Reactive Molecular Dynamics Study of the Thermal Decomposition of Phenolic Resins
Abstract
:1. Introduction
2. Materials and Methods
2.1. Automatic Construction of Models
2.2. Heat Capacity Simulation
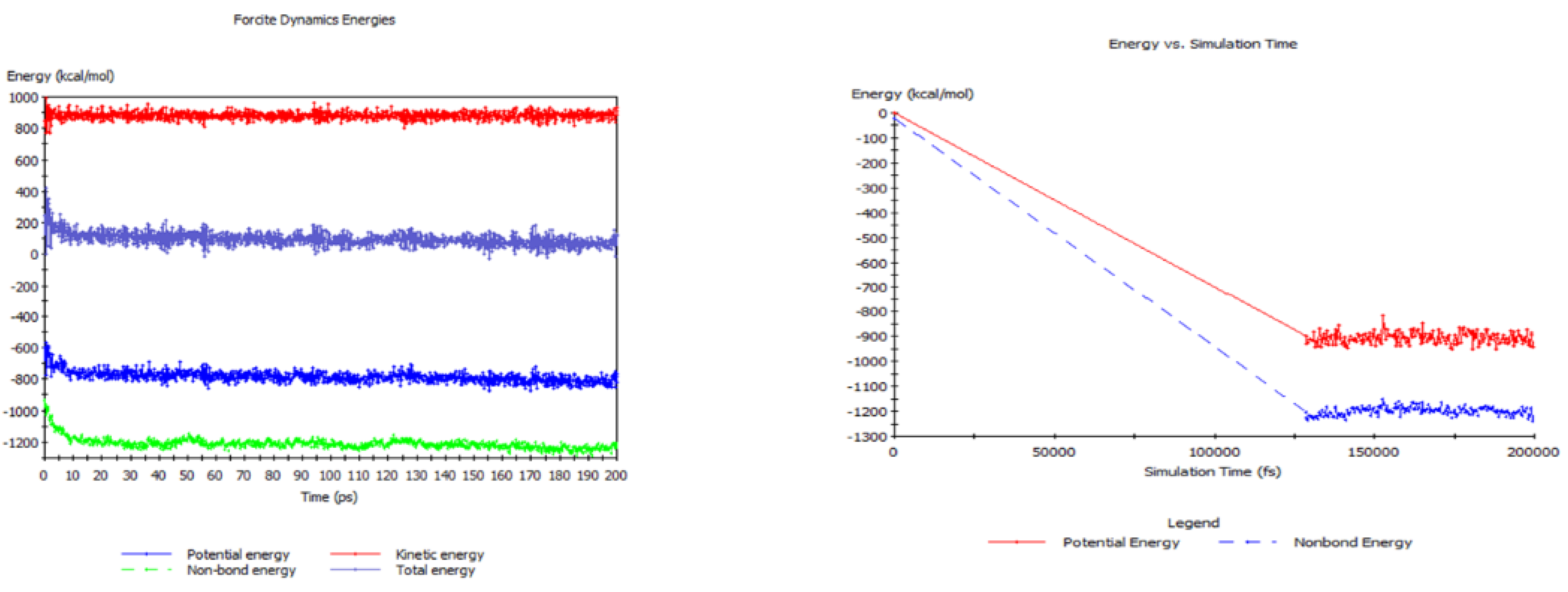
2.3. RMD Simulation of Polyphenolics
3. Results
3.1. RMD Simulations
3.1.1. The 33% Cured 986 Atom Model Simulated at 1250 K
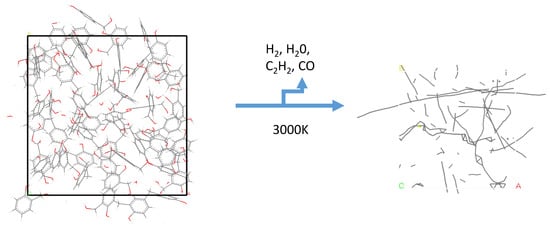
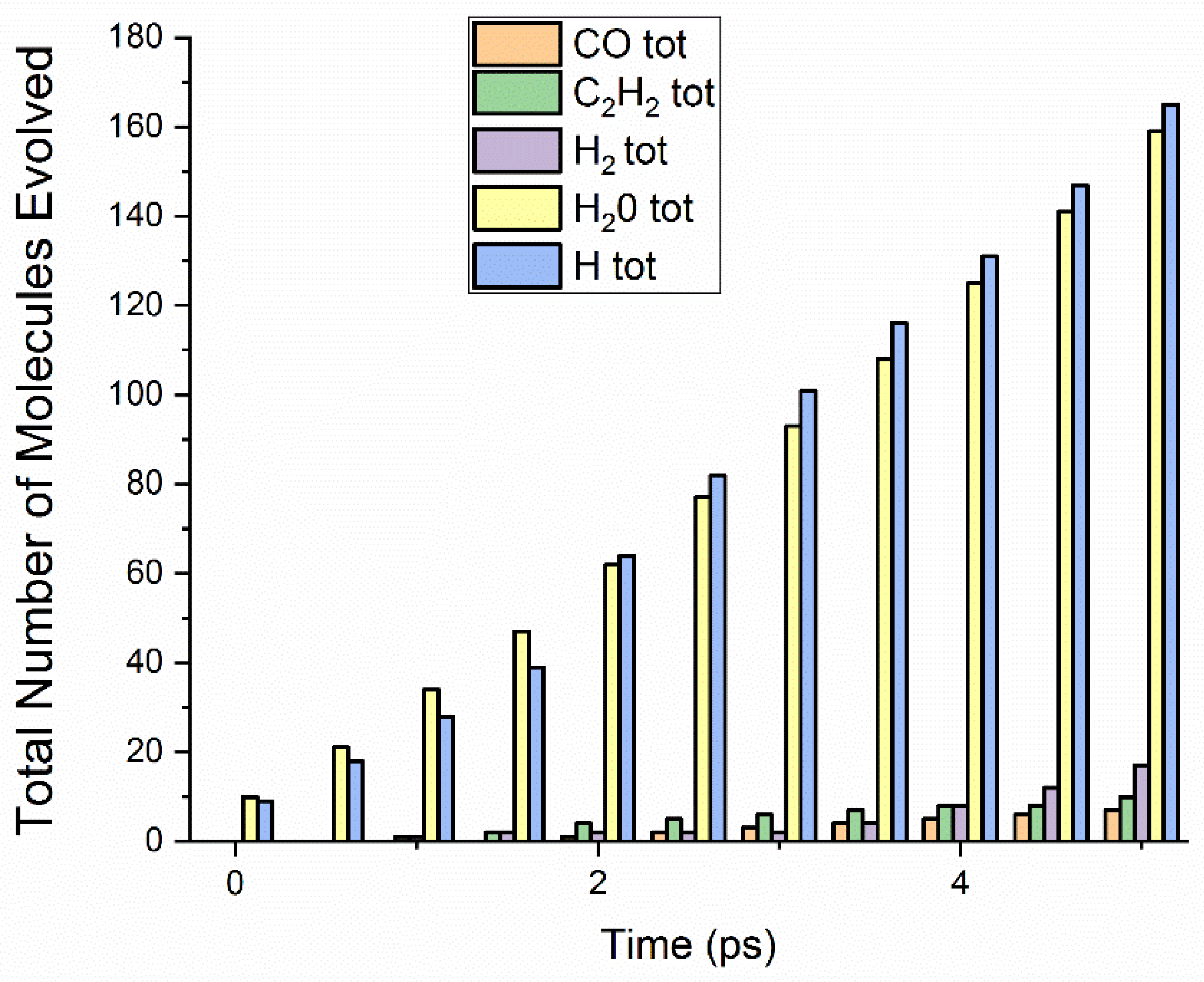
3.1.2. The 33% Cured 986 Atom Model Simulated at 3000 K
3.1.3. The 66% Cured 986 Atom Model Simulated at 1250 K
3.1.4. The 66% Cured 986 Atom Model Simulated at 3000 K
3.1.5. The 100% Cured 986 Atom Model Simulated at 1250 K
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Stoliarov, S.; Nyden, M.; Lyon, R.E. Reactive dynamics model of thermal decomposition in polymers. In Proceedings of the 48th International SAMPE Symposium, Long Beach, CA, USA, 11–15 May 2003. [Google Scholar]
- Sanjukta, B.; Tamal, B.; Mohanty, K. Molecular dynamic simulation of spontaneous combustion and pyrolysis of brown coal using ReaxFF. FUEL 2014, 136, 326–333. [Google Scholar]
- Zhan, J.H.; Wu, R.; Liu, X.; Gao, S.; Xu, G. Preliminary understanding of initial reaction process for subbituminous coal pyrolysis with molecular dynamics simulation. FUEL 2014, 134, 283–292. [Google Scholar] [CrossRef]
- Castro-Marcano, F.; Russo Jr, M.F.; Van Duin, A.C.; Mathews, J.P. Pyrolysis of a large-scale molecular model for Illinois no. 6 coal using the ReaxFF reactive force field. J. Anal. Appl. Pyrolysis 2014, 109, 79–89. [Google Scholar] [CrossRef]
- Liu, X.; Li, X.; Liu, J.; Wang, Z.; Kong, B.; Gong, X.; Yang, X.; Lin, W.; Guo, L. Study of High Density polyethylene (HDPE) pyrolysis with reactive Molecular Dynamics. Polym. Degrad. Stab. 2014, 104, 62–70. [Google Scholar] [CrossRef]
- Liu, J.; Li, X.; Guo, L.; Zheng, M.; Han, J.; Yuan, X.; Nie, F.; Liu, X. Reaction analysis and visualization of ReaxFF molecular dynamics simulations. J. Mol. Graph. Model. 2014, 53, 13–22. [Google Scholar] [CrossRef]
- Chen, W.; Duan, H.T.; Hua, M.; Gu, K.L.; Shang, H.F.; Li, J. Comparison of Oxidation Resistance of UHMWPE and POM in H2O2 Solution from ReaxFF Reactive Molecular Dynamics Simulations. J. Phys. Chem. B 2014, 118, 10311–10318. [Google Scholar] [CrossRef]
- Zhou, T.; Liu, L.; Goddard, I.I.I.W.A.; Zybin, S.V.; Huang, F. ReaxFF reactive molecular dynamics on silicon pentaerythritol tetranitrate crystal validates the mechanism for the colossal sensitivity. Phys. Chem. Chem. Phys. 2014, 16, 23779–23791. [Google Scholar] [CrossRef] [Green Version]
- Zhou, T.; Song, H.; Liu, Y.; Huang, F. Shock initiated thermal and chemical responses of HMX crystal from ReaxFF molecular dynamics simulation. Phys. Chem. Chem. Phys. 2014, 16, 13914–13931. [Google Scholar] [CrossRef]
- Cheng, H.; Zhu, Y.A.; Chen, D.; Åstrand, P.O.; Li, P.; Qi, Z.; Zhou, X.G. Evolution of Carbon Nanofiber-Supported Pt Nanoparticles of Different Particle Sizes: A Molecular Dynamics Study. J. Phys. Chem. C 2014, 118, 23711–23722. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Wen, Y.; Xue, X. Self-Enhanced Catalytic Activities of Functionalized Graphene Sheets in the Combustion of Nitromethane: Molecular Dynamic Simulations by Molecular Reactive Force Field. ACS Appl. Mater. Interfaces 2014, 6, 12235–12244. [Google Scholar] [CrossRef]
- Li, G.Y.; Xie, Q.A.; Zhang, H.; Guo, R.; Wang, F.; Liang, Y.H. Pyrolysis Mechanism of Metal-Ion-Exchanged Lignite: A Combined Reactive Force Field and Density Functional Theory Study. Energy Fuels 2014, 28, 5373–5381. [Google Scholar] [CrossRef]
- Zhang, B.; van Duin, A.C.T.; Johnson, J.K. Development of a ReaxFF Reactive Force Field for Tetrabutylphosphonium Glycinate/CO2 Mixtures. J. Phys. Chem. B 2014, 118, 12008–12016. [Google Scholar] [CrossRef]
- Fantauzzi, D.; Bandlow, J.; Sabo, L.; Mueller, J.E.; van Duin, A.C.; Jacob, T. Development of a ReaxFF potential for Pt-O systems describing the energetics and dynamics of Pt-oxide formation. Phys. Chem. Chem. Phys. 2014, 16, 23118–23133. [Google Scholar] [CrossRef]
- Schönfelder, T.; Friedrich, J.; Prehl, J.; Seeger, S.; Spange, S.; Hoffmann, K.H. Reactive force field for electrophilic substitution at an aromatic system in twin polymerization. Chem. Phys. 2014, 440, 119–126. [Google Scholar] [CrossRef]
- Cheng, T.; Jaramillo-Botero, A.; Goddard, I.I.I.W.A.; Sun, H. Adaptive Accelerated ReaxFF Reactive Dynamics with Validation from Simulating Hydrogen Combustion. J. Am. Chem. Soc. 2014, 136, 13467. [Google Scholar] [CrossRef]
- Kylasa, S.B.; Aktulga, H.M.; Grama, A.Y. PuReMD-GPU: A reactive molecular dynamics simulation package for GPUs. J. Comput. Phys. 2014, 272, 343–359. [Google Scholar] [CrossRef]
- Farah, K.; Muller-Plathe, F.; Bohm, M.C. Classical Reactive Molecular Dynamics Implementations: State of the Art. Minireview. ChemPhysChem 2012, 13, 1127–1151. [Google Scholar] [CrossRef]
- Xiao, H.; Shi, X.; Hao, F.; Liao, X.; Zhang, Y.; Chen, Y. Development of a Transferable Reactive Force Field of P/H Systems: Application to the Chemical and Mechanical Properties of Phosphorene. J. Phys. Chem. A 2017, 121, 6135–6149. [Google Scholar] [CrossRef] [Green Version]
- Gardziella, A.; Knop, A.; Pilato, L.A. Phenolic Resins, 2nd ed.; Springer: Berlin, Germany, 2000; pp. 83–90. [Google Scholar]
- Fakirov, S. Fundamentals of Polymer Science for Engineers; Wiley-VCH: Weinheim, Germany, 2017. [Google Scholar]
- Hall, S.A.; Howlin, B.J.; Hamerton, I.; Baidak, A.; Billaud, C.; Ward, S. Solving the Problem of Building Models of Crosslinked Polymers: An Example Focussing on Validation of the Properties of Crosslinked Epoxy Resins. PloS ONE 2012. [Google Scholar] [CrossRef]
- Jensen, F. Introduction to Computational Chemistry, 2nd ed.; Wiley: Chichester, UK, 2007; pp. 445–471. [Google Scholar]
- Woodcock, L.V. Isothermal molecular dynamics calculations for liquid salts. Chem. Phys. Lett. 1971, 10, 257–261. [Google Scholar] [CrossRef]
- Hünenberger, P.H. Thermostat Algorithms for Molecular Dynamics Simulations. Adv. Polym. Sci. 2005, 173, 105–149. [Google Scholar]
- Andersen, H.C.J. Molecular dynamics simulations at constant pressure and/or temperature. Chem. Phys. 1980, 72, 2384–2393. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Morishita, T.J. Fluctuation formulas in molecular-dynamics simulations with the weak coupling heat bath. Chem. Phys. 2000, 113, 2976–2982. [Google Scholar] [CrossRef]
- Nosé, S. A molecular dynamics method for simulations in the canonical ensemble. Mol. Phys. 1984, 52, 255–268. [Google Scholar] [CrossRef]
- Nosé, S.J. A unified formulation of the constant temperature molecular dynamics methods. Chem. Phys. 1984, 81, 511–519. [Google Scholar]
- Hoover, W.G. Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A 1985, 31, 1695–1697. [Google Scholar] [CrossRef]
- Allen, M.P.; Tildesley, D.J. Computer Simulation of Liquids; Oxford University Press: New York, NY, USA, 1989. [Google Scholar]
- McNaught, A.D.; Wilkinson, A. Compendium of Chemical Terminology, 2nd ed.; Blackwell Scientific Publications: Oxford, UK, 1997. [Google Scholar]
- Fyfe, C.A.; McKinnon, A.R.; Tchir, W.J. Investigation of the Mechanism of the Thermal Decomposition of Cured Phenolic Resins by High-Resolution 13C CP/MAS Solid-state NMR Spectroscopy. Macromolecules 1983, 16, 1216–1219. [Google Scholar] [CrossRef]
- Erä, V.A.; Mattila, A.; Lindberg, J. Determination of specific heat of phenol formaldehyde resol resins by differential scanning calorimetry. J. Angew. Makromol. Chem. 1977, 64, 235–238. [Google Scholar] [CrossRef]
- Trick, K.A.; Saliba, T.E. Mechanisms of the pyrolysis of phenolic resin in a carbon/phenolic composite. Carbon 1995, 33, 1509–1515. [Google Scholar] [CrossRef]




| tsim (ps) | Cure % of 1000 Atom Model | Forcite | Discover | ||
|---|---|---|---|---|---|
| Tavg (K) | E (kJ·mol−1) | Tavg (K) | E (kJ·mol−1) | ||
| 5 | 33% Cured | 299.728 | 843 | 277.924 | 617 |
| 50 | 33% Cured | 300.002 | 318 | 275.732 | 257 |
| 200 | 33% Cured | 299.997 | 229 | 276.660 | 183 |
| 5 | 66% Cured | 299.714 | 707 | 277.059 | 661 |
| 50 | 66% Cured | 300.014 | 310 | 276.983 | 277 |
| 200 | 66% Cured | 300.004 | 245 | 276.000 | 186 |
| 5 | 100% Cured | 297.883 | 632 | 276.527 | 677 |
| 50 | 100% Cured | 300.001 | 285 | 277.012 | 272 |
| 200 | 100% Cured | 300.005 | 196 | 275.565 | 186 |
| tsim (ps) | Cure % of 1000 Atom Model | Forcite | Discover | ||
|---|---|---|---|---|---|
| Cv/N (J·mol−1·K−1) | S (kJ·kg−1·K−1) | Cv/N (J·mol−1·K−1) | S (kJ·kg−1·K−1) | ||
| 5 | 33% Cure | 951.496 | 125.525 | 592.973 | 78.227 |
| 50 | 33% Cure | 135.554 | 17.883 | 104.376 | 13.770 |
| 200 | 33% Cure | 70.386 | 9.286 | 52.864 | 6.974 |
| 5 | 66% Cure | 670.403 | 88.442 | 685.086 | 90.379 |
| 50 | 66% Cure | 128.226 | 16.916 | 120.336 | 15.875 |
| 200 | 66% Cure | 80.072 | 10.563 | 54.822 | 7.232 |
| 5 | 100% Cure | 541.234 | 71.402 | 721.952 | 95.243 |
| 50 | 100% Cure | 108.441 | 14.306 | 116.422 | 15.359 |
| 200 | 100% Cure | 51.573 | 6.804 | 54.924 | 7.246 |
| Region 1 (573–823 K) | Region 2 (673–1073 K) | Region 3 (833–1173 K) | |||
|---|---|---|---|---|---|
| Species | Molar % | Species | Molar % | Species | Molar % |
| H | 0 | H | 59.4 | H | 0 |
| H2 | 0 | H2 | 0 | H2 | 85.7 |
| H2O | 49.8 | H2O2 | 12.7 | H2O | 4.70 |
| CO | 0 | CO | 12.7 | CO | 9.5 |
| O2 | 0.1 | O2 | 0 | O2 | 0 |
| CO2 | 0 | CO2 | 0.2 | CO2 | 0.1 |
| CH4 | 0 | CH4 | 14.9 | CH4 | 0 |
| C2H6 | 0 | C2H6 | 0.1 | C2H6 | 0 |
| Phenol, Cresol | 50.1 | Phenol, Cresol | 0 | Phenol, Cresol | 0 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Purse, M.; Edmund, G.; Hall, S.; Howlin, B.; Hamerton, I.; Till, S. Reactive Molecular Dynamics Study of the Thermal Decomposition of Phenolic Resins. J. Compos. Sci. 2019, 3, 32. https://doi.org/10.3390/jcs3020032
Purse M, Edmund G, Hall S, Howlin B, Hamerton I, Till S. Reactive Molecular Dynamics Study of the Thermal Decomposition of Phenolic Resins. Journal of Composites Science. 2019; 3(2):32. https://doi.org/10.3390/jcs3020032
Chicago/Turabian StylePurse, Marcus, Grace Edmund, Stephen Hall, Brendan Howlin, Ian Hamerton, and Stephen Till. 2019. "Reactive Molecular Dynamics Study of the Thermal Decomposition of Phenolic Resins" Journal of Composites Science 3, no. 2: 32. https://doi.org/10.3390/jcs3020032