogal, M.L.; de Buitrago, G.G.; de Yébenes, V.G.; Salas, M.L.; Revilla, Y.; Carrascosa, A.L. The C-type lectin homologue gene (EP153R) of African swine fever virus inhibits apoptosis both in virus infection and in heterologous expression. Virology 2004, 326, 160–170. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.-N.; Jin, X.-K.; Li, S.; Yu, A.-Q.; Wu, M.-H.; Tan, S.-J.; Zhu, Y.-T.; Li, W.-W.; Zhang, P.; Wang, Q. A novel C-type lectin from Eriocheir sinensis functions as a pattern recognition receptor with antibacterial activity. Fish Shellfish. Immunol. 2013, 35, 1554–1565. [Google Scholar] [CrossRef]
- Hosono, M.; Sugawara, S.; Ogawa, Y.; Kohno, T.; Takayanagi, M.; Nitta, K. Purification, characterization, cDNA cloning, and expression of asialofetuin-binding C-type lectin from eggs of shishamo smelt (Osmerus [Spirinchus] lanceolatus). Biochim. Biophys. Acta—Gen. Subj. 2005, 1725, 160–173. [Google Scholar] [CrossRef] [PubMed]
- Espinosa, E.P.; Allam, B. Food quality and season affect gene expression of the mucosal lectin MeML and particle sorting in the blue mussel Mytilus edulis. Mar. Biol. 2013, 160, 1441–1450. [Google Scholar] [CrossRef]
- Shi, Y.; Zhao, X.; Wang, Z.; Shao, Y.; Zhang, W.; Bao, Y.; Li, C. Novel Ca2+-independent C-type lectin involved in immune defense of the razor clam Sinonovacula constricta. Fish Shellfish. Immunol. 2019, 84, 502–508. [Google Scholar] [CrossRef]
- Espinosa, E.P.; Allam, B. Reverse genetics demonstrate the role of mucosal C-type lectins in food particle selection in the oyster Crassostrea virginica. J. Exp. Biol. 2018, 221, jeb174094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Espinosa, E.P.; Perrigault, M.; Allam, B. Identification and molecular characterization of a mucosal lectin (MeML) from the blue mussel Mytilus edulis and its potential role in particle capture. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2010, 156, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Alhoshy, M.; Shehata, A.I.; Habib, Y.J.; Abdel-Latif, H.M.; Wang, Y.; Zhang, Z. Nutrigenomics in crustaceans: Current status and future prospects. Fish Shellfish. Immunol. 2022, 129, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Nie, H.; Dong, S.; Huo, Z.; Yan, X. Molecular cloning and expression analysis of C-type lectin (RpCTL) in Manila clam Ruditapes philippinarum after lipopolysaccharide challenge. Fish Shellfish. Immunol. 2019, 86, 981–993. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Lou, Y.; Lai, W.; Xu, J.; Zhou, C.; Yan, X. Effect of polyamines on amplification of several marine microalgae. J. Ningbo. Univ. 2018, 31, 38–43. [Google Scholar]
- Espinosa, E.P.; Hassan, D.; Ward, J.E.; Shumway, S.E.; Allam, B. Role of Epicellular Molecules in the Selection of Particles by the Blue Mussel, Mytilus edulis. Biol. Bull. 2011, 219, 50–60. [Google Scholar] [CrossRef]
- Sancho, D.; e Sousa, C.R. Sensing of cell death by myeloid C-type lectin receptors. Curr. Opin. Immunol. 2013, 25, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Deniaud, D.; Julienne, K.; Gouin, S.G. Insights in the rational design of synthetic multivalent glycoconjugates as lectin ligands. Org. Biomol. Chem. 2011, 9, 966–979. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Huang, M.; Zhang, H.; Song, L. The immune role of C-type lectins in molluscs. Invertebr. Surviv. J. 2011, 8, 241–246. [Google Scholar]
- Zhang, H.; Wang, H.; Wang, L.; Song, L.; Song, X.; Zhao, J.; Li, L.; Qiu, L. Cflec-4, a multidomain C-type lectin involved in immune defense of Zhikong scallop Chlamys farreri. Dev. Comp. Immunol. 2009, 33, 780–788. [Google Scholar] [CrossRef] [PubMed]
- Zelensky, A.N.; Gready, J.E. C-type lectin-like domains in Fugu rubripes. BMC genomics 2004, 5, 51. [Google Scholar] [CrossRef] [Green Version]
- Espinosa, E.P.; Perrigault, M.; Ward, J.E.; Shumway, S.E.; Allam, B. Lectins Associated with the Feeding Organs of the Oyster Crassostrea virginica Can Mediate Particle Selection. Biol. Bull. 2009, 217, 130–141. [Google Scholar] [CrossRef] [Green Version]
- Jia, Z.; Zhang, H.; Jiang, S.; Wang, M.; Wang, L.; Song, L. Comparative study of two single CRD C-type lectins, CgCLec-4 and CgCLec-5, from pacific oyster Crassostrea gigas. Fish Shellfish. Immunol. 2016, 59, 220–232. [Google Scholar] [CrossRef] [Green Version]
- Huang, M.; Zhang, H.; Jiang, S.; Wang, L.; Liu, R.; Yi, Q.; Song, L. An EPD/WSD motifs containing C-type lectin from Argopectens irradians recognizes and binds microbes with broad spectrum. Fish Shellfish. Immunol. 2015, 43, 287–293. [Google Scholar] [CrossRef]
- Cambi, A.; Koopman, M.; Figdor, C.G. How C-type lectins detect pathogens. Cell. Microbiol. 2005, 7, 481–488. [Google Scholar] [CrossRef]
- Beninger, P.; St-Jean, S. Particle processing on the labial palps of Mytilus edulis and Placopecten magellanicus (Mollusca:Bivalvia). Mar. Ecol. Prog. Ser. 1997, 147, 117–127. [Google Scholar] [CrossRef] [Green Version]
- Ward, J.E.; Levinton, J.S.; Shumway, S.E.; Cucci, T. Particle sorting in bivalves: In vivo determination of the pallial organs of selection. Mar. Biol. 1998, 131, 283–292. [Google Scholar] [CrossRef]
- Ward, J.E.; Levinton, J.S.; Shumway, S.E.; Cucci, T. Site of particle selection in a bivalve mollusc. Nature 1997, 390, 131–132. [Google Scholar] [CrossRef]
- Espinosa, E.P.; Perrigault, M.; Ward, J.E.; Shumway, S.E.; Allam, B. Microalgal Cell Surface Carbohydrates as Recognition Sites for Particle Sorting in Suspension-Feeding Bivalves. Biol. Bull. 2010, 218, 75–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hegaret, H.; Wikfors, G.H.; Shumway, S.E. Diverse feeding responses of five species of bivalve mollusc when exposed to three species of harmful algae. J. Shellfish. Res. 2007, 26, 549–559. [Google Scholar] [CrossRef]
- Turner, A.D.; Lewis, A.M.; Bradley, K.; Maskrey, B.H. Marine invertebrate interactions with Harmful Algal Blooms—Implications for One Health. J. Invertebr. Pathol. 2021, 186, 107555. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Yin, B.; Wang, Y.; Huai, B. Significantly decreasing harmful algal blooms in China seas in the early 21st century. Mar. Pollut. Bull. 2019, 139, 270–274. [Google Scholar] [CrossRef]
- McNamee, S.E.; Medlin, L.K.; Kegel, J.; McCoy, G.R.; Raine, R.; Barra, L.; Ruggiero, M.V.; Kooistra, W.H.; Montresor, M.; Hagstrom, J.; et al. Distribution, occurrence and biotoxin composition of the main shellfish toxin producing microalgae within European waters: A comparison of methods of analysis. Harmful Algae 2016, 55, 112–120. [Google Scholar] [CrossRef]
- Karlson, B.; Andersen, P.; Arneborg, L.; Cembella, A.; Eikrem, W.; John, U.; West, J.J.; Klemm, K.; Kobos, J.; Lehtinen, S.; et al. Harmful algal blooms and their effects in coastal seas of Northern Europe. Harmful Algae 2021, 102, 101989. [Google Scholar] [CrossRef] [PubMed]
- Robbins, H.M.; Bricelj, V.M.; Ward, J.E. In vivo Effects of Brown Tide on the Feeding Function of the Gill of the Northern Quahog Mercenaria mercenaria (Bivalvia: Veneridae). Biol. Bull. 2010, 219, 61–71. [Google Scholar] [CrossRef] [Green Version]
- Matsuyama, Y.; Uchida, T.; Honjo, T. Toxic effects of the dinoffagellate Heterocapsa circularisquama on clearance rate of the blue mussel Mytilus galloprovincialis. Oceanogr. Lit. Rev. 1997, 7, 731. [Google Scholar]
- Shumway, S.E. A Review of the Effects of Algal Blooms on Shellfish and Aquaculture. J. World Aquac. Soc. 1990, 21, 65–104. [Google Scholar] [CrossRef]
- Drickamer, K. Engineering galactose-binding activity into a C-type mannose-binding protein. Nature 1992, 360, 183–186. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Hu, Z.; Deng, Y.; Tang, Y.Z. Evidence for resting cyst production in the cosmopolitan toxic dinoflagellate Karlodinium veneficum and the cyst distribution in the China seas. Harmful Algae 2020, 93, 101788. [Google Scholar] [CrossRef] [PubMed]


| Gene | Primer | Primer Sequences (5′–3′) | Aim |
|---|---|---|---|
| Rpcl | Forward primer | GTGACTGGAGGCACTATCCG | qRT-PCR |
| ß-actin | Reverse primer | TTGTTGGTGCATGGTTCGTC | |
| Forward primer | CTCCCTTGAGAAGAGCTACGA | ||
| Rpcl-NdeI | Reverse primer | GATACCAGCAGATTCCATACCC | ORF amplification |
| Forward primer | CGCCATATGGAAGAGAAGTCGTGCTCGCCT | ||
| Rpcl-BamHI | Reverse primer | CGCGGATCCTTATTCACTTGAGTTGAGTTTCATTTC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.; Shi, P.; Feng, Q.; Qiu, X.; Xu, J.; Yan, X.; Zhou, C. A Novel C-Type Lectin and Its Potential Role in Feeding and Feed Selection in Ruditapes philippinarum. Fishes 2023, 8, 62. https://doi.org/10.3390/fishes8020062
Chen S, Shi P, Feng Q, Qiu X, Xu J, Yan X, Zhou C. A Novel C-Type Lectin and Its Potential Role in Feeding and Feed Selection in Ruditapes philippinarum. Fishes. 2023; 8(2):62. https://doi.org/10.3390/fishes8020062
Chicago/Turabian StyleChen, Sentao, Peng Shi, Qingkai Feng, Xiaoting Qiu, Jilin Xu, Xiaojun Yan, and Chengxu Zhou. 2023. "A Novel C-Type Lectin and Its Potential Role in Feeding and Feed Selection in Ruditapes philippinarum" Fishes 8, no. 2: 62. https://doi.org/10.3390/fishes8020062





