Using qPCR to Identify Potential Effects of Thermal Conditions during Embryogenesis on Mitochondrial DNA Copy Number in Juvenile Brown Trout Salmo trutta
Abstract
:1. Introduction
2. Materials and Methods
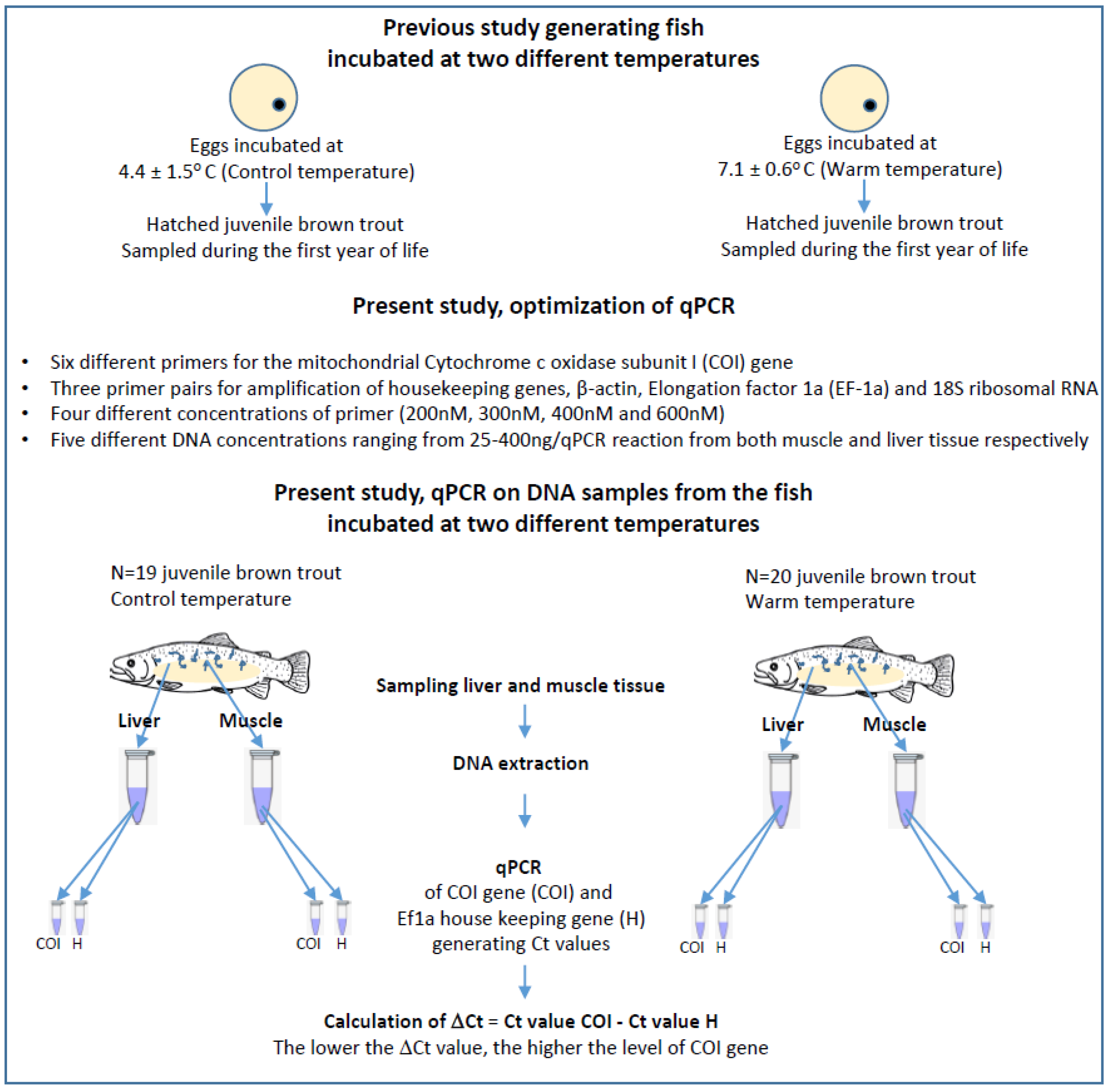
2.1. Rearing of Fish
2.2. Tissue Sampling and DNA Extraction
2.3. Optimization of the qPCR for Mitochondrial DNA Quantification
2.4. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jonsson, B.; Jonsson, N. A review of the likely effects of climate change on anadromous Atlantic salmon Salmo salar and brown trout Salmo trutta, with particular reference to water temperature and flow. J. Fish Biol. 2009, 75, 2381–2447. [Google Scholar] [CrossRef]
- Durtsche, R.D.; Jonsson, B.; Greenberg, L.A. Thermal conditions during embryogenesis influence metabolic rates of juvenile brown trout Salmo trutta. Ecosphere 2021, 12, e0337. [Google Scholar] [CrossRef]
- Blier, P.U.; Lemieux, H.; Pichaud, N. Holding our breath in our modern world: Will mitochondria keep the pace with climate changes? Can. J. Zool. 2014, 92, 591–601. [Google Scholar] [CrossRef]
- Ørtenblad, N. Mitochondrial increase in volume density with exercise training: More, larger or better? Acta Physiol. 2018, 222, e12976. [Google Scholar] [CrossRef]
- Lee, H.C.; Wei, Y.H. Mitochondrial role in life and death of the cell. J. Biomed. Sci. 2000, 7, 2–15. [Google Scholar] [CrossRef]
- Li, Z.H.; Zhang, P.; Ma, H.K.; Xu, W.Y.; Sun, J.Q.; Yan, B.L.; Zhang, Q.Q.; Gao, H. Effect of temperature and salinity on mtDNA copy number of the ridgetail white prawn, Palaemon carinicauda Holthuis, 1950 (Decapoda, Palaemonidae). Crustaceana 2018, 91, 1061–1072. [Google Scholar] [CrossRef]
- Hartmann, N.; Reichwald, K.; Wittig, I.; Dröse, S.; Schmeisser, S.; Lück, C.; Hahn, C.; Graf, M.; Gausmann, U.; Terzibasi, E.; et al. Mitochondrial DNA copy number and function decrease with age in the short-lived fish Nothobranchius furzeri. Aging Cell 2011, 10, 824–831. [Google Scholar] [CrossRef]
- Pieters, N.; Koppen, G.; Smeets, K.; Napierska, D.; Plusquin, M.; De Prins, S.; Van De Weghe, H.; Nelen, V.; Cox, B.; Cuypers, A.; et al. Decreased mitochondrial DNA content in association with exposure to polycyclic aromatic hydrocarbons in house dust during wintertime: From a population enquiry to cell culture. PLoS ONE 2013, 8, e63208. [Google Scholar] [CrossRef]
- Liao, K.; Yan, J.; Mai, K.; Ai, Q. Dietary lipid concentration affects liver mitochondrial DNA copy number, gene expression and DNA methylation in large yellow croaker (Larimichthys crocea). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2016, 193, 25–32. [Google Scholar] [CrossRef]
- Picca, A.; Guerra, F.; Calvani, R.; Coelho-Júnior, H.J.; Leeuwenburgh, C.; Bucci, C.; Marzetti, E. The contribution of mitochondrial DNA alterations to aging, cancer, and neurodegeneration. Exp. Gerontol. 2023, 178, 112203. [Google Scholar] [CrossRef]
- Consuegra, S.; John, E.; Verspoor, E.; De Leaniz, C.G. Patterns of natural selection acting on the mitochondrial genome of a locally adapted fish species. Genet. Sel. Evol. 2015, 47, 58. [Google Scholar] [CrossRef]
- Álvarez, D.; Cano, J.M.; Nicieza, A.G. Microgeographical variation in metabolic rate and energy storage of brown trout: Countergradient selection or thermal sensitivity. Evol. Ecol. 2006, 20, 345–363. [Google Scholar] [CrossRef]
- Wright, T.G.; Dawson, B.; Jalleh, G.; Guelfi, K.J. Influence of hormonal profile on resting metabolic rate in normal, overweight and obese individuals. Ann. Nutr. Metab. 2015, 66, 162–167. [Google Scholar] [CrossRef]
- Dalvin, S.; Glover, K.A.; Sørvik, A.G.; Seliussen, B.B.; Taggart, J.B. Forensic identification of severely degraded Atlantic salmon (Salmo salar) and rainbow trout (Oncorhynchus mykiss) tissues. Investig. Genet. 2010, 1, 12. [Google Scholar] [CrossRef]
- Hsieh, A.Y.Y.; Budd, M.; Deng, D.; Gadawska, I.; Côté, H.C.F. A monochrome multiplex real-time quantitative PCR assay for the measurement of mitochondrial DNA content. J. Mol. Diagn. 2018, 20, 612–620. [Google Scholar] [CrossRef]
- Yin, Y.; Shen, H. Common methods in mitochondrial research (Review). Int. J. Mol. Med. 2022, 50, 126. [Google Scholar] [CrossRef]
- Refinetti, P.; Warren, D.; Morgenthaler, S.; Ekstrøm, P.O. Quantifying mitochondrial DNA copy number using robust regression to interpret real time PCR results. BMC Res. Notes 2017, 10, 593. [Google Scholar] [CrossRef]
- Atkinson, S.; Carlsson, J.E.L.; Ball, B.; Egan, D.; Kelly-Quinn, M.; Whelan, K.; Carlsson, J. A quantitative PCR-based environmental DNA assay for detecting Atlantic salmon (Salmo salar L.). Aquat. Conserv. Mar. Freshw. Ecosyst. 2018, 28, 1238–1243. [Google Scholar] [CrossRef]
- Jonsson, B.; Jonsson, N. Differences in growth between offspring of anadromous and freshwater brown trout Salmo trutta. J. Fish Biol. 2021, 99, 18–24. [Google Scholar] [CrossRef]
- Jonsson, B.; Greenberg, L. Egg incubation temperature influences the population-specific outmigration rate of juvenile brown trout Salmo trutta. Fish Biol. 2022, 100, 909–917. [Google Scholar] [CrossRef]
- Greenberg, L.; Filipsson, K.; Bergman, E.; Jonsson, B. The effects of egg incubation temperature and parental cross on the swimming activity of juvenile brown trout Salmo trutta. Behav. Ecol. Sociobiol. 2023, 77, 114. [Google Scholar] [CrossRef]
- Kalayci, G.; Ozturk, R.C.; Capkin, E.; Altinok, I. Genetic and molecular evidence that brown trout Salmo Trutta belonging to the Danubian lineage are a single biological species. J. Fish Biol. 2018, 93, 792–804. [Google Scholar] [CrossRef]
- Rezaei, A. Studies on protein structure of cytochrome C oxidase subunit II gene in Salmo trutta caspius. J. Adv. Zool. 2015, 36, 1–10. [Google Scholar] [CrossRef]
- Capo, E.; Spong, G.; Königsson, H.; Byström, P. Effects of filtration methods and water volume on the quantification of brown trout (Salmo trutta) and Arctic char (Salvelinus alpinus) EDNA Concentrations via Droplet Digital PCR. Environ. DNA 2020, 2, 152–160. [Google Scholar] [CrossRef]
- Hansen, B.H.; Rømma, S.; Garmo, Ø.A.; Pedersen, S.A.; Olsvik, P.A.; Andersen, R.A. Induction and activity of oxidative stress-related proteins during waterborne Cd/Zn-exposure in brown trout (Salmo trutta). Chemosphere 2007, 67, 2241–2249. [Google Scholar] [CrossRef]
- Churova, M.V.; Meshcheryakova, O.V.; Ruchev, M.; Nemova, N.N. Age- and stage-dependent variations of muscle-specific gene expression in brown trout Salmo trutta L. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2017, 211, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Tinoco, A.B.; Näslund, J.; Delgado, M.J.; de Pedro, N.; Johnsson, J.I.; Jönsson, E. Ghrelin increases food intake, swimming activity and growth in juvenile brown trout (Salmo trutta). Physiol. Behav. 2014, 124, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Cominassi, L.; Ressel, K.N.; Brooking, A.A.; Marbacher, P.; Ransdell-Green, E.C.; O’Brien, K.M. Metabolic rate increases with acclimation temperature and is associated with mitochondrial function in some tissues of threespine stickleback. J. Exp. Biol. 2022, 225, jeb244659. [Google Scholar] [CrossRef]
- Parr, R.L.; Maki, J.; Reguly, B.; Dakubo, G.D.; Aguirre, A.; Wittock, R.; Robinson, K.; Jakupciak, J.P.; Thayer, R.E. The pseudo-mitochondrial genome influences mistakes in heteroplasmy interpretation. BMC Genom. 2006, 7, 185. [Google Scholar] [CrossRef]
- Dawnay, N.; Ogden, R.; McEwing, R.; Carvalho, G.R.; Thorpe, R.S. Validation of the barcoding gene COI for use in forensic genetic species identification. Forensic Sci. Int. 2007, 173, 1–6. [Google Scholar] [CrossRef]
- Takatsu, K.; Selz, O.M.; Brodersen, J. Temperature regime during embryogenesis alters subsequent behavioural phenotypes of juvenile brown trout. Biol. Lett. 2022, 18, 20220369. [Google Scholar] [CrossRef] [PubMed]
- Bærum, K.M.; Finstad, A.G.; Ulvan, E.M.; Haugen, T.O. Population consequences of climate change through effects on functional traits of lentic brown trout in the sub-Arctic. Sci. Rep. 2021, 11, 15246. [Google Scholar] [CrossRef] [PubMed]


| Primer (Target) | Forward Sequence 5′-3′ | Reverse Sequence 5′-3′ | Reference |
|---|---|---|---|
| Dalvin_COI030 | GAATAGTCGGCACCGCCCTAAGTCTCT | CGCCAGGCTGGCTGAGTTCTGCT | [14] |
| Ward_COI | TCAACCAACCACAAAGACATTGGCAC | TAGACTTCTGGGTGGCCAAAGAATCA | [22] |
| Rezaei_COI | GGCAATCACACGATGATTTTT | TCGTTAACTTGCTTGTACTTGGA | [23] |
| Atkinson_COI | CGCCCTAAGTCTCTTGATTCGA | CGTTATAAATTTGGTCATCTCCCAGA | [18] |
| Dalvin_COI334 | CTACCCCCCTCTAGCAGGTAATCTT | GGGAAAAAATAGTTAAGTCAACGGAA | [14] |
| Capo_COI | TCAACCAACCACAAAGACATTGGCAC | AGTGTTTCACAGTGTGTAGGC | [24] |
| Primer (housekeeping) | Forward Sequence 5′-3′ | Reverse Sequence 5′-3′ | Reference |
| β-actin | CCAAAGCCAACAGGGAGAAG | AGGGACAACACTGCCTGGAT | [25] |
| Elongation factor 1a | GCAAGAACGACCCTCCAATG | CAGGCGATGTGAGCAGTATG | [26] |
| 18S ribosomal RNA | TGGCCGTTCTTAGTTGGT | CTCTAAGAAGTTGGACGCCG | [27] |
| Primer (Target) | Range PCR Efficiency (%) | Specificity of Primer (Melting Curve) | Reference |
|---|---|---|---|
| Dalvin_COI030 | Poor efficiency | - | [14] |
| Ward_COI | Poor efficiency | - | [22] |
| Rezaei_COI | Poor efficiency | - | [23] |
| Atkinson_COI | Poor efficiency | - | [18] |
| Dalvin_COI334 | 103–114% | One peak | [14] |
| Cap_COI | Poor efficiency | - | [24] |
| Primer (housekeeping) | Range PCR efficiency (%) | Specificity of primer (melting curve) | Reference |
| β-actin | 96–314% | One peak | [25] |
| Elongation factor 1a | 100–111% | One peak | [26] |
| 18S ribosomal RNA | 93–104% | One peak | [27] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Erlandsson, A.; Ašmonaitė, G.; Jonsson, B.; Greenberg, L. Using qPCR to Identify Potential Effects of Thermal Conditions during Embryogenesis on Mitochondrial DNA Copy Number in Juvenile Brown Trout Salmo trutta. Fishes 2024, 9, 142. https://doi.org/10.3390/fishes9040142
Erlandsson A, Ašmonaitė G, Jonsson B, Greenberg L. Using qPCR to Identify Potential Effects of Thermal Conditions during Embryogenesis on Mitochondrial DNA Copy Number in Juvenile Brown Trout Salmo trutta. Fishes. 2024; 9(4):142. https://doi.org/10.3390/fishes9040142
Chicago/Turabian StyleErlandsson, Ann, Giedrė Ašmonaitė, Bror Jonsson, and Larry Greenberg. 2024. "Using qPCR to Identify Potential Effects of Thermal Conditions during Embryogenesis on Mitochondrial DNA Copy Number in Juvenile Brown Trout Salmo trutta" Fishes 9, no. 4: 142. https://doi.org/10.3390/fishes9040142







