- Article
Prolactin and 17β-Estradiol Are Epigenetic Regulators That Modify the Effector Response of Bovine Macrophages During Staphylococcus aureus Challenge
- Marco Antonio Barajas-Mendiola,
- Josmarth Remigio-Hernández and
- Alejandra Ochoa-Zarzosa
- + 2 authors
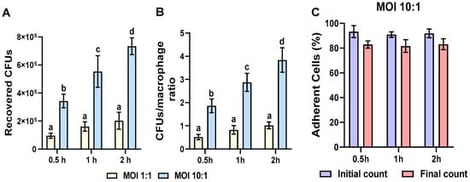
Staphylococcus aureus (S. aureus) is the most prevalent pathogen associated with subclinical mastitis, which significantly impacts dairy farming worldwide. Fluctuations in reproductive hormones, such as bovine prolactin (bPRL) and 17β-estradiol (E2), are known to compromise the innate immune response (IIR) of the mammary gland (MG). In this study, we evaluated the effects of bPRL and E2 on the effector response of primary bovine macrophages, isolated from lactating Holstein cows, challenged with S. aureus. We demonstrated that physiological concentrations of bPRL (5 ng/mL) and E2 (50 pg/mL) induced differential changes in the expression of pro-inflammatory (TNF-α, IL-6, and IL-1β) and anti-inflammatory (IL-10) cytokines, chemokines (IL-8), antimicrobial peptides (BNBD10 and S100A7), and miRNAs (miR-451, miR-155, miR-7863, miR-146a, miR-21a, Let-7a-5p, miR-30b, and miR-23a) in S. aureus-challenged macrophages. Moreover, these hormones promoted global histone H3 acetylation and the epigenetic H3K9ac mark without affecting H3K9me2 levels. Hormonal treatment also modulated histone deacetylase (HDAC) activity. Furthermore, hormonal treatment altered macrophage chemotaxis and phagocytosis. In conclusion, bPRL and E2 modulate the effector functions of bovine macrophages during S. aureus infection. This process could be associated with the regulation of histone H3 modifications, such as H3K9ac, in IIR-related genes.
3 March 2026