L-Alanine Prototrophic Suppressors Emerge from L-Alanine Auxotroph through Stress-Induced Mutagenesis in Escherichia coli
Abstract
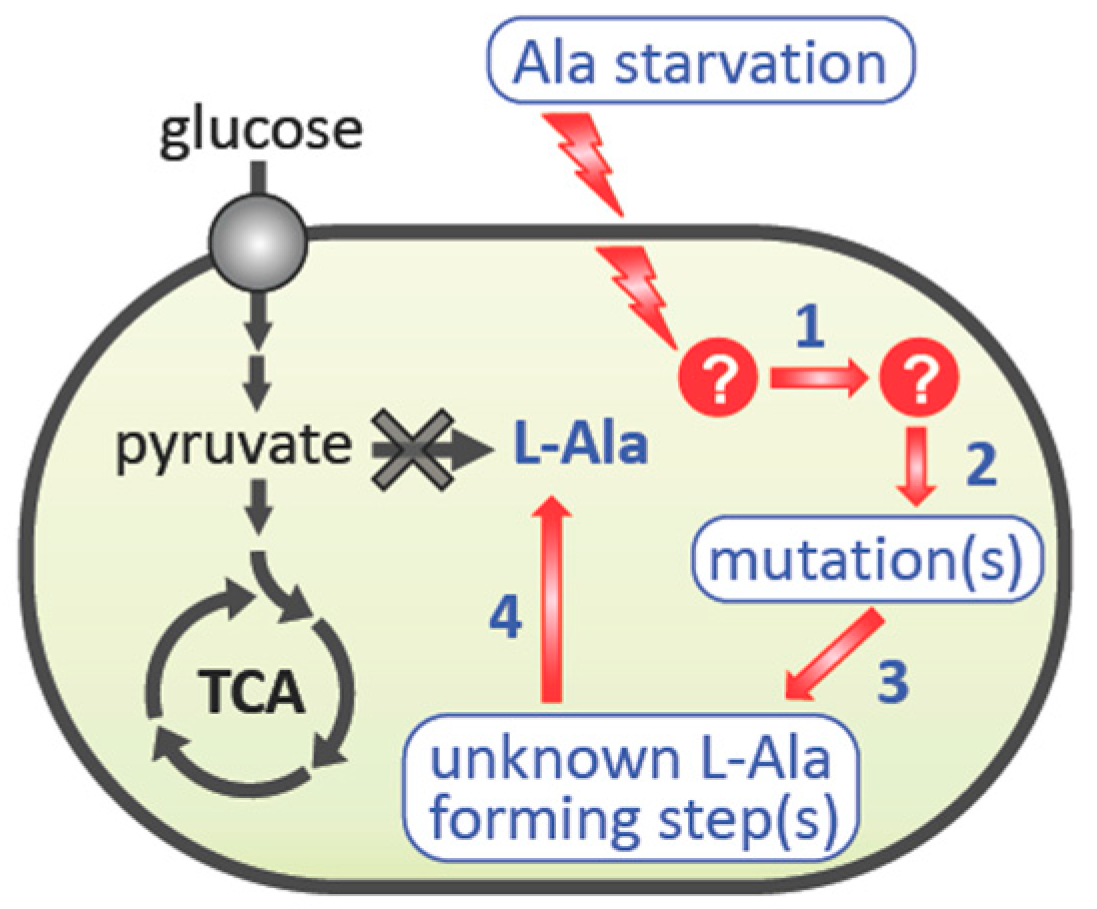
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Culture Conditions
2.2. Isolation of L-Alanine Prototrophic Clones
2.3. Frequencies of Mutants Grown in Minimal Medium with or without L-Alanine Supplementation
2.4. DNA Sequencing and Annotation of Mutations
2.5. Construction of Isogenic Mutants
2.6. Crude Cell Extract Preparation
2.7. Enzyme Activity Assay
2.8. Protein Concentration Measurement
2.9. Statistics
3. Results
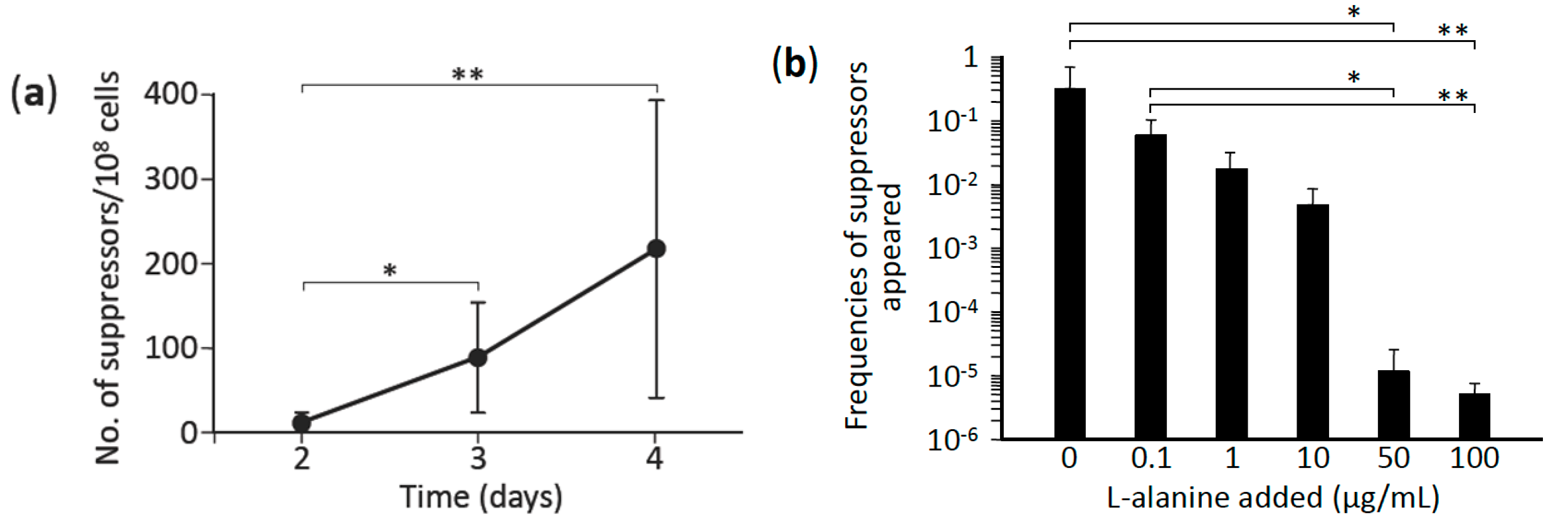
3.1. L-Alanine Prototrophs Emerge from L-Alanine Auxotroph through Suppressor Mutation
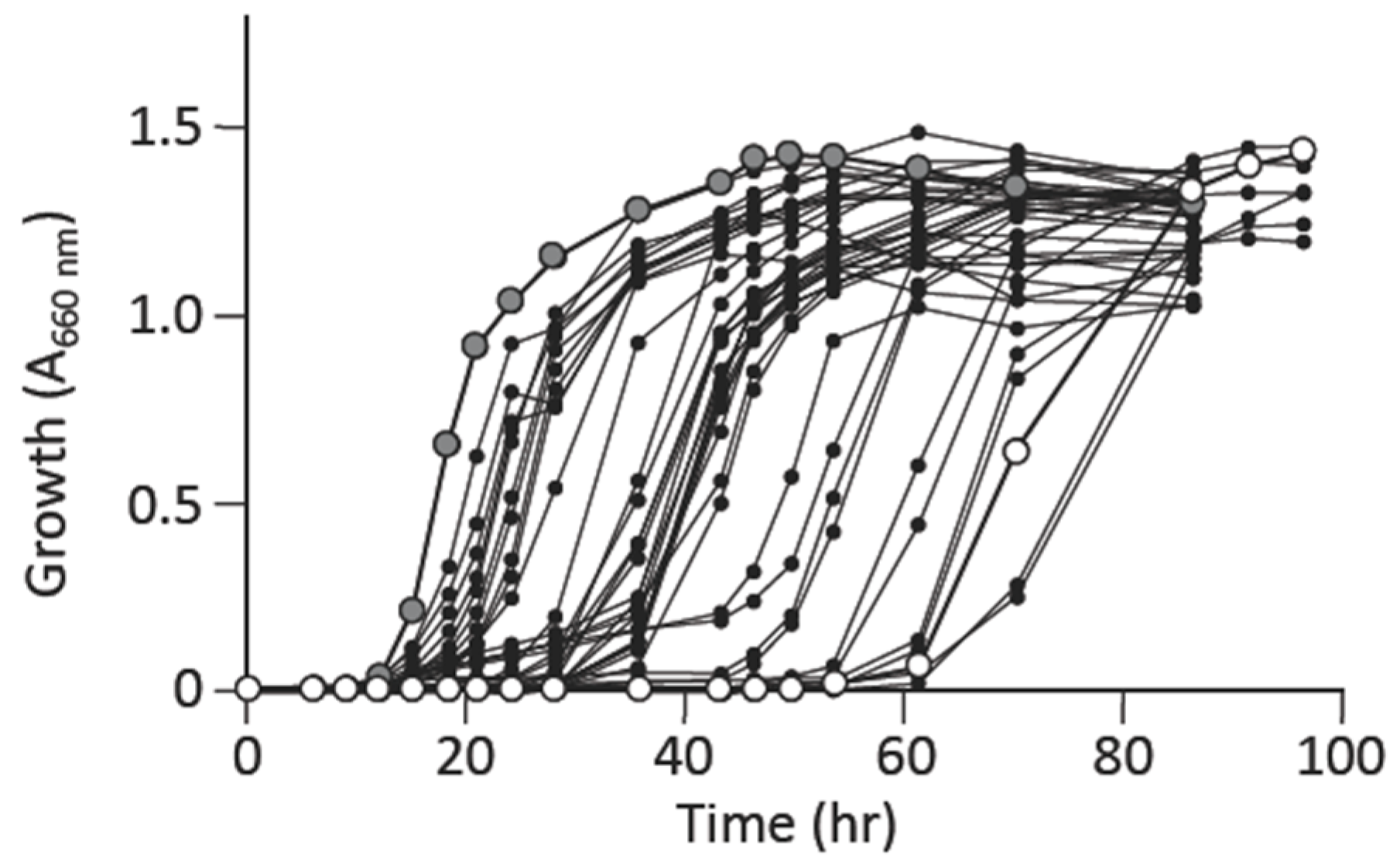
3.2. L-Alanine Prototrophic Suppressor Mutants are Generated through Stress-Induced Mutagenesis
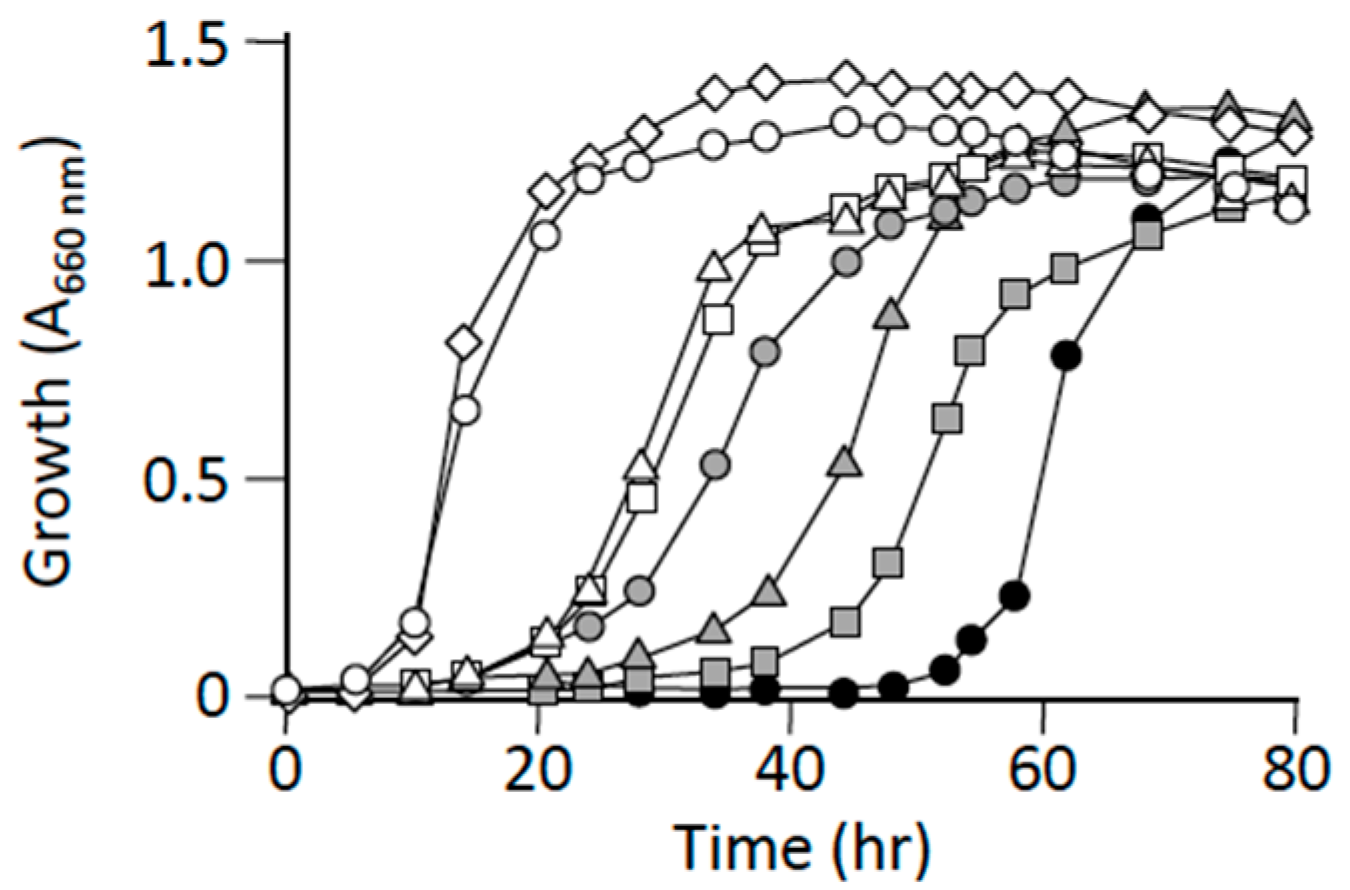
3.3. L-Alanine Prototrophic Suppressors Show Distinct Levels of Growth Recovery
3.4. Factors Supporting Growth of L-Alanine Auxotroph in Minimal Medium
3.5. Identification of Suppressor Mutations that Cause L-Alanine Auxotroph to Grow without L-Alanine Supplementation
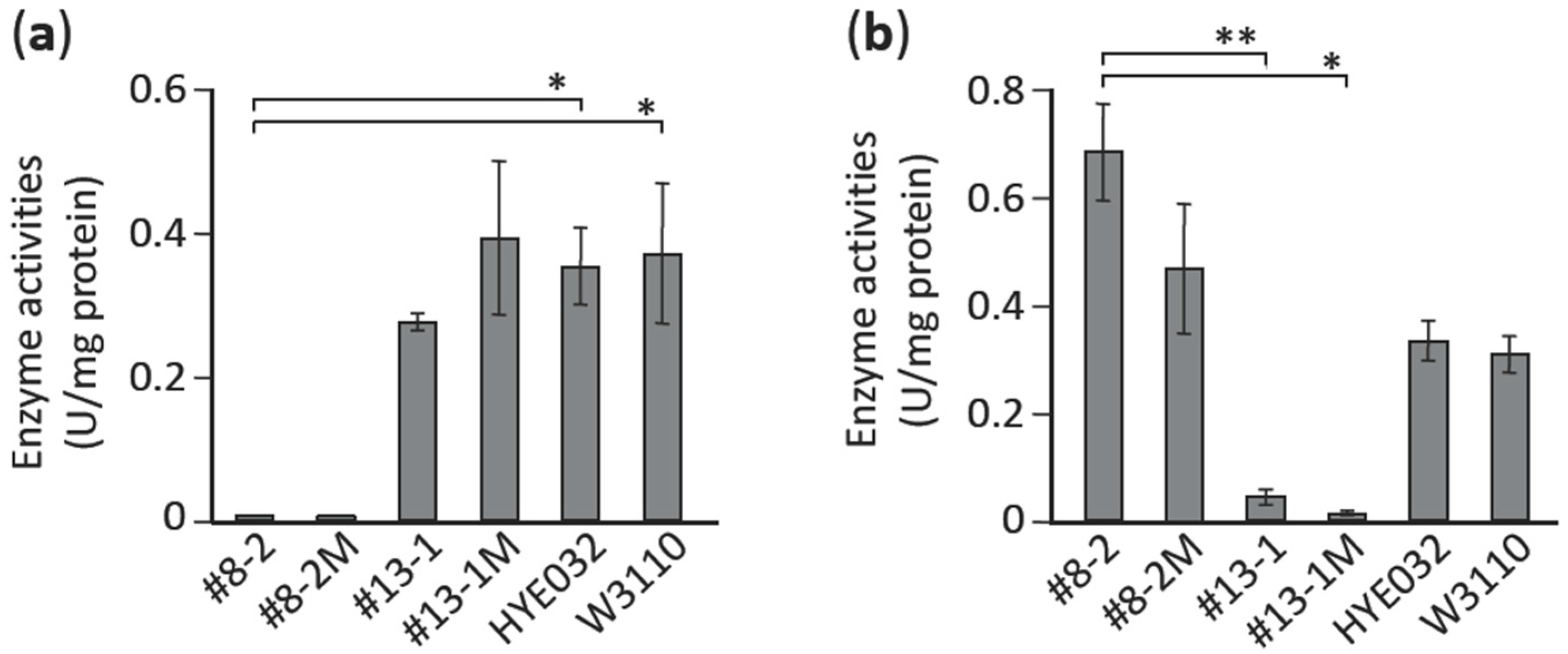
3.6. Characterization of Mutations Identified in pta and aceE
3.7. Impact of Extracellular Pyruvate and Anaerobic Culture Conditions on Emergence of L-Alanine Prototrophic Suppressors
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Reed, J.L.; Vo, T.D.; Schilling, C.H.; Palsson, B.O. An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR). Genome Biol. 2003, 4, R54. [Google Scholar] [CrossRef] [Green Version]
- Umbarger, H.E. Amino Acid Biosynthesis and its Regulation. Annu. Rev. Biochem. 1978, 47, 533–606. [Google Scholar] [CrossRef] [PubMed]
- Reitzer, L.J. Ammonia Assimilation and the Biosynthesis of Glutamine, Glutamate, Aspartate, Asparagine, L-Alanine, and D-Alanine. In Escherichia Coli and Salmonella: Cellular and Molecular Biology; Frederick, C.N., John, L.I., Boris, M., Brooks, K.L., Moselio, S., Edwin, U.H., Eds.; ASM Press: Washington, DC, USA, 1996; pp. 391–407. [Google Scholar]
- Pittard, A.J. Biosynthesis of the Aromatic Amino Acids. In Escherichia Coli and Salmonella: Cellular and Molecular Biology; Frederick, C.N., John, L.I., Boris, M., Brooks, K.L., Moselio, S., Edwin, U.H., Eds.; ASM Press: Washington, DC, USA, 1996; pp. 458–484. [Google Scholar]
- Patte, J.-C. Biosynthesis of Threonine and Lysine. In Escherichia Coli and Salmonella: Cellular and Molecular Biology; Frederick, C.N., John, L.I., Boris, M., Brooks, K.L., Moselio, S., Edwin, U.H., Eds.; ASM Press: Washington, DC, USA, 1996. [Google Scholar]
- Yoneyama, H.; Hori, H.; Lim, S.-J.; Murata, T.; Ando, T.; Isogai, E.; Katsumata, R. Isolation of a Mutant Auxotrophic for L-Alanine and Identification of Three Major Aminotransferases That SynthesizeL-Alanine in Escherichia coli. Biosci. Biotechnol. Biochem. 2011, 75, 930–938. [Google Scholar] [CrossRef]
- Kim, S.H.; Schneider, B.L.; Reitzer, L. Genetics and Regulation of the Major Enzymes of Alanine Synthesis in Escherichia coli. J. Bacteriol. 2010, 192, 5304–5311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patrick, W.M.; Quandt, E.M.; Swartzlander, D.B.; Matsumura, I. Multicopy Suppression Underpins Metabolic Evolvability. Mol. Biol. Evol. 2007, 24, 2716–2722. [Google Scholar] [CrossRef] [Green Version]
- Notebaart, A.R.; Kintses, B.; Feist, A.M.; Papp, B. Underground metabolism: Network-level perspective and biotechnological potential. Curr. Opin. Biotechnol. 2018, 49, 108–114. [Google Scholar] [CrossRef]
- Cocks, G.T.; Aguilar, J.; Lin, E.C.C. Evolution of L-1, 2-propanediol catabolism in Escherichia coli by recruitment of enzymes for L-fucose and L-lactate metabolism. J. Bacteriol. 1974, 118, 83–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, E.J.S.; Mortlock, R.P. Natural and altered induction of the L-fucose catabolic enzymes in Klebsiella aerogenes. J. Bacteriol. 1976, 127, 91–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, B.G. The EBG system of E. coli: Origin and evolution of a novel beta-galactosidase for the metabolism of lactose. Genetica 2003, 118, 143–156. [Google Scholar] [CrossRef]
- Kang, L.; Shaw, A.C.; Xu, D.; Xia, W.; Zhang, J.; Deng, J.; Woldike, H.F.; Liu, Y.; Su, J. Upregulation of MetC is essential for D-alanine-independent growth of an alr/dadX-deficient Escherichia coli strain. J. Bacteriol. 2011, 193, 1098–1106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fisher, R.; Tuli, R.; Haselkorn, R. A cloned cyanobacterial gene for glutamine synthetase functions in Escherichia coli, but the enzyme is not adenylylated. Proc. Natl. Acad. Sci. USA 1981, 78, 3393–3397. [Google Scholar] [CrossRef] [Green Version]
- Hashimoto-Gotoh, T.; Yamaguchi, M.; Yasojima, K.; Tsujimura, A.; Wakabayashi, Y.; Watanabe, Y. A set of temperature sensitive-replication/-segregation and temperature resistant plasmid vectors with different copy numbers and in an isogenic background (chloramphenicol, kanamycin, lacZ, repA, par, polA). Gene 2000, 241, 185–191. [Google Scholar] [CrossRef]
- Hori, H.; Yoneyama, H.; Tobe, R.; Ando, T.; Isogai, E.; Katsumata, R. Inducible L-alanine exporter encoded by the novel gene ygaW (alaE) in Escherichia coli. Appl. Environ. Microbiol. 2011, 77, 4027–4034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Garrison, E.; Marth, G. Haplotype-based variant detection from short-read sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; Depristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hinman, L.M.; Blass, J.P. An NADH-linked spectrophotometric assay for pyruvate dehydrogenase complex in crude tissue homogenates. J. Biol. Chem. 1981, 256, 6583–6586. [Google Scholar] [CrossRef]
- Ferry, J.G. Acetate Kinase and Phosphotransacetylase. Methods Enzymol. 2011, 494, 219–231. [Google Scholar] [CrossRef] [PubMed]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Yanga, Y.-T.; Bennett, G.N.; San, K.-Y. The Effects of Feed and Intracellular Pyruvate Levels on the Redistribution of Metabolic Fluxes in Escherichia coli. Metab. Eng. 2001, 3, 115–123. [Google Scholar] [CrossRef]
- Wang, Q.; Ou, M.S.; Kim, Y.; Ingram, L.O.; Shanmugam, K.T. Metabolic Flux Control at the Pyruvate Node in an Anaerobic Escherichia coli Strain with an Active Pyruvate Dehydrogenase. Appl. Environ. Microbiol. 2010, 76, 2107–2114. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Alonso, A.P.; Allen, D.K.; Reed, J.L.; Shachar-Hill, Y. Synergy between 13C-metabolic flux analysis and flux balance analysis for understanding metabolic adaption to anaerobiosis in E. coli. Metab. Eng. 2011, 13, 38–48. [Google Scholar] [CrossRef]
- Cairns, J.; Foster, P.L. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics 1991, 128, 695–701. [Google Scholar] [CrossRef]
- Bridges, B.A. Starvation-associated mutation in Escherichia coli: A spontaneous lesion hypothesis for “directed” mutation. Mutat. Res. Mol. Mech. Mutagen. 1994, 307, 149–156. [Google Scholar] [CrossRef]
- Godoy, V.G.; Gizatullin, F.S.; Fox, M.S. Some features of the mutability of bacteria during nonlethal selection. Genetics 2000, 154, 49–59. [Google Scholar]
- Yokota, A.; Shimizu, H.; Terasawa, Y.; Takaoka, N.; Tomita, F. Pyruvic acid production by a lipoic acid auxotroph of Escherichia coliW1485. Appl. Microbiol. Biotechnol. 1994, 41, 638–643. [Google Scholar] [CrossRef]
- Wu, J.; Li, Y.; Cai, Z.; Jin, Y. Pyruvate-Associated Acid Resistance in Bacteria. Appl. Environ. Microbiol. 2014, 80, 4108–4113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomar, A.; Eiteman, M.A.; Altman, E. The effect of acetate pathway mutations on the production of pyruvate in Escherichia coli. Appl. Microbiol. Biotechnol. 2003, 62, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, D.S.; Liu, Z.; Domagalski, N.; Koepsel, R.R.; Ataai, M.M.; Domach, M.M. Pyruvate Kinase-Deficient Escherichia coli Exhibits Increased Plasmid Copy Number and Cyclic AMP Levels. J. Bacteriol. 2009, 191, 3041–3049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hogema, B.M.; Arents, J.C.; Bader, R.; Eijkemans, K.; Yoshida, H.; Takahashi, H.; Aiba, H.; Postma, P.W. Inducer exclusion in Escherichia coli by non-PTS substrates: The role of the PEP to pyruvate ratio in de-termining the phosphorylation state of enzyme IIAGlc. Mol. Microbiol. 1998, 30, 487–498. [Google Scholar] [CrossRef]
- Luria, S.E.; Delbruck, M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics 1943, 28, 491–511. [Google Scholar] [PubMed]
- Lederberg, J.; Lederberg, E.M. Replica Plating and Indirect Selection of Bacterial Mutants. J. Bacteriol. 1952, 63, 399–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cairns, J.; Overbaugh, J.; Miller, S. The origin of mutants. Nat. Cell Biol. 1988, 335, 142–145. [Google Scholar] [CrossRef]
- Hall, B.G. Spontaneous point mutations that occur more often when advantageous than when neutral. Genetics 1990, 126, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Rosche, W.A.; Foster, P.L. The role of transient hypermutators in adaptive mutation in Escherichia coli. Proc. Natl. Acad. Sci. USA 1999, 96, 6862–6867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torkelson, J.; Harris, R.S.; Lombardo, M.J.; Nagendran, J.; Thulin, C.; Rosenberg, S.M. Genome-wide hypermutation in a subpopulation of stationary-phase cells underlies recombina-tion-dependent adaptive mutation. EMBO J. 1997, 16, 3303–3311. [Google Scholar] [CrossRef] [PubMed]


| Strain or Plasmid | Relevant Properties | Source |
|---|---|---|
| Strain | ||
| Escherichia coli W3110 | Wild-type | Laboratory strain |
| HYE032 | avtA::GM, yfbQ::KM, yfdZ::FRT, L-alanine auxotroph | [6] |
| JM109 | recA1, endA1, gyrA96, thi-1, hsdR17 (rk−mk+), e14−(mcrA−), supE44, relA1, Δ (lac-proAB)/F’[traD36, proAB+, laclq, lacZ Δ M15] | Laboratory strain |
| #8-2M | Isogenic mutant of HYE032 with a point-mutation found in a suppressor mutant #8-2 in pta gene | This study |
| #13-1M | Isogenic mutant of HYE032 with a point-mutation found in a suppressor mutant #13-1 in aceE gene | This study |
| #3 to #70 * | L-alanine prototrophic derivatives isolated from HYE032 | This study |
| Plasmid | ||
| pTH18cs1 | cat1, lacZ’, repAts1, derivative of pSC101 | [15] |
| pTH8-2 | pTH18cs1 harboring 0.5-kb PCR fragment of pta gene | This study |
| pTH13-1 | pTH18cs1 harboring 1.8-kb PCR fragment of aceE gene | This study |
| Primer | Nucleotide Sequence (5′-3′) | Comment |
|---|---|---|
| pta-Fwd | tgtctagaCACTACCGCAAACACCATCC | XbaI tag * |
| pta-Rev | gttctagaACCAACGTATCGGGCATTGC | XbaI tag * |
| aceE-Fwd | attctagaATTCGCGTCGCAATTGCTCT | XbaI tag * |
| aceE-Rev | tttctagaAACGAAAGCTTCAGAGCACG | XbaI tag * |
| pta-seq | GATGACGAGATTACTGCTGC | for sequencing |
| aceE-seq | AAAAGACCTCGAACTGGGC | for sequencing |
| Suppressor Strain | Days of Isolation | Gene | Nucleotide Change | Amino Acid Change |
|---|---|---|---|---|
| #3 | 2 | aceE | T110A | I37N |
| flgJ | A123 > AA *1 | 42 frameshift | ||
| #4-2 | 2 | aceE | T110A | I37N |
| #5-1 | 2 | aceE | A775G | N259D |
| #8-2 | 2 | pta | A1967C | D656A |
| #10-1 | 2 | aceE | T110A | I37N |
| cyaA | C1957T | R653C | ||
| #11-2 | 2 | aceF | A1504C | T502P |
| #12-2 | 2 | aceE | A775C | N259H |
| #13-1 | 2 | aceE | G440A | G147D |
| #14-1 | 2 | aceE | C802T | P268S |
| cpxP | T269G | F90C | ||
| yhbO | T5A | S2R | ||
| #15-1 | 2 | pta | A1967C | D656A |
| gspE | A1381C | K461Q | ||
| #16-2 | 2 | pta | A1967C | D656A |
| #17-2 | 2 | pta | A1967C | D656A |
| plsB | G2198T | R733L | ||
| #18-1 | 2 | pta | A1967C | D656A |
| #19 | 2 | pta | A1967C | D656A |
| #20 | 2 | pta | G1420T | E474stop |
| #21 | 2 | pta | A1967C | D656A |
| #23 | 2 | pta | A1967C | D656A |
| ydhK | C305A | A102E | ||
| #24 | 2 | pta | T2A*2 | No initiation |
| #25 | 2 | pta | A1967C | D656A |
| #27 | 2 | pta | A1967C | D656A |
| #29 | 2 | pta | A1967C | D656A |
| #31-1 | 3 | lpd | C963A | H322Q |
| #32-1 | 3 | aceE | G1193T | G398V |
| yhiX | C831T *3 | Silent | ||
| #33-1 | 3 | entE | C827T | T276I |
| #35-1 | 3 | pta | A1967C | D656A |
| #37-1 | 3 | aceF | C1700A | A567E |
| #39-2 | 3 | aceE | C617T | A206V |
| aceE | G810A *3 | silent | ||
| #40-2 | 3 | pta | C1884G | D628E |
| #43-1 | 3 | lpd | T956G | L320R |
| #44-1 | 3 | cyaA | C1957T | R653C |
| #45-1 | 3 | pta | A1967C | D656A |
| #46-2 | 3 | pta | A1967C | D656A |
| #48-1 | 3 | aceE | G440A | G147D |
| flhC | C115 deletion *4 | 39 frameshift | ||
| #51 | 3 | ynbB | A380C | D127A |
| #54 | 3 | aceE | T584C | M195T |
| ftsI | C1599T*3 | Silent | ||
| #57 | 3 | lpd | T956G | L320R |
| #64-1 | 4 | cyaA | C1957T | R653C |
| #65-2 | 4 | yhjG | T543A | D181E |
| #68-1 | 4 | cyaA | C1957T | R653C |
| glnS | A956T | Q319L | ||
| #70 | 4 | ackA | A878 deletion *4 | 293 frameshift |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mishima, H.; Watanabe, H.; Uchigasaki, K.; Shimoda, S.; Seki, S.; Kumagai, T.; Nochi, T.; Ando, T.; Yoneyama, H. L-Alanine Prototrophic Suppressors Emerge from L-Alanine Auxotroph through Stress-Induced Mutagenesis in Escherichia coli. Microorganisms 2021, 9, 472. https://doi.org/10.3390/microorganisms9030472
Mishima H, Watanabe H, Uchigasaki K, Shimoda S, Seki S, Kumagai T, Nochi T, Ando T, Yoneyama H. L-Alanine Prototrophic Suppressors Emerge from L-Alanine Auxotroph through Stress-Induced Mutagenesis in Escherichia coli. Microorganisms. 2021; 9(3):472. https://doi.org/10.3390/microorganisms9030472
Chicago/Turabian StyleMishima, Harutaka, Hirokazu Watanabe, Kei Uchigasaki, So Shimoda, Shota Seki, Toshitaka Kumagai, Tomonori Nochi, Tasuke Ando, and Hiroshi Yoneyama. 2021. "L-Alanine Prototrophic Suppressors Emerge from L-Alanine Auxotroph through Stress-Induced Mutagenesis in Escherichia coli" Microorganisms 9, no. 3: 472. https://doi.org/10.3390/microorganisms9030472
APA StyleMishima, H., Watanabe, H., Uchigasaki, K., Shimoda, S., Seki, S., Kumagai, T., Nochi, T., Ando, T., & Yoneyama, H. (2021). L-Alanine Prototrophic Suppressors Emerge from L-Alanine Auxotroph through Stress-Induced Mutagenesis in Escherichia coli. Microorganisms, 9(3), 472. https://doi.org/10.3390/microorganisms9030472







