Phytochemicals in Chinese Chive (Allium tuberosum) Induce the Skeletal Muscle Cell Proliferation via PI3K/Akt/mTOR and Smad Pathways in C2C12 Cells
Abstract
:1. Introduction
2. Results and Discussion
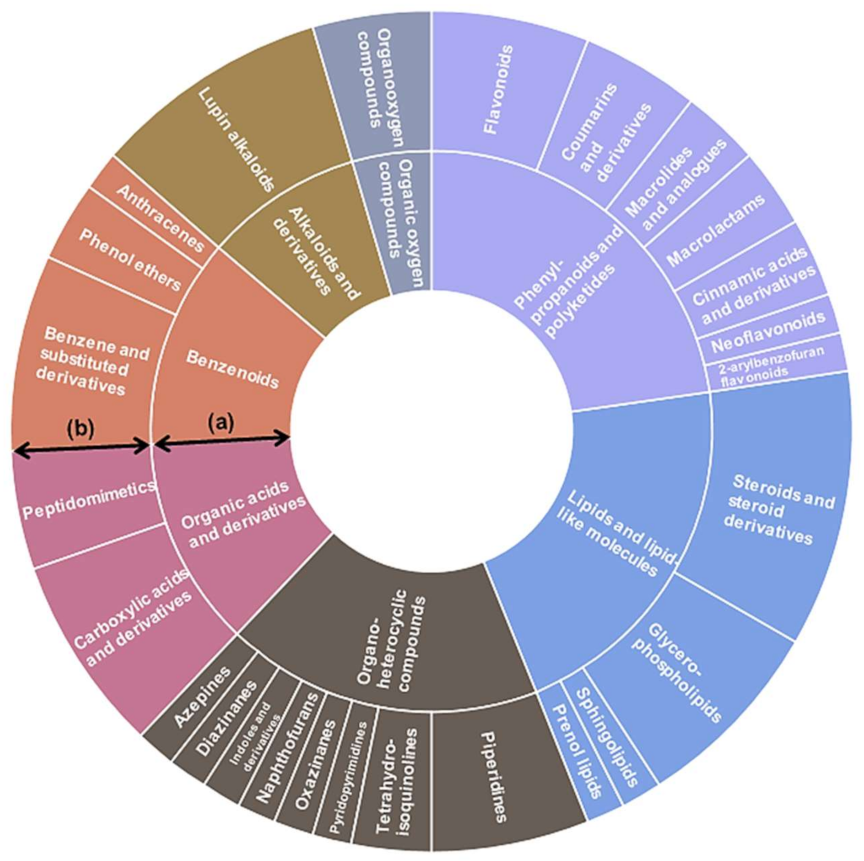
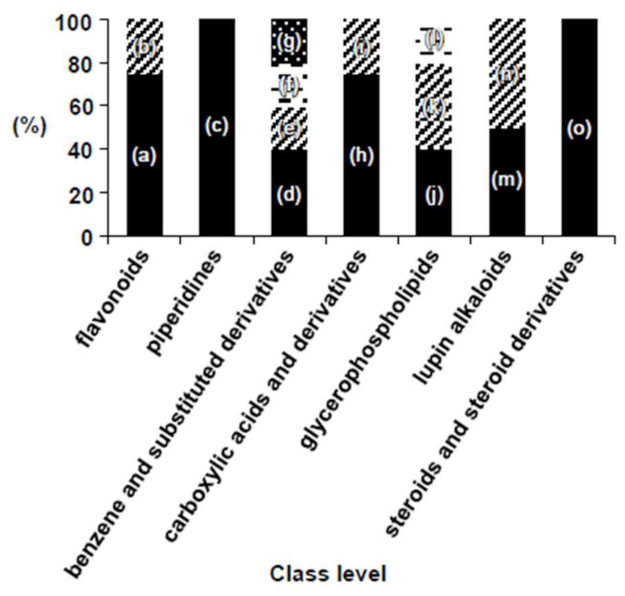
2.1. Chemical Composition of Chinese Chive
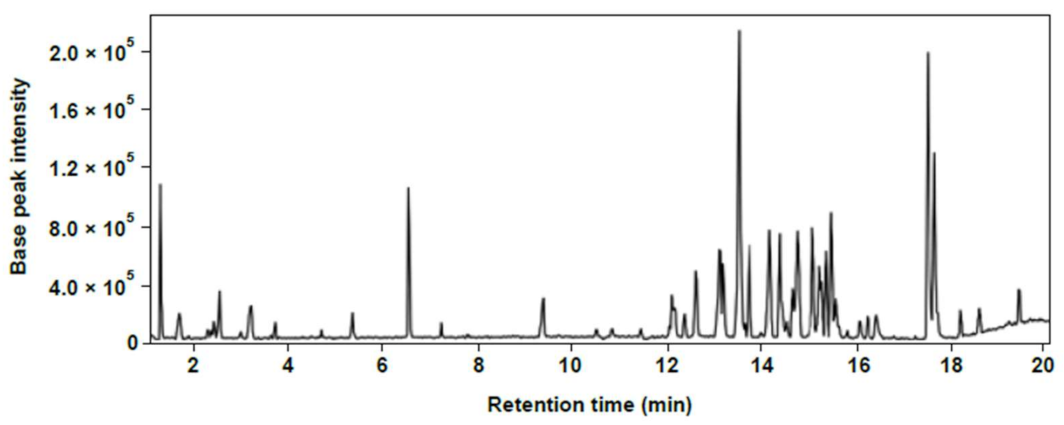
2.2. Isolation and Identification of Phytochemicals in Chinese Chive Extract
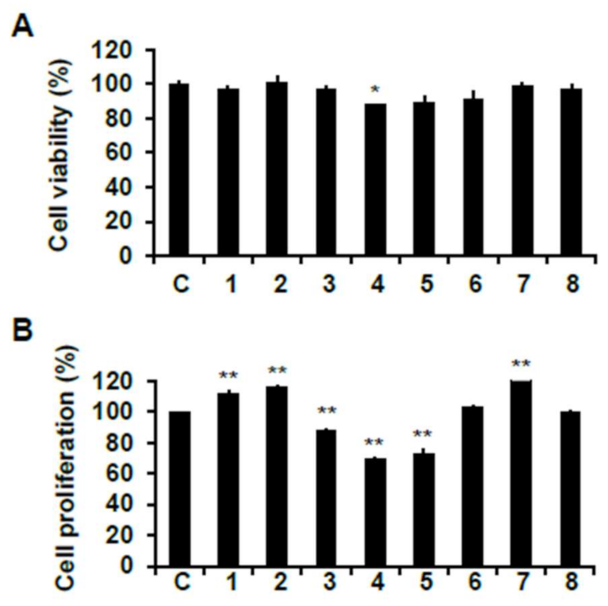
2.3. Skeletal Muscle Cell Proliferation Activities of Phytochemicals from Chinese Chive on C2C12 Cells
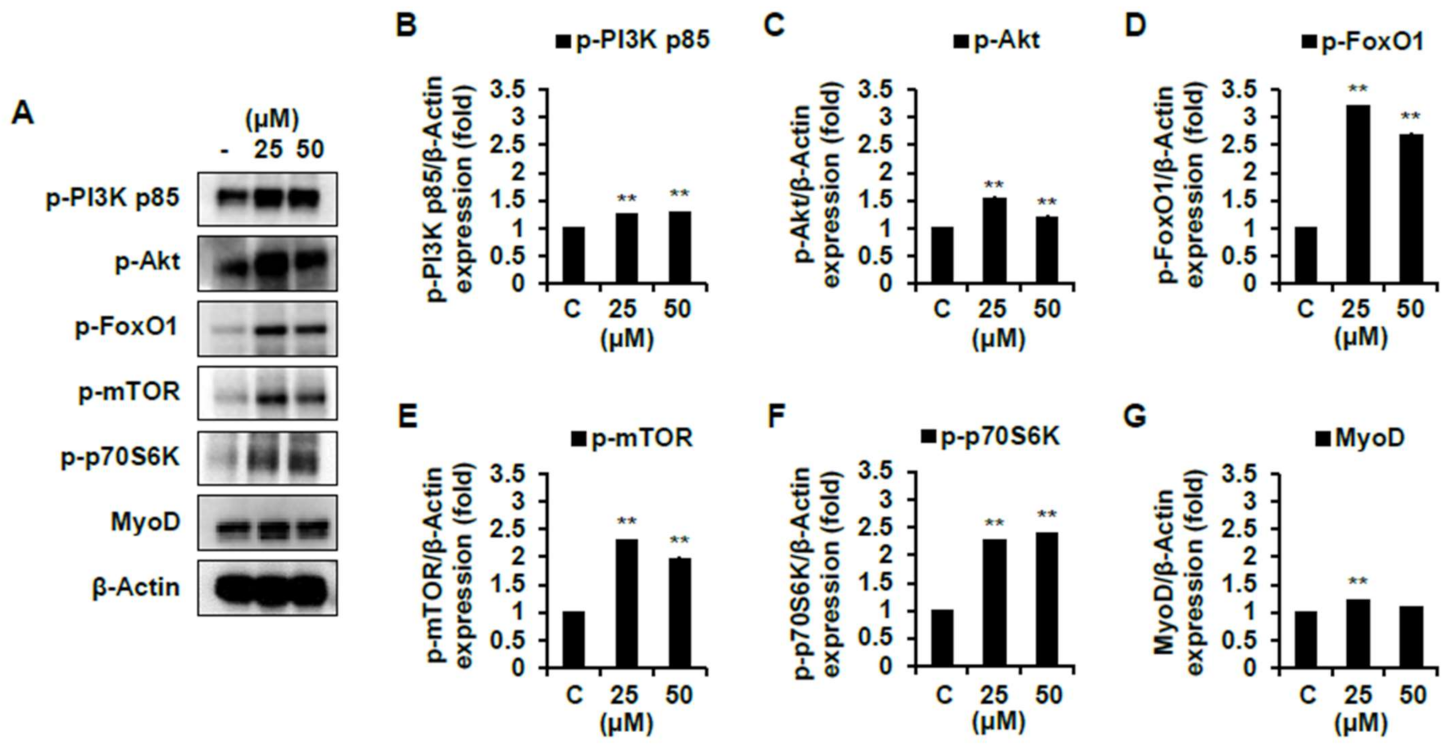
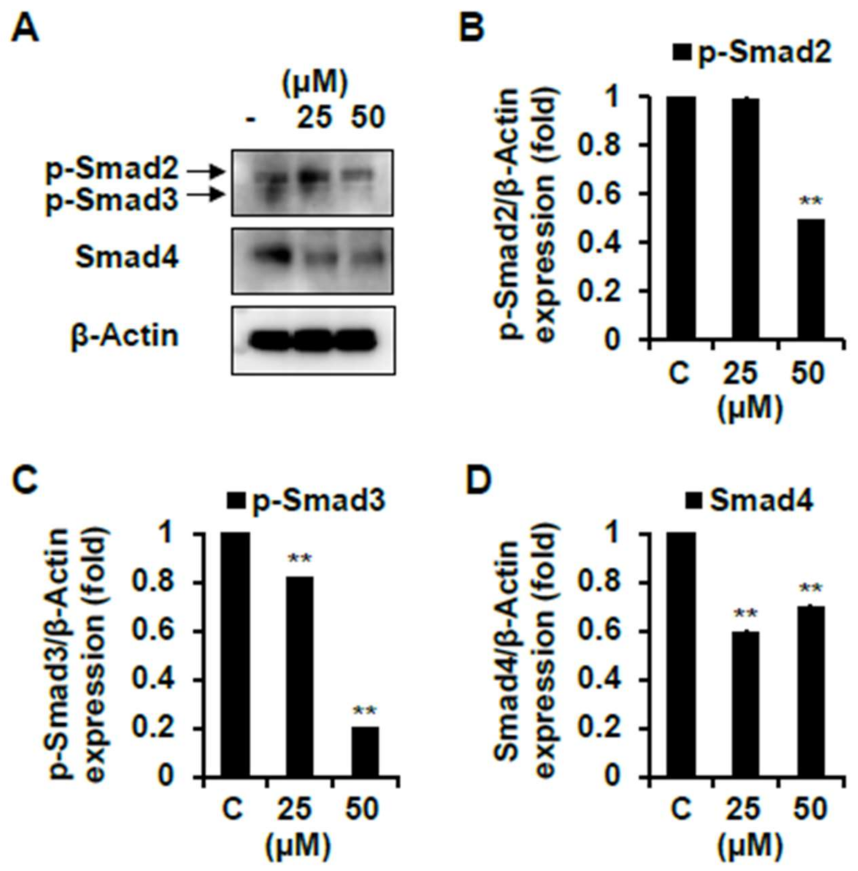
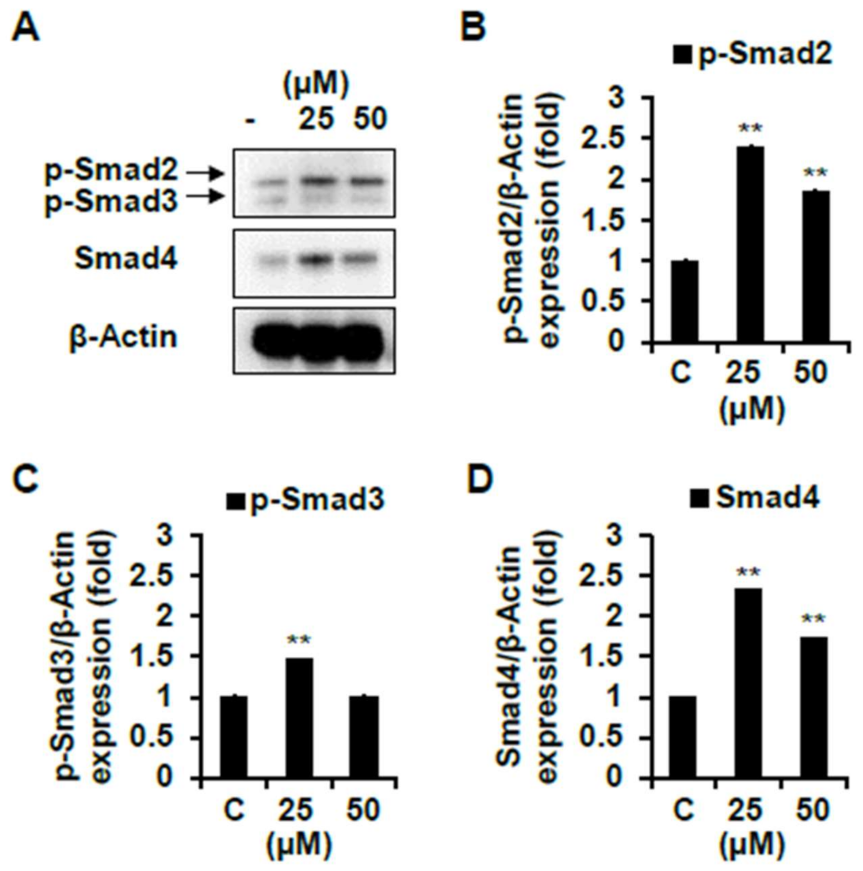
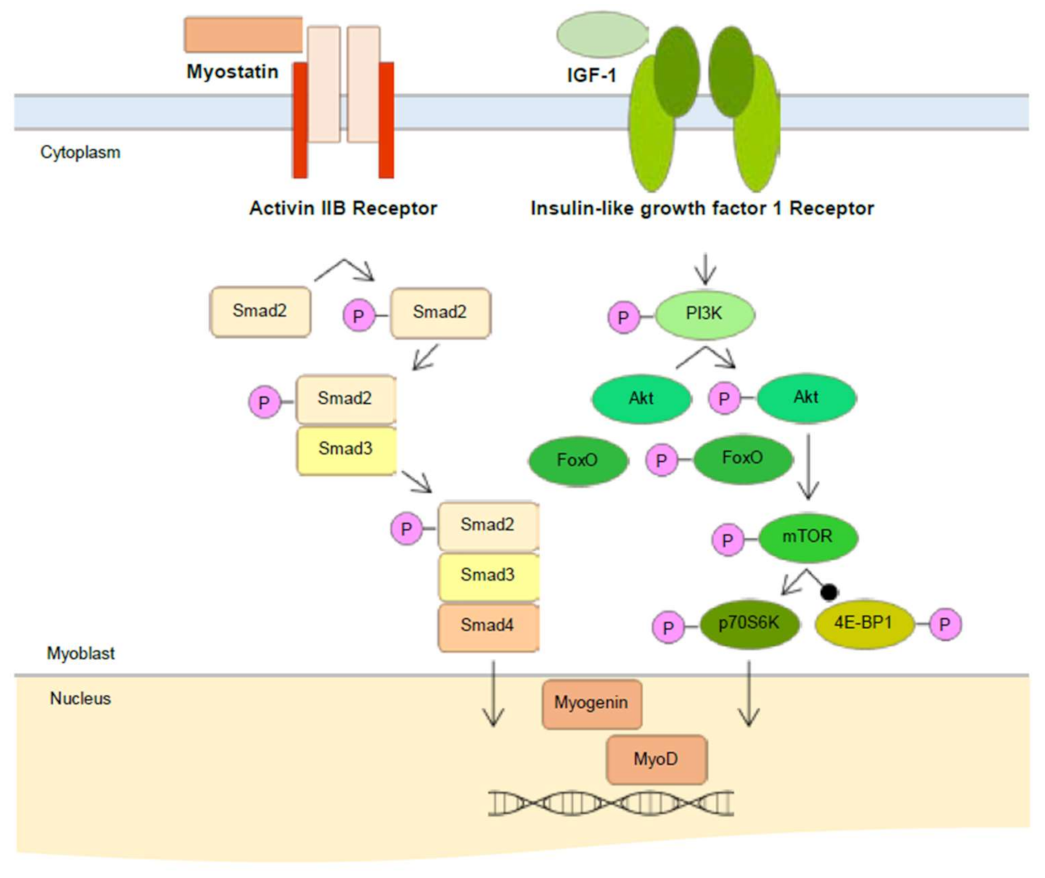
2.4. Effect of Skeletal Muscle Growth Regulation of Compounds 1 and 7 from Chinese Chive via PI3K/Akt/mTOR and/or Smad Signaling Pathways
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Plant Material
3.3. UPLC-QTOF-MS Analysis
3.4. Extraction and Isolation
3.5. Kaempferol-3-O-(6ʺ-feruloyl)-sophoroside (1)
3.6. Acid Hydrolysis of 1
3.7. Materials for Skeletal Muscle Cell Proliferation Activity
3.8. Cell Culture and Differentiation
3.9. Skeletal Muscle Cell Proliferation Activity
3.10. Western Blot Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- McCormick, R.; Vasilaki, A. Age-related changes in skeletal muscle: Changes to life-style as a therapy. Biogerontology 2018, 19, 519–536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, C.H.; Shin, J.-H.; Hwang, S.J.; Choi, Y.H.; Kim, D.-S.; Kim, C.M. Schisandrae fructus enhances myogenic differentiation and inhibits atrophy through protein synthesis in human myotubes. Int. J. Nanomed. 2016, 11, 2407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, Y.D.C.D. The Dictionary of Chinese Drugs; Shanghai Science and Technological Publisher: Shanghai, China, 1985; p. 2383. [Google Scholar]
- Tang, X.; Olatunji, O.J.; Zhou, Y.; Hou, X. Allium tuberosum: Antidiabetic and hepatoprotective activities. Food Res. Int. 2017, 102, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, R. Benificial effect of Allium sativum and Allium tuberosum on experimental hyperlipidemia and atherosclerosis. Pak. J. Physiol. 2008, 4, 7–10. [Google Scholar]
- Park, S.-Y.; Kim, J.-Y.; Park, K.-W.; Kang, K.-S.; Park, K.-H.; Seo, K.-I. Effects of thiosulfinates isolated from Allium tuberosum L. on the growth of human cancer cells. J. Korean Soc. Food Sci. Nutr. 2009, 38, 1003–1007. [Google Scholar] [CrossRef]
- Gautam, S.; Pal, S.; Maurya, R.; Srivastava, A.K. Ethanolic extract of Allium cepa stimulates glucose transporter typ 4-mediated glucose uptake by the activation of insulin signaling. Planta Med. 2015, 81, 208–214. [Google Scholar] [CrossRef] [Green Version]
- Yang, J. Enhanced skeletal muscle for effective glucose homeostasis. Prog. Mol. Biol. Ttransl. Sci. 2014, 121, 133–163. [Google Scholar]
- Xie, W.; Zhang, Y.; Wang, N.; Zhou, H.; Du, L.; Ma, X.; Shi, X.; Cai, G. Novel effects of macrostemonoside A, a compound from Allium macrostemon Bung, on hyperglycemia, hyperlipidemia, and visceral obesity in high-fat diet-fed C57BL/6 mice. Eur. J. Pharmacol. 2008, 599, 159–165. [Google Scholar] [CrossRef]
- Kalhotra, P.; Chittepu, V.C.; Osorio-Revilla, G.; Gallardo-Velazquez, T. Phytochemicals in garlic extract inhibit therapeutic enzyme DPP-4 and induce skeletal muscle cell proliferation: A possible mechanism of action to benefit the treatment of diabetes mellitus. Biomolecules 2020, 10, 305. [Google Scholar] [CrossRef] [Green Version]
- Da Silva, R.R.; Wang, M.; Nothias, L.-F.; van der Hooft, J.J.; Caraballo-Rodríguez, A.M.; Fox, E.; Balunas, M.J.; Klassen, J.L.; Lopes, N.P.; Dorrestein, P.C. Propagating annotations of molecular networks using in silico fragmentation. PLoS Comput. Biol. 2018, 14, e1006089. [Google Scholar] [CrossRef]
- Aron, A.T.; Gentry, E.C.; McPhail, K.L.; Nothias, L.-F.; Nothias-Esposito, M.; Bouslimani, A.; Petras, D.; Gauglitz, J.M.; Sikora, N.; Vargas, F. Reproducible molecular networking of untargeted mass spectrometry data using GNPS. Nat. Protoc. 2020, 15, 1954–1991. [Google Scholar] [CrossRef] [PubMed]
- Feunang, Y.D.; Eisner, R.; Knox, C.; Chepelev, L.; Hastings, J.; Owen, G.; Fahy, E.; Steinbeck, C.; Subramanian, S.; Bolton, E. ClassyFire: Automated chemical classification with a comprehensive, computable taxonomy. J. Cheminform. 2016, 8, 61. [Google Scholar] [CrossRef] [Green Version]
- Markham, K.; Ternai, B.; Stanley, R.; Geiger, H.; Mabry, T. Carbon-13 NMR studies of flavonoids—III: Naturally occurring flavonoid glycosides and their acylated derivatives. Tetrahedron 1978, 34, 1389–1397. [Google Scholar] [CrossRef]
- Xiao, Y.; Wang, Y.L.; Gao, S.X.; Sun, C.; Zhou, Z.Y. Chemical composition of Hydrilla verticillata (L. f.) royle in Taihu lake. Chin. J. Chem. 2007, 25, 661–665. [Google Scholar] [CrossRef]
- Ciuffreda, P.; Casati, S.; Manzocchi, A. Complete 1H and 13C NMR spectral assignment of α-and β-adenosine, 2′-deoxyadenosine and their acetate derivatives. Magn. Reson. Chem. 2007, 45, 781–784. [Google Scholar] [CrossRef]
- Mantsch, H.; Smith, I.C. Fourier-transformed 13C NMR spectra of polyuridylic acid, uridine, and related nucleotides—the use of 31POC13C couplings for conformational analysis. Biochem. Bioph. Res. Commun. 1972, 46, 808–815. [Google Scholar] [CrossRef]
- Flores-Sanchez, I.J.; Peč, J.; Fei, J.; Choi, Y.H.; Dušek, J.; Verpoorte, R. Elicitation studies in cell suspension cultures of Cannabis sativa L. J. Biotechnol. 2009, 143, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Tatsuzawa, F.; Aiba, Y.; Morino, T.; Saito, N.; Shinoda, K.; Kato, K.; Toki, K.; Honda, T. Copigmentation with acylated anthocyanin and kaempferol glycosides in violet and purple flower cultivars of Aubrieta × cultorum (Brassicaceae). J. Jpn. Soc. Hortic. Sci. 2012, 81, 275–284. [Google Scholar] [CrossRef] [Green Version]
- Kamel, M.S.; Mohamed, K.M.; Hassanean, H.A.; Ohtani, K.; Kasai, R.; Yamasaki, K. Acylated flavonoid glycosides from Bassia muricata. Phytochemistry 2001, 57, 1259–1262. [Google Scholar] [CrossRef] [Green Version]
- Tureckova, J.; Wilson, E.M.; Cappalonga, J.L.; Rotwein, P. Insulin-like growth factor-mediated muscle differentiation collaboration between phosphatidylinositol 3-kinase-Akt-signaling pathways and myogenin. J. Biol. Chem. 2001, 276, 39264–39270. [Google Scholar] [CrossRef] [Green Version]
- Jacquemin, V.; Butler-Browne, G.S.; Furling, D.; Mouly, V. IL-13 mediates the recruitment of reserve cells for fusion during IGF-1-induced hypertrophy of human myotubes. J. Cell Sci. 2007, 120, 670–681. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Park, M.Y.; Kim, H.K.; Park, Y.; Whang, K.-Y. Cortisone and dexamethasone inhibit myogenesis by modulating the AKT/mTOR signaling pathway in C2C12. Biosci. Biotech. Bioch. 2016, 80, 2093–2099. [Google Scholar] [CrossRef] [Green Version]
- Elkina, Y.; von Haehling, S.; Anker, S.D.; Springer, J. The role of myostatin in muscle wasting: An overview. J. Cachexia Sarcopenia Muscle 2011, 2, 143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.-K.; Jie, L.; Jin, L.; Baosheng, G.; Leung, A.; Zhang, G.; Zhang, B.-T. Icaritin requires Phosphatidylinositol 3 kinase (PI3K)/Akt signaling to counteract skeletal muscle atrophy following mechanical unloading. Sci. Rep. 2016, 6, 20300. [Google Scholar] [CrossRef] [PubMed]
- Sung, B.; Hwang, S.Y.; Kim, M.J.; Kim, M.; Jeong, J.W.; Kim, C.M.; Chung, H.Y.; Kim, N.D. Loquat leaf extract enhances myogenic differentiation, improves muscle function and attenuates muscle loss in aged rats. Int. J. Mol. Med. 2015, 36, 792–800. [Google Scholar] [CrossRef] [Green Version]
- Zhao, B.; Xiong, Y.; Zhang, Y.; Jia, L.; Zhang, W.; Xu, X. Rutin promotes osteogenic differentiation of periodontal ligament stem cells through the GPR30-mediated PI3K/AKT/mTOR signaling pathway. Exp. Biol. Med. 2020, 245, 552–561. [Google Scholar] [CrossRef]
- Ali, M.Y.; Zaib, S.; Rahman, M.M.; Jannat, S.; Iqbal, J.; Park, S.K.; Chang, M.S. Poncirin, an orally active flavonoid exerts antidiabetic complications and improves glucose uptake activating PI3K/Akt signaling pathway in insulin resistant C2C12 cells with anti-glycation capacities. Bioorg. Chem. 2020, 102, 104061. [Google Scholar]
- Tato, I.; Bartrons, R.; Ventura, F.; Rosa, J.L. Amino acids activate mammalian target of rapamycin complex 2 (mTORC2) via PI3K/Akt signaling. J. Biol. Chem. 2011, 286, 6128–6142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saha, A.; Connelly, S.; Jiang, J.; Zhuang, S.; Amador, D.T.; Phan, T.; Pilz, R.B.; Boss, G.R. Akt phosphorylation and regulation of transketolase is a nodal point for amino acid control of purine synthesis. Mol. Cell 2014, 55, 264–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, M.-S.; Chen, J. Distinct amino acid–sensing mTOR pathways regulate skeletal myogenesis. Mol. Biol. Cell 2013, 24, 3754–3763. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Black, B.L.; Derynck, R. TGF-β inhibits muscle differentiation through functional repression of myogenic transcription factors by Smad3. Gene. Dev. 2001, 15, 2950–2966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.-T.; Chen, L.; Tan, Z.-B.; Fan, H.-J.; Xie, L.-P.; Zhang, W.-T.; Chen, H.-M.; Li, J.; Liu, B.; Zhou, Y.-C. Luteolin inhibits vascular smooth muscle cell proliferation and migration by inhibiting TGFBR1 signaling. Front. Pharmacol. 2018, 9, 1059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, F.; He, X.; Liang, S.; Liu, S.; Liu, H.; He, Q.; Chen, L.; Jiang, H.; Zhang, Y. Quercetin ameliorates podocyte injury via inhibition of oxidative stress and the TGF-β1/Smad pathway in DN rats. RSC Adv. 2018, 8, 35413–35421. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Shan, S.; Wang, J.; Cheng, X.; Yi, B.; Zhou, J.; Li, Q. Galangin inhibits hypertrophic scar formation via ALK5/Smad2/3 signaling pathway. Mol. Cell. Biochem. 2016, 413, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Muthuramalingam, K.; Kim, S.-Y.; Kim, Y.; Kim, H.-S.; Jeon, Y.-J.; Cho, M. Bigbelly seahorse (Hippocampus abdominalis)-derived peptides enhance skeletal muscle differentiation and endurance performance via activated P38MAPK/AKT signalling pathway: An in vitro and in vivo analysis. J. Funct. Foods 2019, 52, 147–155. [Google Scholar] [CrossRef]
- Kim, S.-Y.; Kim, H.-S.; Cho, M.; Jeon, Y.-J. Enzymatic Hydrolysates of Hippocampus abdominalis Regulates the Skeletal Muscle Growth in C2C12 Cells and Zebrafish Model. J. Aquat. Food Prod. Technol. 2019, 28, 264–274. [Google Scholar] [CrossRef]



| Pos. | δCa,b | δHa,c (J in Hz) | Pos. | δCa,b | δHa,c (J in Hz) | ||
|---|---|---|---|---|---|---|---|
| Kaempferol | Glucose B | ||||||
| 2 | 158.5 | 1′′′ | 106.2 | 4.75 (d, 7.5) | |||
| 3 | 135.3 | 2′′′ | 75.7 | 3.66 * | |||
| 4 | 180.0 | 3′′′ | 77.9 | 3.49 * | |||
| 5 | 163.1 | 4′′′ | 72.0 | 3.41 * | |||
| 6 | 99.9 | 6.12 (d, 2.1) | 5′′′ | 76.3 | 3.43 * | ||
| 7 | 165.7 | 6′′′ | 62.4 | 3.64 *, 3.51 * | |||
| 8 | 94.8 | 6.18 (d, 2.1) | |||||
| 9 | 158.3 | Ferulic acid | |||||
| 10 | 105.8 | 1′′′′ | 127.4 | ||||
| 2′′′′ | 111.2 | 6.82 (d, 1.9) | |||||
| 1’ | 122.6 | 3′′′′ | 149.2 | ||||
| 2’ | 132.8 | 8.06 (m) | 4′′′′ | 150.5 | |||
| 3’ | 116.4 | 6.90 (m) | 5′′′′ | 116.3 | 6.65 (d, 8.1) | ||
| 4’ | 161.7 | 6′′′′ | 124.0 | 6.72 (dd, 1.9, 8.2) | |||
| 5’ | 116.4 | α | 146.9 | 7.34 (d, 15.9) | |||
| 6’ | 132.8 | - | β | 115.0 | 6.05 (d, 15.9) | ||
| -COO | 169.0 | ||||||
| Glucose A | -OCH3 | 56.3 | 3.79 (s) | ||||
| 1’’ | 101.3 | 5.12 (d, 7.5) | |||||
| 2’’ | 84.7 | 3.71 * | |||||
| 3’’ | 78.3 | 3.11 * | |||||
| 4’’ | 71.0 | 3.39 * | |||||
| 5’’ | 77.8 | 3.53 * | |||||
| 6’’ | 64.8 | 4.44 *, 4.45 * | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oh, M.; Kim, S.-Y.; Park, S.; Kim, K.-N.; Kim, S.H. Phytochemicals in Chinese Chive (Allium tuberosum) Induce the Skeletal Muscle Cell Proliferation via PI3K/Akt/mTOR and Smad Pathways in C2C12 Cells. Int. J. Mol. Sci. 2021, 22, 2296. https://doi.org/10.3390/ijms22052296
Oh M, Kim S-Y, Park S, Kim K-N, Kim SH. Phytochemicals in Chinese Chive (Allium tuberosum) Induce the Skeletal Muscle Cell Proliferation via PI3K/Akt/mTOR and Smad Pathways in C2C12 Cells. International Journal of Molecular Sciences. 2021; 22(5):2296. https://doi.org/10.3390/ijms22052296
Chicago/Turabian StyleOh, Mira, Seo-Young Kim, SeonJu Park, Kil-Nam Kim, and Seung Hyun Kim. 2021. "Phytochemicals in Chinese Chive (Allium tuberosum) Induce the Skeletal Muscle Cell Proliferation via PI3K/Akt/mTOR and Smad Pathways in C2C12 Cells" International Journal of Molecular Sciences 22, no. 5: 2296. https://doi.org/10.3390/ijms22052296
APA StyleOh, M., Kim, S.-Y., Park, S., Kim, K.-N., & Kim, S. H. (2021). Phytochemicals in Chinese Chive (Allium tuberosum) Induce the Skeletal Muscle Cell Proliferation via PI3K/Akt/mTOR and Smad Pathways in C2C12 Cells. International Journal of Molecular Sciences, 22(5), 2296. https://doi.org/10.3390/ijms22052296







