Tree Species Influence Nitrate and Nitrous Oxide Production in Forested Riparian Soils
Abstract
:1. Introduction
1.1. Nitrogen Pollutants

1.2. Use of the Genes Associated with the N Cycle to Predict Process Rates
1.3. Tree Species Influence on N Cycling
1.4. N Fertilization Influence on N Cycling
- N enrichment would contribute to greater NH4+, NO3−, and N2O concentrations that correspond to greater abundances of corresponding microbial functional genes in the N cycle.
- Soil microbial N cycling gene abundance would differ significantly between tree species, specifically related to mycorrhizal association. We anticipated that the AM-dominant soils would likely contain greater abundances of nirK and NO3− relative to the ECM soils due to high quality, low C:N ratio as compared to the AM litter.
- Soil influenced by tree species associated with AM fungi would contain a higher nirK:nosZ ratio, indicative of greater potential for N2O production due to incomplete nitrous oxide reduction by nosZ relative to trees associated with ECM fungi, especially in areas of high N.
2. Materials and Methods
2.1. Study Area
2.2. Soil Collection, Processing, and Analysis
2.3. Genetic Analysis of nosZ and nirK in Soil Samples
2.4. Data Analysis
3. Results
3.1. Nitrate and Ammonium
3.2. Soil pH, Soil Organic Matter, and Soil Moisture
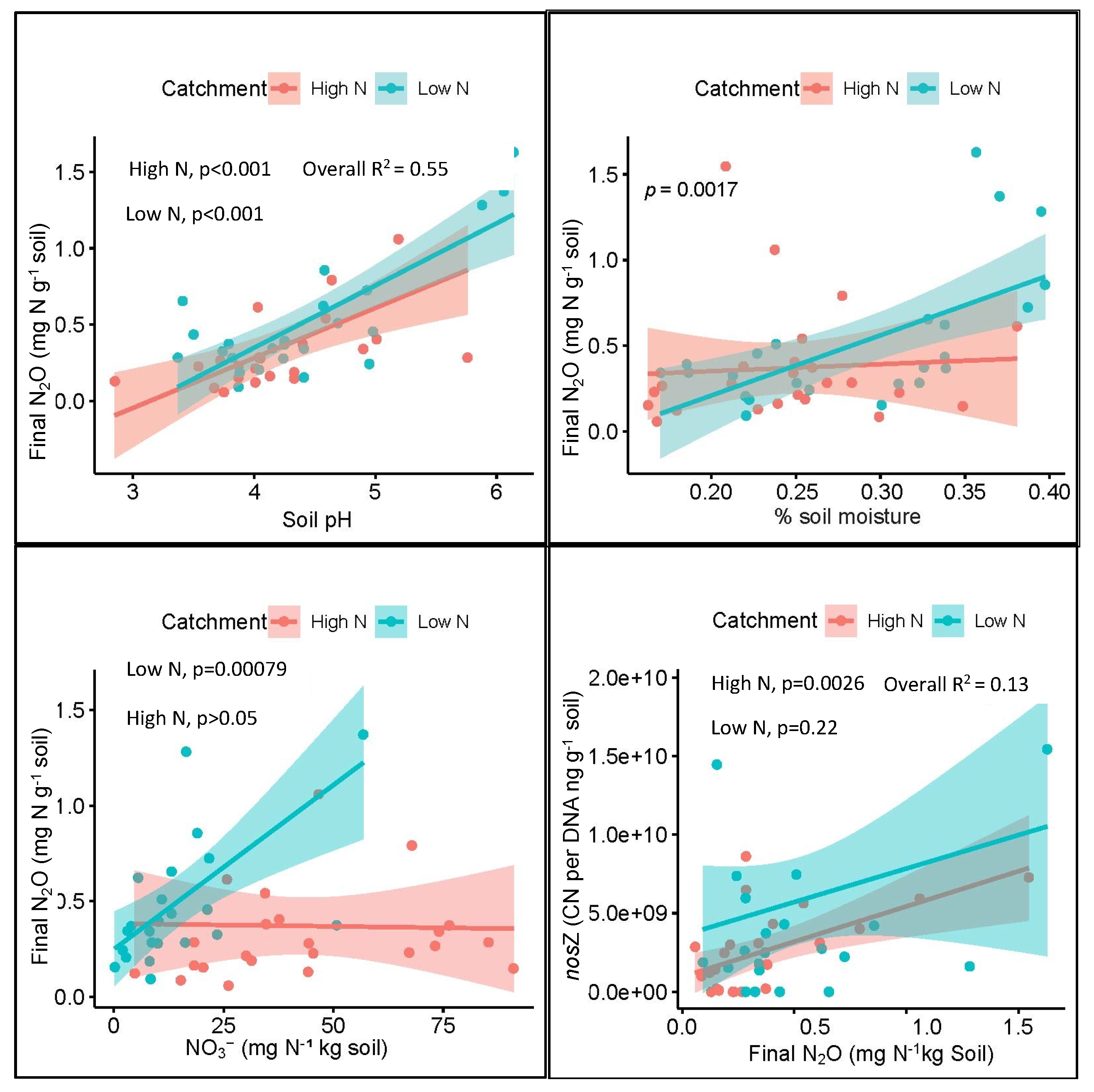
3.3. Potential N2O Production
3.4. Target N Cycling Genes
3.5. Relationships between Gene Abundance and Soil Parameters
4. Discussion
4.1. Relationships of Environmental Variables to Gene Abundance
4.2. Limitations and Future Directions
4.3. Limitations of Quantifying Gene Abundance
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Groffman, P.M.; Gold, A.J.; Addy, K. Nitrous oxide production in riparian zones and its importance to national emission inventories. Chemosphere-Glob. Chang. Sci. 2000, 2, 291–299. [Google Scholar] [CrossRef]
- Wiseman, J.D.; Burchell, M.R.; Grabow, G.L.; Osmond, D.L.; Messer, T.L. Groundwater nitrate concentration reductions in a riparian buffer enrolled in the NC Conservation Reserve Enhancement Program. JAWRA J. Am. Water Resour. Assoc. 2014, 50, 653–664. [Google Scholar] [CrossRef]
- Shepard, J.P. Water quality protection in bioenergy production: The US system of forestry best management practices. Biomass Bioenergy 2006, 30, 378–384. [Google Scholar] [CrossRef]
- Stutter, M.I.; Chardon, W.J.; Kronvang, B. Riparian buffer strips as a multifunctional management tool in agricultural landscapes: Introduction. J. Environ. Qual. 2012, 41, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Cirmo, C.P.; McDonnell, J.J. Linking the hydrologic and biogeochemical controls of nitrogen transport in near-stream zones of temperate-forested catchments: A review. J. Hydrol. 1997, 199, 88–120. [Google Scholar] [CrossRef]
- Sabater, S.; Butturini, A.; Clement, J.-C.; Burt, T.; Dowrick, D.; Hefting, M.; Matre, V.; Pinay, G.; Postolache, C.; Rzepecki, M.; et al. Nitrogen removal by riparian buffers along a European climatic gradient: Patterns and factors of variation. Ecosystems 2003, 6, 0020–0030. [Google Scholar] [CrossRef]
- Hefting, M.M.; Clement, J.-C.; Bienkowski, P.; Dowrick, D.; Guenat, C.; Butturini, A.; Topa, S.; Pinay, G.; Verhoeven, J.T. The role of vegetation and litter in the nitrogen dynamics of riparian buffer zones in Europe. Ecol. Eng. 2005, 24, 465–482. [Google Scholar] [CrossRef]
- Fenn, M.E. Nitrogen excess in North American ecosystems: Predisposing factors, ecosystem responses, and management strategies. Ecol. Appl. 1998, 8, 706–733. [Google Scholar] [CrossRef]
- US Environmental Protection Agency. A Compilation of Cost Data Associated with the Impacts and Control of Nutrient Pollution; EPA 820-F-15-096; Office of Water: Washington, DC, USA, 2015.
- Wuebbles, D.J. Nitrous oxide: No laughing matter. Science 2009, 326, 56–57. [Google Scholar] [CrossRef]
- Ravishankara, A.R.; Daniel, J.S.; Portmann, R.W. Nitrous oxide (N2O): The dominant ozone-depleting substance emitted in the 21st century. Science 2009, 326, 123–125. [Google Scholar] [CrossRef]
- Giles, M.E.; Morley, N.J.; Baggs, E.M.; Daniell, T.J. Soil nitrate reducing processes–drivers, mechanisms for spatial variation, and significance for nitrous oxide production. Front. Microbiol. 2012, 3, 407. [Google Scholar] [CrossRef] [PubMed]
- Bates, B.; Kundzewicz, Z.W.; Wu, S.; Palutikof, J. Climate Change and Water: Technical Paper VI; Intergovernmental Panel on Climate Change (IPCC): Geneva, Switzerland, 2008.
- Richardson, D.; Felgate, H.; Watmough, N.; Thomson, A.; Baggs, E. Mitigating release of the potent greenhouse gas N 2 O from the nitrogen cycle–could enzymic regulation hold the key? Trends Biotechnol. 2009, 27, 388–397. [Google Scholar] [CrossRef] [PubMed]
- Zumft, W.G. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 1997, 61, 533–616. [Google Scholar] [PubMed]
- Firestone, M.K.; Davidson, E.A. Microbiological basis of NO and N2O production and consumption in soil. Exch. Trace Gases Between Terr. Ecosyst. Atmos. 1989, 47, 7–21. [Google Scholar]
- Rochette, P. No-till only increases N2O emissions in poorly-aerated soils. Soil Tillage Res. 2008, 101, 97–100. [Google Scholar] [CrossRef]
- Deslippe, J.R.; Jamali, H.; Jha, N.; Saggar, S. Denitrifier community size, structure and activity along a gradient of pasture to riparian soils. Soil Biol. Biochem. 2014, 71, 48–60. [Google Scholar] [CrossRef]
- Weier, K.L.; Doran, J.W.; Power, J.F.; Walters, D.T. Denitrification and the dinitrogen/nitrous oxide ratio as affected by soil water, available carbon, and nitrate. Soil Sci. Soc. Am. J. 1993, 57, 66–72. [Google Scholar] [CrossRef]
- Liu, B.; Mørkved, P.T.; Frostegård, Å.; Bakken, L.R. Denitrification gene pools, transcription and kinetics of NO, N2O and N2 production as affected by soil pH. FEMS Microbiol. Ecol. 2010, 72, 407–417. [Google Scholar] [CrossRef]
- Liu, B.; Frostegard, A.; Bakken, L. Impaired reduction of N2O to N2 in acid soils is due to a posttranscriptional interference with the expression of nosZ. mBio 2014, 5, e01383-14. [Google Scholar] [CrossRef]
- Philippot, L.; Andert, J.; Jones, C.M.; Bru, D.; Hallin, S. Importance of denitrifiers lacking the genes encoding the nitrous oxide reductase for N2O emissions from soil. Glob. Chang. Biol. 2011, 17, 1497–1504. [Google Scholar] [CrossRef]
- Jones, C.M.; Graf, D.R.; Bru, D.; Philippot, L.; Hallin, S. The unaccounted yet abundant nitrous oxide-reducing microbial community: A potential nitrous oxide sink. ISME J. 2013, 7, 417. [Google Scholar] [CrossRef] [PubMed]
- Tu, Q.; He, Z.; Wu, L.; Xue, K.; Xie, G.; Chain, P.; Reich, P.B.; Hobbie, S.E.; Zhou, J. Metagenomic reconstruction of nitrogen cycling pathways in a CO2-enriched grassland ecosystem. Soil Biol. Biochem. 2017, 106, 99–108. [Google Scholar] [CrossRef]
- Patra, A.K.; Abbadie, L.; Clays-Josserand, A.; Degrange, V.; Grayston, S.J.; Loiseau, P.; Louault, F.; Mahmood, S.; Nazaret, S.; Philippot, L.; et al. Effects of grazing on microbial functional groups involved in soil N dynamics. Ecol. Monogr. 2005, 75, 65–80. [Google Scholar] [CrossRef]
- Throbäck, I.N.; Johansson, M.; Rosenquist, M.; Pell, M.; Hansson, M.; Hallin, S. Silver (Ag+) reduces denitrification and induces enrichment of novel nirK genotypes in soil. FEMS Microbiol. Lett. 2007, 270, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Hallin, S.; Jones, C.M.; Schloter, M.; Philippot, L. Relationship between N-cycling communities and ecosystem functioning in a 50-year-old fertilization experiment. ISME J. 2009, 3, 597. [Google Scholar] [CrossRef] [PubMed]
- Petersen, D.G.; Blazewicz, S.J.; Firestone, M.; Herman, D.J.; Turetsky, M.; Waldrop, M. Abundance of microbial genes associated with nitrogen cycling as indices of biogeochemical process rates across a vegetation gradient in Alaska. Environ. Microbiol. 2012, 14, 993–1008. [Google Scholar] [CrossRef] [PubMed]
- Enwall, K.; Philippot, L.; Hallin, S. Activity and composition of the denitrifying bacterial community respond differently to long-term fertilization. Appl. Environ. Microbiol. 2005, 71, 8335–8343. [Google Scholar] [CrossRef]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; Van Der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789. [Google Scholar] [CrossRef]
- Jha, N.; Saggar, S.; Giltrap, D.; Tillman, R.; Deslippe, J. Soil properties impacting denitrifier community size, structure, and activity in New Zealand dairy-grazed pasture. Biogeosciences 2017, 14, 4243–4253. [Google Scholar] [CrossRef]
- Robertson, G.P. Nitrification in forested ecosystems. Philos. Trans. R. Soc. Lond. 1982, 296, 445–457. [Google Scholar]
- Finzi, A.C.; Canham, C.D. Non-additive effects of litter mixtures on net N mineralization in a southern New England forest. For. Ecol. Manag. 1998, 105, 129–136. [Google Scholar] [CrossRef]
- Stark, J.M.; Hart, S.C. High rates of nitrification and nitrate turnover in undisturbed coniferous forests. Nature 1997, 385, 61–65. [Google Scholar] [CrossRef]
- Fitzhugh, R.D.; Lovett, G.M.; Venterea, R.T. Biotic and abiotic immobilization of ammonium, nitrite, and nitrate in soils developed under different tree species in the Catskill Mountains, New York, USA. Glob. Chang. Biol. 2003, 9, 1591–1601. [Google Scholar] [CrossRef]
- Lovett, G.M.; Weathers, K.C.; Arthur, M.A.; Schultz, J.C. Nitrogen cycling in a northern hardwood forest: Do species matter? Biogeochemistry 2004, 67, 289–308. [Google Scholar] [CrossRef]
- Christenson, L.M.; Lovett, G.M.; Weathers, K.C.; Arthur, M.A. The influence of tree species, nitrogen fertilization, and soil C to N ratio on gross soil nitrogen transformations. Soil Sci. Soc. Am. J. 2009, 73, 638–646. [Google Scholar] [CrossRef]
- Kelly, C.N.; Schoenholtz, S.H.; Adams, M.B. Soil properties associated with net nitrification following watershed conversion from Appalachian hardwoods to Norway spruce. Plant Soil 2011, 344, 361–376. [Google Scholar] [CrossRef]
- Phillips, R.P.; Brzostek, E.; Midgley, M.G. The mycorrhizal-associated nutrient economy: A new framework for predicting carbon–nutrient couplings in temperate forests. New Phytol. 2013, 199, 41–51. [Google Scholar] [CrossRef]
- Peterjohn, W.T.; Harlacher, M.A.; Chris, M.J.; Adams, M.B. Testing associations between tree species and nitrate availability: Do consistent patterns exist across spatial scales? For. Ecol. Manag. 2015, 358, 335–343. [Google Scholar] [CrossRef]
- Menyailo, O.V.; Huwe, B. Activity of denitrification and dynamics of N2O release in soils under six tree species and grassland in central Siberia. J. Plant Nutr. Soil Sci. 1999, 162, 533–538. [Google Scholar] [CrossRef]
- Melillo, J.M.; Naimen, R.J.; Aber, J.D.; Eschleman, K.N. The influence of substrate quality and stream size on wood decomposition dynamics. Oecologia 1983, 58, 281–285. [Google Scholar] [CrossRef]
- Binkley, D. The influence of tree species on forest soils: Processes and patterns. In Proceedings of the Trees and Soil Workshop, Lincoln University, 28 February–2 March 1994; Mead, D.J., Cornforth, I.S., Eds.; Agronomy Society of New Zealand Special Publication No. 10. Lincoln University Press: Canterbury, UK, 1995. [Google Scholar]
- Sun, T.; Hobbie, S.E.; Berg, B.; Zhang, H.; Wang, Q.; Wang, Z.; Hättenschwiler, S. Contrasting dynamics and trait controls in first-order root compared with leaf litter decomposition. Proc. Natl. Acad. Sci. USA 2018, 115, 10392–10397. [Google Scholar] [CrossRef] [PubMed]
- Melillo, J.M.; Aber, J.D.; Linkins, A.E.; Ricca, A.; Fry, B.; Nadelhoffer, K.J. Carbon and nitrogen dynamics along the decay continuum: Plant litter to soil organic matter. Plant Soil 1989, 115, 189–198. [Google Scholar] [CrossRef]
- Rotkin-Ellman, M.; Addy, K.; Gold, A.J.; Groffman, P.M. Tree species, root decomposition and subsurface denitrification potential in riparian wetlands. Plant Soil 2004, 263, 335–344. [Google Scholar] [CrossRef]
- Templer, P.H.; Lovett, G.M.; Weathers, K.C.; Findlay, S.E.; Dawson, T.E. Influence of tree species on forest nitrogen retention in the Catskill Mountains, New York, USA. Ecosystems 2005, 8, 1–16. [Google Scholar] [CrossRef]
- Vesterdal, L.; Clarke, N.; Sigurdsson, B.D.; Gundersen, P. Do tree species influence soil carbonstocks in temperate and boreal forests? For. Ecol. Manag. 2013, 309, 4–18. [Google Scholar] [CrossRef]
- Averill, C.; Turner, B.L.; Finzi, A.C. Mycorrhiza-mediated competition between plants and decomposers drives soil carbon storage. Nature 2014, 505, 543. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Wheeler, E.; Phillips, R.P. Root-induced changes in nutrient cycling in forests depend on exudation rates. Soil Biol. Biochem. 2014, 78, 213–221. [Google Scholar] [CrossRef]
- Matson, P.A.; Vitousek, P.M. Cross-system comparisons of soil nitrogen transformations and nitrous oxide flux in tropical forest ecosystems. Glob. Biogeochem. Cycles 1987, 1, 163–170. [Google Scholar] [CrossRef]
- Davidson, E.A.; Keller, M.; Erickson, H.E.; Verchot, L.V.; Veldkamp, E. Testing a conceptual model of soil emissions of nitrous and nitric oxides: Using two functions based on soil nitrogen availability and soil water content, the hole-in-the-pipe model characterizes a large fraction of the observed variation of nitric oxide and nitrous oxide emissions from soils. AIBS Bull. 2000, 50, 667–680. [Google Scholar]
- Nuccio, E.E.; Hodge, A.; Pett-Ridge, J.; Herman, D.J.; Weber, P.K.; Firestone, M.K. An arbuscular mycorrhizal fungus significantly modifies the soil bacterial community and nitrogen cycling during litter decomposition. Environ. Microbiol. 2013, 15, 1870–1881. [Google Scholar] [CrossRef]
- Burton, A.J.; Pregitzer, K.S.; Crawford, J.N.; Zogg, G.P.; Zak, D.R. Simulated chronic NO3−deposition reduces soil respiration in northern hardwood forests. Glob. Chang. Biol. 2004, 10, 1080–1091. [Google Scholar] [CrossRef]
- Janssens, I.A.; Dieleman, W.; Luyssaert, S.; Subke, J.A.; Reichstein, M.; Ceulemans, R.; Ciais, P.; Dolman, A.J.; Grace, J.; Matteucci, G.; et al. Reduction of forest soil respiration in response to nitrogen deposition. Nat. Geosci. 2010, 3, 315. [Google Scholar] [CrossRef]
- Wallenstein, M.D.; McNulty, S.; Fernandez, I.J.; Boggs, J.; Schlesinger, W.H. Nitrogen fertilization decreases forest soil fungal and bacterial biomass in three long-term experiments. For. Ecol. Manag. 2006, 222, 459–468. [Google Scholar] [CrossRef]
- Nemergut, D.R.; Townsend, A.R.; Sattin, S.R.; Freeman, K.R.; Fierer, N.; Neff, J.C.; Bowman, W.D.; Schadt, C.W.; Weintraub, M.N.; Schmidt, S.K. The effects of chronic nitrogen fertilization on alpine tundra soil microbial communities: Implications for carbon and nitrogen cycling. Environ. Microbiol. 2008, 10, 3093–4105. [Google Scholar] [CrossRef] [PubMed]
- Eisenlord, S.D.; Zak, D.R. Simulated atmospheric nitrogen deposition alters actinobacterial community composition in forest soils. Soil Sci. Soc. Am. J. 2010, 74, 1157–1166. [Google Scholar] [CrossRef]
- Fierer, N.; Lauber, C.L.; Ramirez, K.S.; Zaneveld, J.; Bradford, M.A.; Knight, R. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 2012, 6, 1007. [Google Scholar] [CrossRef] [PubMed]
- Allison, S.D.; Hanson, C.A.; Treseder, K.K. Nitrogen fertilization reduces diversity and alters community structure of active fungi in boreal ecosystems. Soil Biol. Biochem. 2007, 39, 1. [Google Scholar] [CrossRef]
- Edwards, I.P.; Zak, D.R.; Kellner, H.; Eisenlord, S.D.; Pregitzer, K.S. Simulated atmospheric N deposition alters fungal community composition and suppresses ligninolytic gene expression in a northern hardwood forest. PLoS ONE 2011, 6, e20421. [Google Scholar] [CrossRef]
- Carrara, J.E.; Walter, C.A.; Hawkins, J.S.; Peterjohn, W.T.; Averill, C.; Brzostek, E.R. Interactions among plants, bacteria, and fungi reduce extracellular enzyme activities under long-term N fertilization. Glob. Chang. Biol. 2018, 24, 2721–2734. [Google Scholar] [CrossRef]
- Freedman, Z.; Eisenlord, S.D.; Zak, D.R.; Xue, K.; He, Z.; Zhou, J. Towards a molecular understanding of N cycling in northern hardwood forests under future rates of N deposition. Soil Biol. Biochem. 2013, 66, 130–138. [Google Scholar] [CrossRef]
- Freedman, Z.B.; Upchurch, R.A.; Zak, D.R.; Cline, L.C. Anthropogenic N deposition slows decay by favoring bacterial metabolism: Insights from metagenomic analyses. Front. Microbiol. 2016, 7, 259. [Google Scholar] [CrossRef] [PubMed]
- Lilleskov, E.A.; Fahey, T.J.; Horton, T.R.; Lovett, G.M. Belowground ectomycorrhizal fungal community change over a nitrogen deposition gradient in Alaska. Ecology 2002, 83, 104–115. [Google Scholar] [CrossRef]
- Avrahami, S.; Conrad, R.; Braker, G. Effect of soil ammonium concentration on N2O release and on the community structure of ammonia oxidizers and denitrifiers. Appl. Environ. Microbiol. 2002, 68, 5685–5692. [Google Scholar] [CrossRef] [PubMed]
- Kellner, E.; Hubbart, J.; Stephan, K.; Morrissey, E.; Freedman, Z.; Kutta, E.; Kelly, C. Characterization of sub-watershed-scale stream chemistry regimes in an Appalachian mixed-land-use watershed. Environ. Monit. Assess. 2018, 190, 586. [Google Scholar] [CrossRef] [PubMed]
- Estavillo, J.; Merino, P.; Pinot, M.; Yamulki, S.; Gebauer, G.; Sapek, A.; Corre, W. Short term effect of ploughing permanent pasture on N2O production from nitrification and denitrification. Plant Soil 2002, 239, 253–265. [Google Scholar] [CrossRef]
- Meyer, R.L.; Allen, D.E.; Schmidt, S. Nitrification and denitrification as sources of sediment nitrous oxide production: A microsensor approach. Mar. Chem. 2008, 110, 68–76. [Google Scholar] [CrossRef]
- Butterbach-Bahl, K.; Baggs, E.M.; Dannenmann, M.; Kiese, R.; Zechmeister-Boltenstern, S. Nitrous oxide emissions from soils: How well do we understand the processes and theircontrols? Phil. Trans. R. Soc. B 2013, 368, 20130122. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Hu, H.; Suter, H.; Hayden, H.L.; He, J.; Mele, P.; Chen, D. Nitrification is a primary driver of nitrous oxide production in laboratory microcosms from different land-use soils. Front. Microbiol. 2016, 7, 1373. [Google Scholar] [CrossRef]
- Kaden, U.S.; Fuchs, E.; Hecht, C.; Hein, T.; Rupp, H.; Scholz, M.; Schulz-Zunkel, C. Advancement of the Acetylene Inhibition Technique Using Time Series Analysis on Air-Dried Floodplain Soils to Quantify Denitrification Potential. Geosciences 2020, 10, 431. [Google Scholar] [CrossRef]
- Henry, S.; Bru, D.; Stres, B.; Hallet, S.; Philippot, L. Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl. Environ. Microbiol. 2006, 72, 5181–5189. [Google Scholar] [CrossRef]
- Jung, J.; Choi, S.; Jung, H.; Scow, K.M.; Park, W. Primers for amplification of nitrous oxide reductase genes associated with Firmicutes and Bacteroidetes in organic-compound-rich soils. Microbiology 2013, 159 Pt 2, 307. [Google Scholar] [CrossRef] [PubMed]
- Rotthauwe, J.H.; Witzel, K.P.; Liesack, W. The ammonia monooxygenase structural gene amoA as a functional marker: Molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 1997, 63, 4704–4712. [Google Scholar] [CrossRef] [PubMed]
- Levy-Booth, D.J.; Prescott, C.E.; Grayston, S.J. Microbial functional genes involved in nitrogen fixation, nitrification and denitrification in forest ecosystems. Soil Biol. Biochem. 2014, 75, 11–25. [Google Scholar] [CrossRef]
- Holmes, A.J.; Costello, A.; Lidstrom, M.E.; Murrell, J.C. Evidence that participate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol. Lett. 1995, 132, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Jensen, S.; Holmes, A.; Olsen, R.; Murrell, J.C. Detection of Methane Oxidizing Bacteria in Forest Soil by Monooxygenase PCR Amplification. Microb. Ecol. 2000, 39, 282–289. [Google Scholar] [PubMed]
- Meinhardt, K.A.; Bertagnolli, A.; Pannu, M.W.; Strand, S.E.; Brown, S.L.; Stahl, D.A. Evaluation of revised polymerase chain reaction primers for more inclusive quantification of ammonia-oxidizing archaea and bacteria. Environ. Microbiol. 2015, 7, 354–363. [Google Scholar] [CrossRef] [PubMed]
- Michotey, V.; Méjean, V.; Bonin, P. Comparison of methods for quantification of cytochrome cd 1-denitrifying bacteria in environmental marine samples. Appl. Environ. Microbiol. 2000, 66, 1564–1571. [Google Scholar] [CrossRef]
- Throbäck, I.N.; Enwall, K.; Jarvis, Å.; Hallin, S. Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol. Ecol. 2004, 49, 401–417. [Google Scholar] [CrossRef]
- Kelly, C.N.; Koos, J.; Griggs, T.; Freedman, Z.B. Prescribed defoliation strategies influence soil carbon and nitrous oxide potential in pastures. Agron. J. 2022, 114, 2264–2279. [Google Scholar] [CrossRef]
- Hurlbert, S.H. Pseudoreplication and the design of ecological field experiments. Ecol. Monogr. 1984, 54, 187–211. [Google Scholar] [CrossRef]
- Matson, P.; Lohse, K.A.; Hall, S.J. The globalization of nitrogen deposition: Consequences for terrestrial ecosystems. Ambio 2002, 31, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Sobota, D.J.; Compton, J.E.; McCrackin, M.L.; Singh, S. Cost of reactive nitrogen release from human activities to the environment in the United States. Environ. Res. Lett. 2015, 10, 025006. [Google Scholar] [CrossRef]
- Suddick, E.C.; Six, J. An estimation of annual nitrous oxide emissions and soil quality following the amendment of high temperature walnut shell biochar and compost to a small-scale vegetable crop rotation. Sci. Total Environ. 2013, 465, 298–307. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.; Dang, C.; Morrissey, E.; Hubbart, J.; Kellner, E.; Kelly, C.; Stephan, K.; Freedman, Z. Stream sediment bacterial communities exhibit temporally-consistent and distinct thresholds to land use change in a mixed-use watershed. FEMS Microbiol. 2021, 97, fiaa256. [Google Scholar] [CrossRef] [PubMed]
- Kelly, C.N.; Schwaner, G.W.; Cumming, J.R.; Driscoll, T.P. Metagenomic reconstruction of nitrogen and carbon cycling pathways in forest soil: Influence of different hardwood tree species. Soil Biol. Biochem. 2021, 156, 108226. [Google Scholar] [CrossRef]
- Eisenhut, S.E.; Holásková, I.; Stephan, K. Role of Tree Species, the Herb Layer and Watershed Characteristics in Nitrate Assimilation in a Central Appalachian Hardwood Forest. Nitrogen 2022, 3, 333–352. [Google Scholar] [CrossRef]
- Royo, A.A.; Vickers, L.A.; Long, R.P.; Ristau, T.E.; Stoleson, S.H.; Stout, S.L. The forest of unintended consequences: Anthropogenic actions trigger the rise and fall of black cherry. BioScience 2021, 71, 683–696. [Google Scholar] [CrossRef]
- Schwaner, G.W.; Kelly, C.N. American chestnut soil carbon and nitrogen dynamics: Implications for ecosystem response following restoration. Pedobiologia 2019, 75, 24–33. [Google Scholar] [CrossRef]
- Brenzinger, K.; Dörsch, P.; Braker, G. pH-driven shifts in overall and transcriptionally active denitrifiers control gaseous product stoichiometry in growth experiments with extracted bacteria from soil. Front. Microbiol. 2015, 6, 961. [Google Scholar] [CrossRef]
- Grennfelt, P.; Hultberg, H. Effects of nitrogen deposition on the acidification of terrestrial and aquatic ecosystems. Water Air Soil Pollut. 1986, 30, 945–963. [Google Scholar] [CrossRef]
- Morales, S.E.; Cosart, T.; Holben, W.E. Bacterial gene abundances as indicators of greenhouse gas emission in soils. ISME J. 2010, 4, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Wang, B.; Xu, M.; Zhang, H.; Zhang, L.; Gao, S. Nitrification and acidification from urea application in red soil (Ferralic Cambisol) after different long-term fertilization treatments. J. Soils Sediments 2014, 14, 1526–1536. [Google Scholar] [CrossRef]
- Bergaust, L.; Mao, Y.; Bakken, L.R.; Frostegard, A. Denitrification response patterns during the transition to anoxic respiration and posttranscriptional effects of suboptimal pH on nitrogen oxide reductase in Paracoccus denitrificans. Appl. Environ. Microbiol. 2010, 76, 6387–6396. [Google Scholar] [CrossRef] [PubMed]
- Gilliam, F.S.; Welch, N.T.; Phillips, A.H.; Billmyer, J.H.; Peterjohn, W.T.; Fowler, Z.K.; Walter, C.A.; Burnham, M.B.; May, J.D.; Adams, M.B. Twenty-five-year response of the herbaceous layer of a temperate hardwood forest to elevated nitrogen deposition. Ecosphere 2016, 7, e01250. [Google Scholar] [CrossRef]
- Sterngren, A.E.; Hallin, S.; Bengtson, P. Archaeal ammonia oxidizers dominate in numbers, but bacteria drive gross nitrification in N-amended grassland soil. Front. Microbiol. 2015, 6, 1350. [Google Scholar] [CrossRef] [PubMed]
- Nesme, J.; Achouak, W.; Agathos, S.N.; Bailey, M.; Baldrian, P.; Brunel, D.; Frostegård, Å.; Heulin, T.; Jansson, J.K.; Jurkevitch, E.; et al. Back to the future of soil metagenomics. Front. Microbiol. 2016, 7, 73. [Google Scholar] [CrossRef] [PubMed]
- Wertz, S.; Leigh, A.K.; Grayston, S.J. Effects of long-term fertilization of forest soils on potential nitrification and on the abundance and community structure of ammonia oxidizers and nitrite oxidizers. FEMS Microbiol. Ecol. 2012, 79, 142–154. [Google Scholar] [CrossRef]
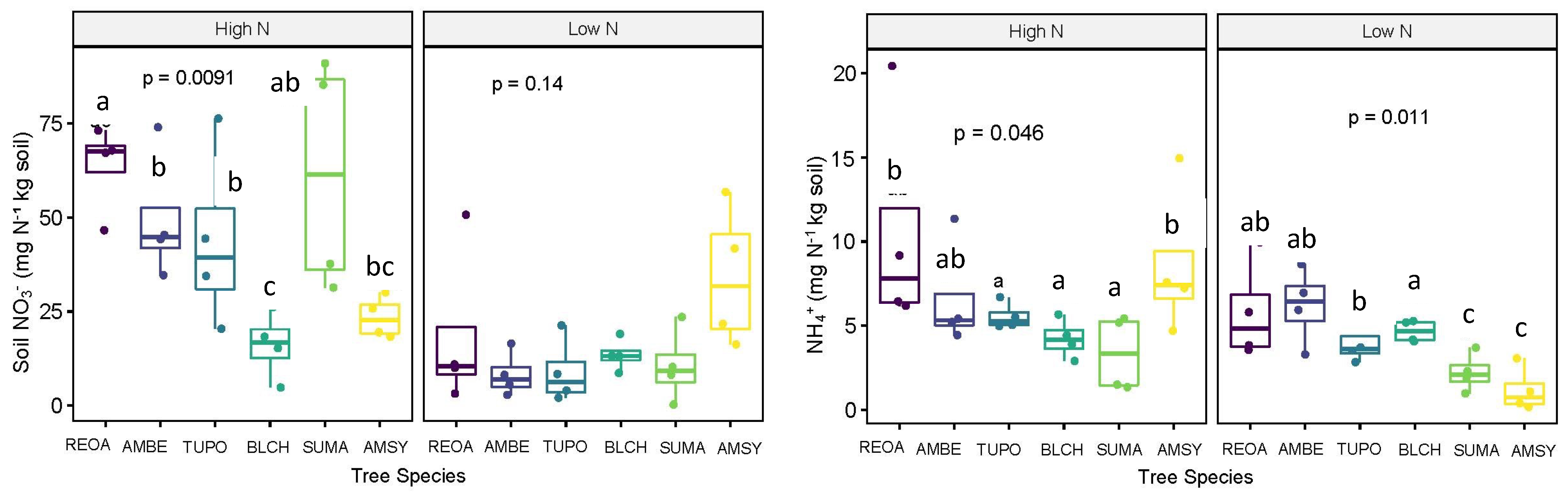


| Low N | High N | |
|---|---|---|
| Soil NO3− (mg N/kg soil) * | 15.66 (3.02) | 43.01 (5.02) |
| Soil NH4+ (mg N/kg soil) * | 4.03 (0.50) | 6.49 (0.84) |
| Soil pHCaCl2 | 4.41 (0.16) | 4.27 (0.13) |
| Soil OMLOI (%) | 13.19 (0.67) | 15.59 (1.08) |
| Soil moisture (%) * | 28.75 (1.47) | 24.49 (1.14) |
| nirK (1012 copy number) | 5.64 (1.48) | 3.65 (1.20) |
| nosZ (109 copy number) | 5.78 (1.34) | 2.61 (0.53) |
| N2O production (mg/kg/30 h) | 0.51 (0.08) | 0.37 (0.07) |
| Predicted Dependent Variable | Explanatory Independent Variable | F Ratio | Parameter p-Value | Model Adjusted R2 | Model p-Value |
|---|---|---|---|---|---|
| Soil NO3− (mg N/kg soil) | Tree species | 2.72 | 0.033 | 0.442 | <0.001 |
| Location | 33.89 | <0.001 | |||
| Soil moisture | 7.37 | 0.009 | |||
| Soil NH4+ (mg N/kg soil) | Tree species | 2.19 | 0.075 | 0.444 | |
| nosZ gene abundance | 3.93 | ||||
| Soil pH | 5.78 | 0.021 | |||
| Soil OM | 6.44 | 0.015 | |||
| Potential N2O (mg N/kg soil) | Tree species | 3.31 | 0.015 | 0.686 | <0.001 |
| Location | 4.73 | 0.036 | |||
| Soil pH | 63.77 | <0.001 | |||
| Soil OM | 6.33 | 0.016 |
| Predicted Dependent Variable | Tree Species | Mean Response |
|---|---|---|
| Soil NO3− (mg N/kg soil) | Sycamore (AMSY) | 28.77 ab |
| Cherry (BLCH) | 14.79 b | |
| Oak (REOA) | 41.21 a | |
| Beech (AMBE) | 28.91 ab | |
| Poplar (TUPO) | 26.41 ab | |
| Sugar maple (SUMA) | 35.94 ab | |
| Soil NH4+ (mg N/kg soil) | Sycamore (AMSY) | 4.90 ab |
| Cherry (BLCH) | 4.45 b | |
| Oak (REOA) | 8.17 a | |
| Beech (AMBE) | 6.40 ab | |
| Poplar (TUPO) | 4.83 ab | |
| Sugar maple (SUMA) | 2.79 ab | |
| Potential N2O (mg N/kg soil/30 h) | Sycamore (AMSY) | 0.83 a |
| Cherry (BLCH) | 0.33 ab | |
| Oak (REOA) | 0.48 ab | |
| Beech (AMBE) | 0.44 ab | |
| Poplar (TUPO) | 0.31 ab | |
| Sugar maple (SUMA) | 0.26 b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kelly, C.N.; Matejczyk, E.A.; Fox-Fogle, E.G.; Hubbart, J.A.; Driscoll, T.P. Tree Species Influence Nitrate and Nitrous Oxide Production in Forested Riparian Soils. Nitrogen 2023, 4, 311-330. https://doi.org/10.3390/nitrogen4040023
Kelly CN, Matejczyk EA, Fox-Fogle EG, Hubbart JA, Driscoll TP. Tree Species Influence Nitrate and Nitrous Oxide Production in Forested Riparian Soils. Nitrogen. 2023; 4(4):311-330. https://doi.org/10.3390/nitrogen4040023
Chicago/Turabian StyleKelly, Charlene N., Elizabeth A. Matejczyk, Emma G. Fox-Fogle, Jason A. Hubbart, and Timothy P. Driscoll. 2023. "Tree Species Influence Nitrate and Nitrous Oxide Production in Forested Riparian Soils" Nitrogen 4, no. 4: 311-330. https://doi.org/10.3390/nitrogen4040023





