Association, Conformational Rearrangements and the Reverse Process of Aggregates Dissociation during Apomyoglobin Amyloid Formation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Protein Expression and Isolation
2.2. Absorption Spectroscopy
2.3. Aggregation Conditions
2.4. Fluorescence Spectroscopy
2.5. Electron Microscopy
2.6. Fourier Transforms Infrared Spectroscopy (FTIR)
2.7. Non-Denaturing Electrophoresis
2.8. Amyloids Purification
3. Results
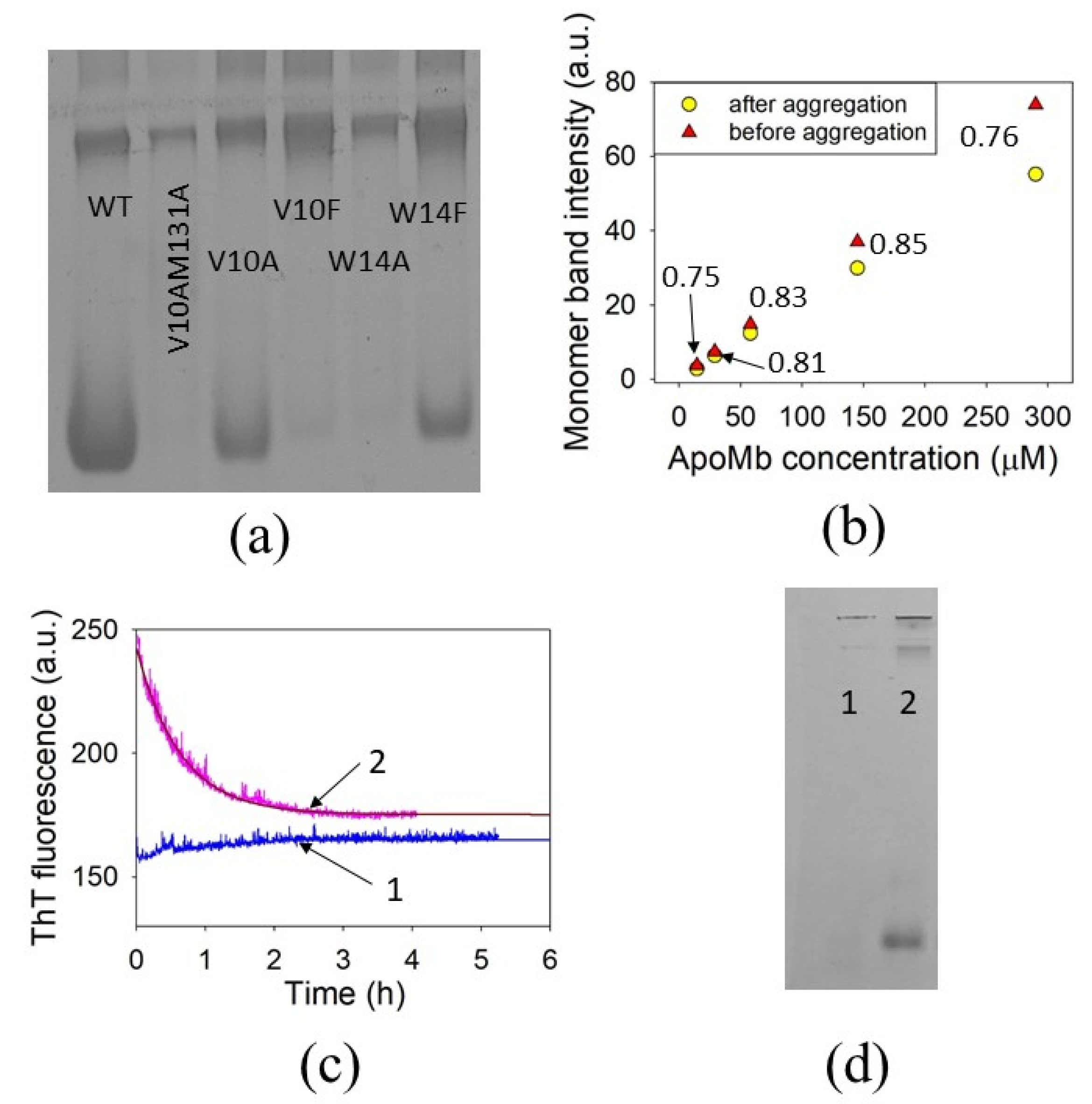
3.1. Selection of Amino Acid Residues for Substitutions
3.2. The Stages of ApoMb Amyloid Formation
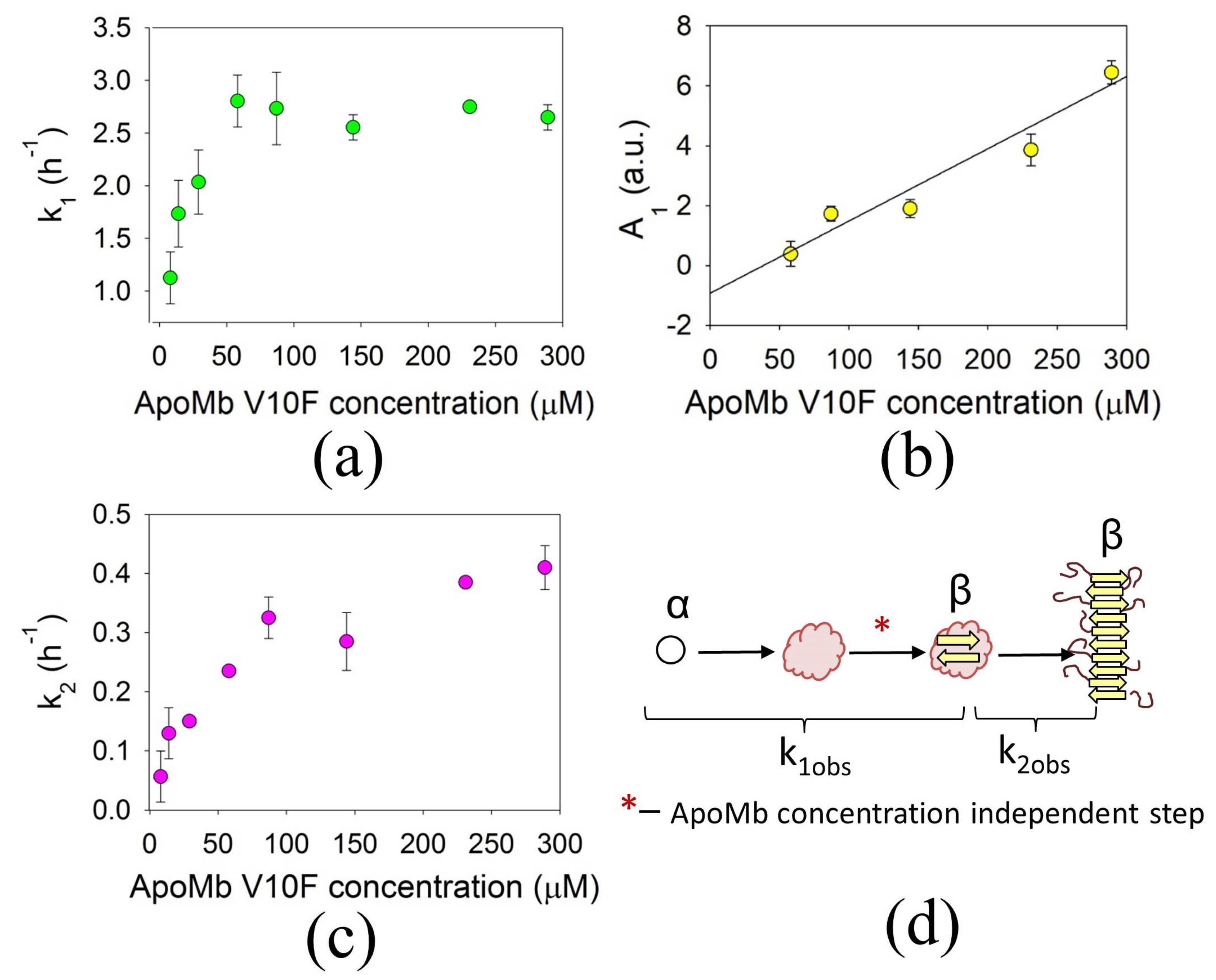
3.3. Concentration Dependences of the ApoMb Aggregation Rates
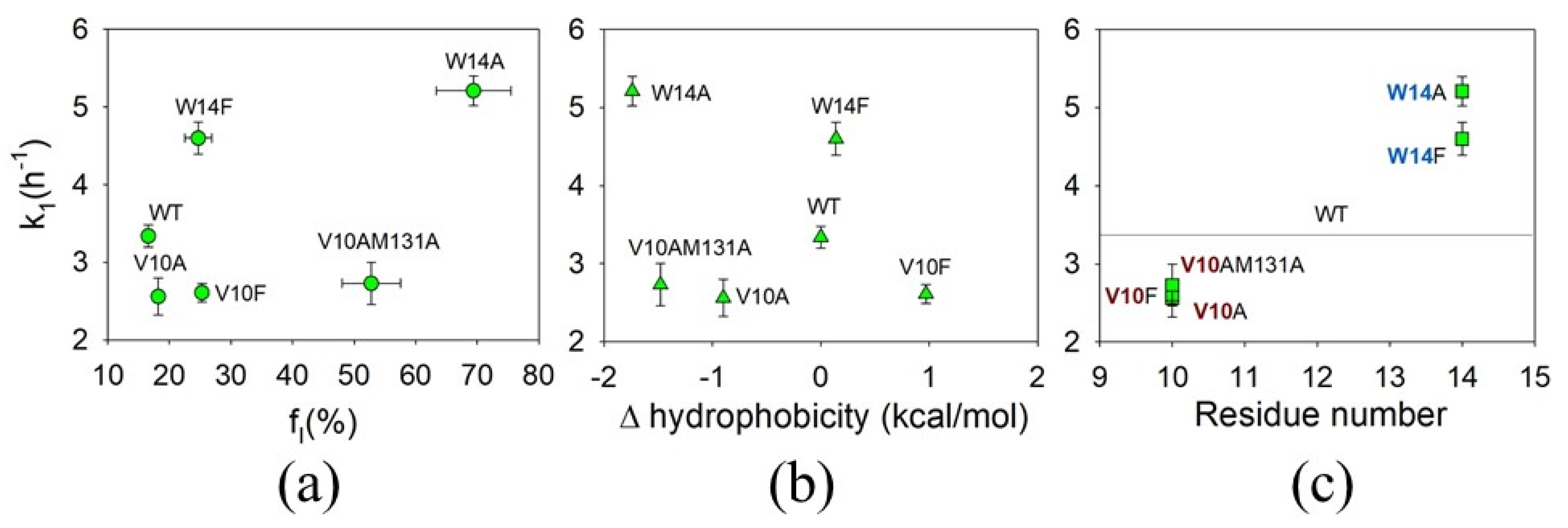
3.4. The Effect of Mutations on the First Rate of ApoMb Amyloid Aggregation
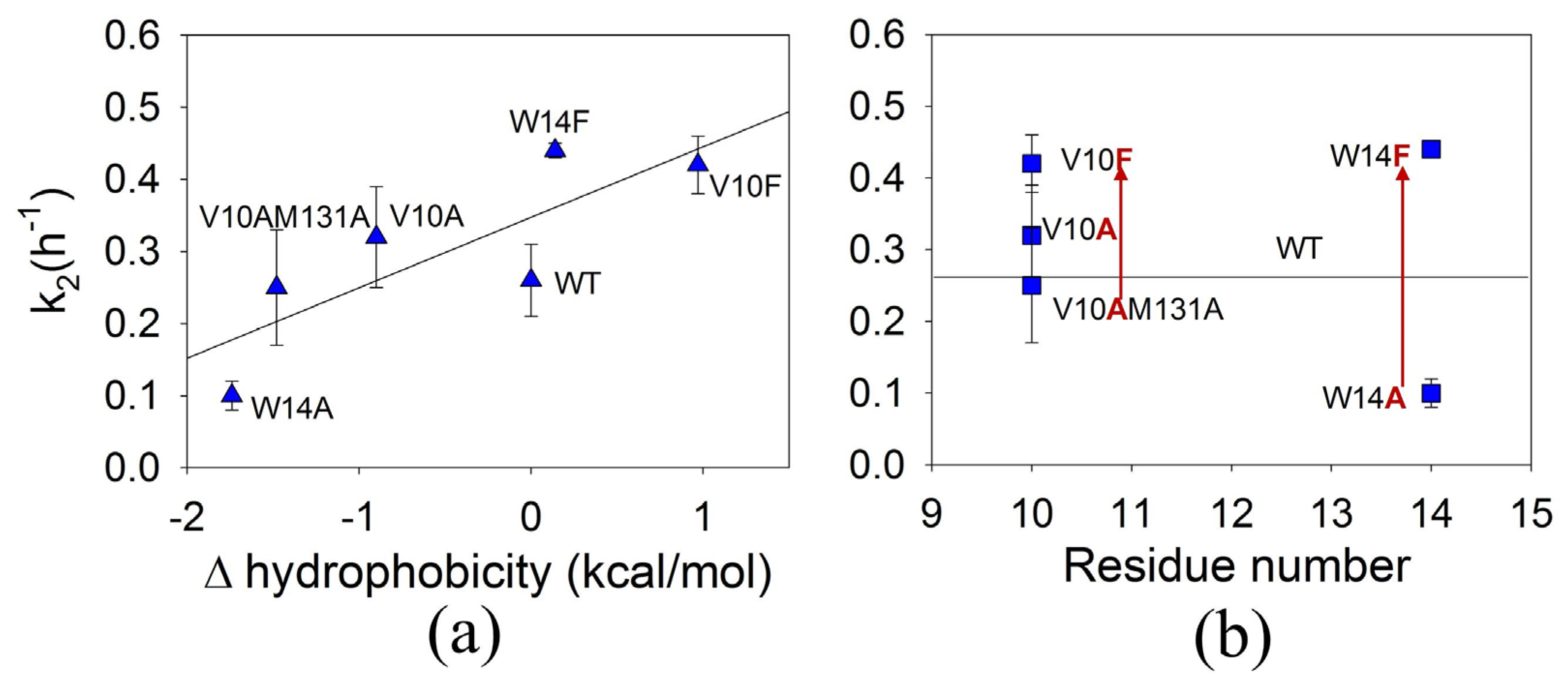
3.5. The Effect of Mutations on the Second Rate of ApoMb Amyloid Aggregation
3.6. The Amyloid Formation Reversibility
4. Discussion
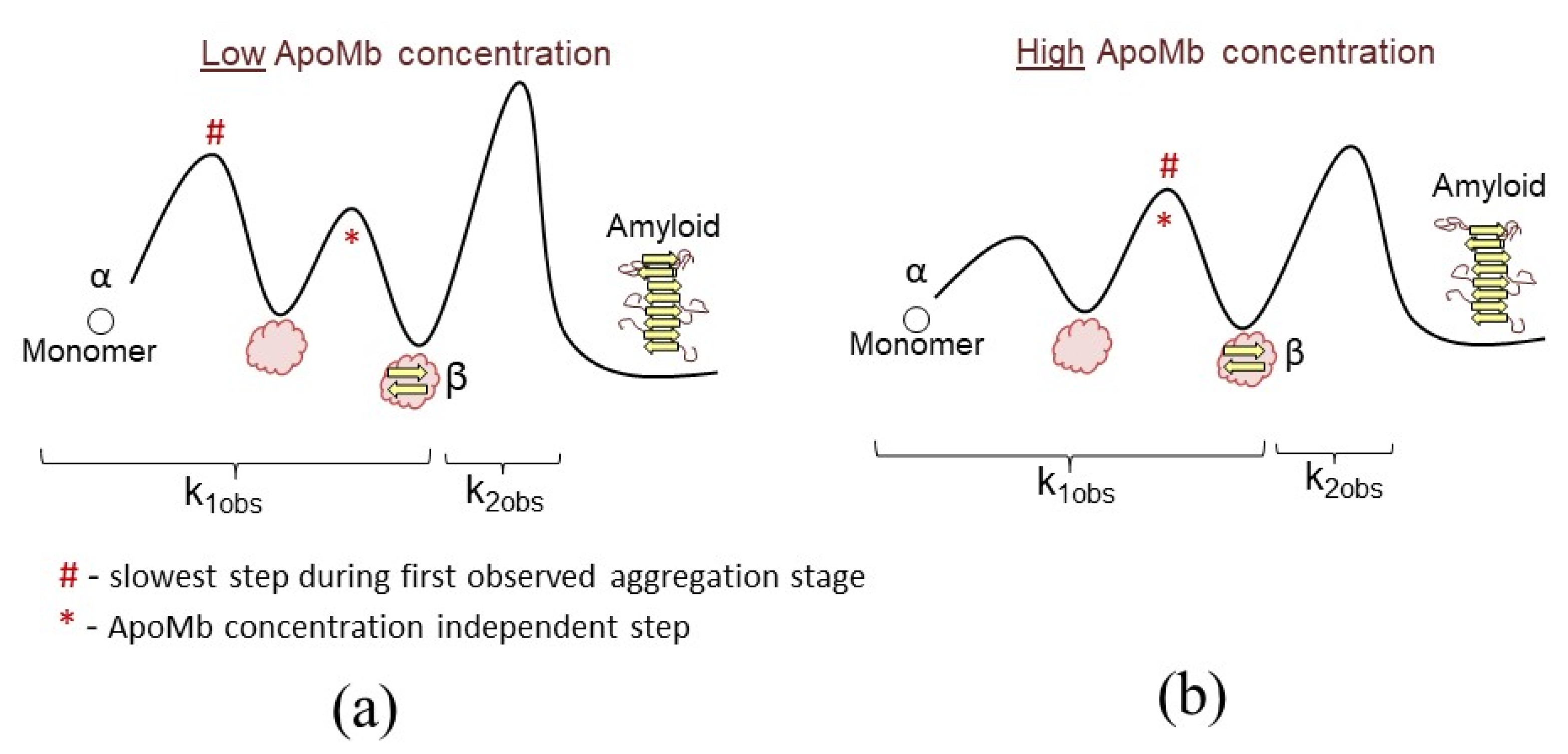
4.1. Association and Conformational Rearrangements during Amyloid Aggregation
4.2. The Reversibility of ApoMb Amyloid Formation
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Makin, O.S.; Atkins, E.; Sikorski, P.; Johansson, J.; Serpell, L.C. Molecular basis for amyloid fibril formation and stability. Proc. Natl. Acad. Sci. USA 2005, 102, 315–320. [Google Scholar] [CrossRef]
- Knowles, T.P.; Vendruscolo, M.; Dobson, C.M. The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell. Biol. 2014, 15, 384–396. [Google Scholar] [CrossRef]
- Chiti, F.; Dobson, C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006, 75, 333–366. [Google Scholar] [CrossRef] [Green Version]
- Salahuddin, P.; Fatima, M.T.; Abdelhameed, A.S.; Nusrat, S.; Khan, R.H. Structure of amyloid oligomers and their mechanism of toxicities: Targeting amyloid oligomers using novel therapeutic approaches. Eur. J. Med. Chem. 2016, 23, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Bucciantini, M.; Giannoni, E.; Chiti, F.; Baroni, F.; Formigli, L.; Zurdo, J.; Taddei, N.; Ramponi, G.; Dobson, C.M.; Stefani, M. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 2002, 416, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Winner, B.; Jappelli, R.; Maji, S.K.; Desplats, P.A.; Boyer, L.; Aigner, S.; Hetzer, C.; Lo-her, T.; Vilar, M.; Campioni, S.; et al. In vivo demonstration that alpha-synuclein oligomers are toxic. Proc. Natl. Acad. Sci. USA 2011, 108, 4194–4199. [Google Scholar] [CrossRef] [PubMed]
- Habchi, J.; Chia, S.; Limbocker, R.; Manini, B.; Ahh, M.; Perni, M.; Hansson, O.; Arosio, P.; Kumita, J.R.; Challa, P.K.; et al. Systematic development of small molecules to inhibit specific microscopic steps of Aβ42 aggregation in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2017, 114, E200–E208. [Google Scholar] [CrossRef]
- Goldstein, R.F.; Stryer, L. Cooperative polymerization reactions. Analytical approximations, numerical examples, and experimental strategy. Biophys. J. 1986, 50, 583–599. [Google Scholar] [CrossRef] [Green Version]
- Cohen, S.I.A.; Vendruscolo, M.; Dobson, C.M.; Knowles, T.P.J. From macroscopic measurements to microscopic mechanisms of protein aggregation. J. Mol. Biol. 2012, 421, 160–171. [Google Scholar] [CrossRef]
- Lomakin, A.; Chung, D.S.; Benedek, G.B.; Kirschner, D.A.; Teplow, D.B. On the nucleation and growth of amyloid beta-protein fibrils: Detection of nuclei and quantitation of rate constants. Proc. Natl. Acad. Sci. USA 1996, 93, 1125–1129. [Google Scholar] [CrossRef]
- Cohen, S.I.A.; Linse, S.; Luheshi, L.M.; Hellstrand, E.; White, D.A.; Rajah, L.; Otzen, D.E.; Vendruscolo, M.; Dobson, C.M.; Knowles, T.P.J. Proliferation of amyloid-β42 aggregates occurs through a secondary nucleation mechanism. Proc. Natl. Acad. Sci. USA 2013, 110, 9758–9763. [Google Scholar] [CrossRef]
- Gillam, J.E.; MacPhee, C.E. Modelling amyloid fibrils formation kinetics: Mechanism of nucleation growth. J. Phys. Condens. Matter. 2013, 25, 373101. [Google Scholar] [CrossRef]
- Chiti, F.; Taddei, N.; Baroni, F.; Capanni, C.; Stefani, M.; Ramponi, G.; Dobson, C.M. Kinetic partitioning of protein folding and aggregation. Nat. Struct. Biol. 2002, 9, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Calamai, M.; Taddei, N.; Stefani, M.; Ramponi, G.; Chiti, F. Relative influence of hydrophobicity and net charge in the aggregation of two homologous proteins. Biochemistry 2003, 42, 15078–15083. [Google Scholar] [CrossRef] [PubMed]
- Chiti, F.; Stefani, M.; Taddei, N.; Ramponi, G.; Dobson, C.M. Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature 2003, 424, 805–808. [Google Scholar] [CrossRef]
- Tjernberg, L.; Hosia, W.; Bark, N.; Thyberg, J.; Johansson, J. Charge attraction and beta propensity are necessary for amyloid fibril formation from tetrapeptides. J. Biol. Chem. 2002, 277, 43243–43246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plakoutsi, G.; Bemporad, F.; Monti, M.; Pagnozzi, D.; Pucci, P.; Chiti, F. Exploring the mechanism of formation of native-like and precursor amyloid oligomers for the native acylphosphatase from Sulfolobus solfataricus. Structure 2006, 14, 993–1001. [Google Scholar] [CrossRef] [Green Version]
- Marin-Argany, M.; Guell-Bosch, J.; Blancas-Mejia, L.M.; Villegas, S.; Ramirez-Alvarado, M. Mutations can cause light chains to be too stable or too unstable to form amyloid fibrils. Protein Sci. 2015, 24, 1829–1840. [Google Scholar] [CrossRef] [Green Version]
- Cerda-Costa, N.; Esteras-Chopo, A.; Aviles, F.X.; Serrano, L.; Villegas, V. Early kinetics of amyloid fibril formation reveals conformational reorganization of initial aggregates. J. Mol. Biol. 2007, 366, 1351–1363. [Google Scholar] [CrossRef]
- Serio, T.R.; Cashikar, A.G.; Kowal, A.S.; Sawicki, G.J.; Moslehi, J.J.; Serpell, L.; Arnsdorf, M.F.; Lindquist, S.L. Nucleated conformational conversion and the replication of conformational information by a prion. Science 2000, 289, 1317–1321. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Mohanty, S.K.; Udgaonkar, J.B. Mechanism of formation of protofibrils of barstar from soluble oligomers: Evidence for multiple steps and lateral association coupled to conformational conversion. J. Mol. Biol. 2007, 367, 1186–1204. [Google Scholar] [CrossRef] [PubMed]
- Leonil, J.; Henry, G.; Jouanneau, D.; Delage, M.M.; Forge, V.; Putaux, J.L. Kinetics of fibril formation of bovine kappa-casein indicate a conformational rearrangement as a critical step in the process. J. Mol. Biol. 2008, 381, 1267–1280. [Google Scholar] [CrossRef] [PubMed]
- Zandomeneghi, G.; Krebs, M.R.H.; McCammon, M.G.; Fandrich, M. FTIR reveals structural differences between native beta-sheet proteins and amyloid fibrils. Protein Sci. 2004, 13, 3314–3321. [Google Scholar] [CrossRef] [Green Version]
- Biancalana, M.; Makabe, K.; Koide, A.; Koide, S. Molecular mechanism of thioflavin-T binding to the surface of beta-rich peptide self-assemblies. J. Mol. Biol. 2009, 385, 1052–1063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vignaud, H.; Bobo, C.; Lascu, I.; Sorgjerd, M.K.; Zako, T.; Maeda, M.; Salin, B.; Lecomte, S.; Cullin, C. A structure-toxicity study of Aβ42 reveals a new anti-parallel aggregation pathway. PLoS ONE 2013, 8, e80262. [Google Scholar] [CrossRef] [Green Version]
- Celej, M.S.; Sarroukh, R.; Goormaghtigh, E.; Fidelio, G.D.; Rysschaert, J.-M.; Raussens, V. Toxic prefibrillar α-synuclein amyloid oligomers adopt distinctive antiparallel β-sheet structure. Biochem. J. 2012, 443, 719–726. [Google Scholar] [CrossRef]
- Hoshino, M. Fibril formation from the amyloid β-peptide is governed by a dynamic equilibrium involving association and dissociation of the monomer. Biophys. Rev. 2017, 9, 9–16. [Google Scholar] [CrossRef] [Green Version]
- Gazit, E. The “Correctly Folded” state of proteins: Is it a metastable state? Angew. Chem. Int. Ed. Engl. 2002, 41, 257–259. [Google Scholar] [CrossRef]
- Buell, A.K. Stability matters, too—the thermodynamics of amyloid fibril formation. Chem. Sci. 2022, 13, 10177–10192. [Google Scholar] [CrossRef]
- Pashley, C.L.; Hewitt, E.W.; Radford, S.E. Comparison of the aggregation of homologous β2-microglobulin variants reveals protein solubility as a key determinant of amyloid formation. J. Mol. Biol. 2016, 428, 631–643. [Google Scholar] [CrossRef] [Green Version]
- Williams, A.D.; Portelius, E.; Kheterpal, I.; Guo, J.-T.; Cook, K.D.; Xu, Y.; Wetzel, R. Mapping abeta amyloid fibril secondary structure using scanning proline mutagenesis. J. Mol. Biol. 2004, 335, 833–842. [Google Scholar] [CrossRef] [PubMed]
- O’Nuallain, B.; Shivaprasad, S.; Kheterpal, I.; Wetzel, R. Thermodynamics of A beta(1–40) amyloid fibril elongation. Biochemistry 2005, 44, 12709–12718. [Google Scholar] [CrossRef] [PubMed]
- Katina, N.S.; Balobanov, V.A.; Ilyina, N.B.; Vasiliev, V.D.; Marchenkov, V.V.; Glukhov, A.S.; Nikulin, A.D.; Bychkova, V.E. sw ApoMb amyloid aggregation under non-denaturing conditions: The role of native structure stability. Biophys. J. 2017, 113, 991–1001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eliezer, D.; Wright, P.E. Is apomyoglobin a molten globule? Structural characterization by NMR. J. Mol. Biol. 1996, 263, 531–538. [Google Scholar] [CrossRef]
- Griko, Y.V.; Privalov, P.L. Thermodynamic puzzle of apomyoglobin unfolding. J. Mol. Biol. 1994, 235, 1318–1325. [Google Scholar] [CrossRef]
- Nishimura, C.; Dyson, H.J.; Wright, P.E. Identification of native and non-native structure in kinetic folding intermediates of apomyoglobin. J. Mol. Biol. 2006, 355, 139–156. [Google Scholar] [CrossRef]
- Fandrich, M.; Forge, V.; Buder, K.; Kittler, M.; Dobson, C.M.; Diekmann, S. Myoglobin forms amyloid fibrils by association of unfolded polypeptide segments. Proc. Natl. Acad. Sci. USA 2003, 100, 15463–15468. [Google Scholar] [CrossRef]
- Sirangelo, I.; Malmo, C.; Iannuzzi, C.; Mezzogiorno, A.; Bianco, M.R.; Papa, M.; Irace, G. Fibrillogenesis and cytotoxic activity of the amyloid-forming apomyoglobin mutant W7FW14F. J. Biol. Chem. 2004, 279, 13183–13189. [Google Scholar] [CrossRef] [Green Version]
- Picotti, P.; Franceschi, G.D.; Frare, E.; Spolaore, B.; Zambonin, M.; Chiti, F.; Polverino de Laureto, P.; Fontana, A. Amyloid fibril formation and disaggregation of fragment 1-29 of apomyoglobin: Insight into the effect of pH on protein fibrillogenesis. J. Mol. Biol. 2007, 367, 1237–1245. [Google Scholar] [CrossRef]
- Samatova, E.N.; Katina, N.S.; Balobanov, V.A.; Melnik, B.S.; Dolgikh, D.A.; Bychkova, V.E.; Finkelstein, A.V. How strong are side chain interactions in the folding intermediate? Protein Sci. 2009, 18, 2152–2159. [Google Scholar] [CrossRef] [Green Version]
- Gill, S.C.; von Hippel, P.H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989, 182, 319–326. [Google Scholar] [CrossRef]
- Marchenkov, V.; Ivashina, T.; Marchenko, N.; Ryabova, N.; Selivanova, O.; Timchenko, A.; Kihara, H.; Ksenzenko, V.; Semisotnov, G. In vivo incorporation of photoproteins into GroEL chaperonin retaining structural and functional properties. Molecules 2023, 28, 1901. [Google Scholar] [CrossRef] [PubMed]
- Katina, N.S.; Ilyina, N.B.; Kashparov, I.A.; Balobanov, V.A.; Vasiliev, V.D.; Bychkova, V.E. Apomyoglobin mutants with single point mutations at val10 can form amyloid structures at permissive temperature. Biochemistry 2011, 76, 555–563. [Google Scholar] [CrossRef]
- Pollard, T.D.; De La Cruz, E.M. Take advantage of time in your experiments: A guide to simple, informative kinetics assays. Mol. Biol. Cell 2013, 24, 1103–1110. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, A.V.; Dovidchenko, N.V.; Galzitskaya, O.V. What is responsible for atypical dependence of the rate of amyloid formation on protein concentration: Fibril-catalyzed initiation of new fibrils or competition with oligomers? J. Phys. Chem. Lett. 2018, 9, 1002–1006. [Google Scholar] [CrossRef]
- DuBay, K.F.; Pawar, A.P.; Chiti, F.; Zurdo, J.; Dobson, C.M.; Vendruscolo, M. Prediction of the absolute aggregation rates of amyloidogenic polypeptide chains. J. Mol. Biol. 2004, 341, 1317–1326. [Google Scholar] [CrossRef] [PubMed]
- Tartaglia, G.G.; Cavalli, A.; Pellarin, R.; Caflish, A. The role of aromaticity, exposed surface, and dipole moment in determining protein aggregation rates. Protein Sci. 2004, 13, 1939–1941. [Google Scholar] [CrossRef] [PubMed]
- Rawat, P.; Prabakaran, R.; Kumar, S.; Gromiha, M.M. AbsoluRATE: An in-silico method to predict the aggregation kinetics of native proteins. Biochim. Biophys. Acta Proteins Proteom. 2021, 1896, 140682. [Google Scholar] [CrossRef]
- Harper, J.D.; Lansbury, P.T. Models of amyloid seeding in Alzheimer’s disease and scrapie: Mechanistic truths and physiological consequences of the time-dependent solubility of amyloid proteins. Annu Rev. Biochem. 1997, 66, 385–407. [Google Scholar] [CrossRef]


| ApoMb Variant | k1obs (h−1) | k2obs (h−1) | fI (%) a | Δhydrophobicity (kcal/mol) b | Monomer Band Intensity (a.u.) c |
|---|---|---|---|---|---|
| W14A | 5.21 ± 0.09 | 0.10 ± 0.02 | 69.4 ± 6.1 | −1.74 | 0.08 ± 0.01 |
| V10AM131A | 2.73 ± 0.27 | 0.25 ± 0.08 | 52.8 ± 4.7 | −1.48 | 0.09 ± 0.02 |
| V10A | 2.56 ± 0.24 | 0.32 ± 0.07 | 18.2 ± 0.9 | −0.90 | 0.49 ± 0.01 |
| WT | 3.34 ± 0.14 | 0.26 ± 0.05 | 16.6 ± 0.6 | 0 | 1.00 ± 0 |
| W14F | 4.60 ± 0.01 | 0.44 ± 0.01 | 24.7 ± 2.2 | 0.14 | 0.34 ± 0.02 |
| V10F | 2.61 ± 0.12 | 0.42 ± 0.04 | 25.3 ± 0.6 | 0.97 | 0.11 ± 0.02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marchenkov, V.; Balobanov, V.; Majorina, M.; Ilyina, N.; Kashparov, I.; Glukhov, A.; Ryabova, N.; Katina, N. Association, Conformational Rearrangements and the Reverse Process of Aggregates Dissociation during Apomyoglobin Amyloid Formation. Physchem 2023, 3, 304-318. https://doi.org/10.3390/physchem3030021
Marchenkov V, Balobanov V, Majorina M, Ilyina N, Kashparov I, Glukhov A, Ryabova N, Katina N. Association, Conformational Rearrangements and the Reverse Process of Aggregates Dissociation during Apomyoglobin Amyloid Formation. Physchem. 2023; 3(3):304-318. https://doi.org/10.3390/physchem3030021
Chicago/Turabian StyleMarchenkov, Victor, Vitaly Balobanov, Mariya Majorina, Nelly Ilyina, Ivan Kashparov, Anatoly Glukhov, Natalya Ryabova, and Natalya Katina. 2023. "Association, Conformational Rearrangements and the Reverse Process of Aggregates Dissociation during Apomyoglobin Amyloid Formation" Physchem 3, no. 3: 304-318. https://doi.org/10.3390/physchem3030021





