Use of Information Measures and Their Approximations to Detect Predictive Gene-Gene Interaction
Abstract
:1. Introduction
2. Measures of Interaction
2.1. Interaction Information Measure
2.2. Other Nonparametric Measures of Interaction
2.3. Estimation of the Interaction Measures
3. Modeling Gene-Gene Interactions
3.1. Logistic Modeling of Gene-Gene Interactions
3.2. ANOVA Model for Binary Outcome
3.3. Behavior of Interaction Indices for Logistic Models
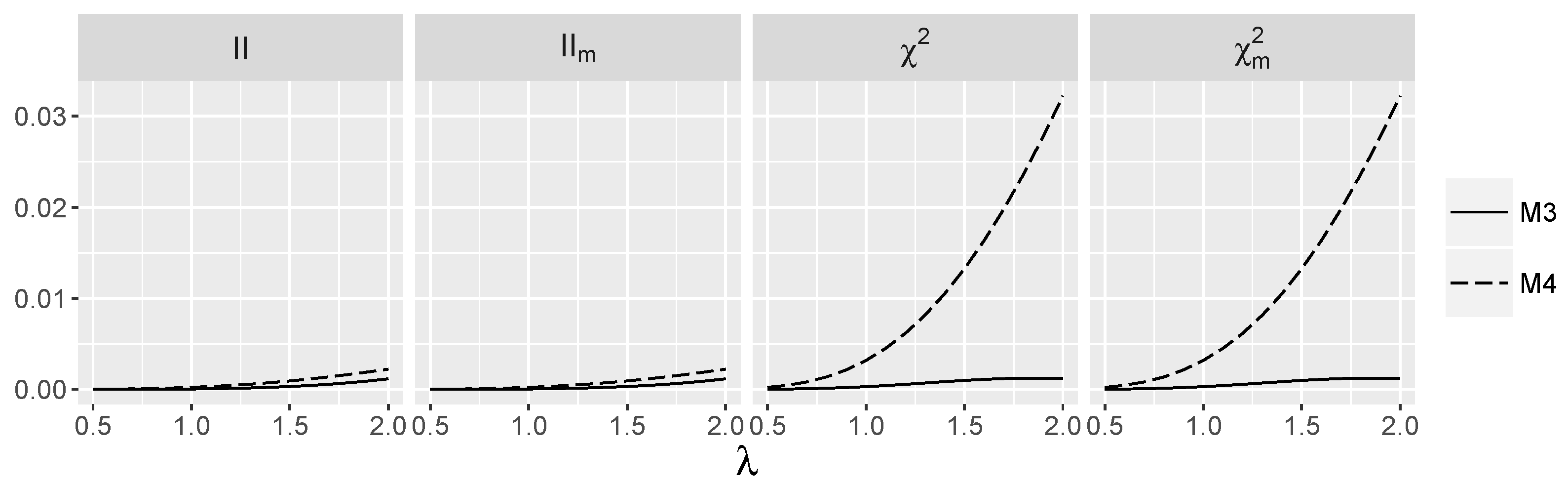
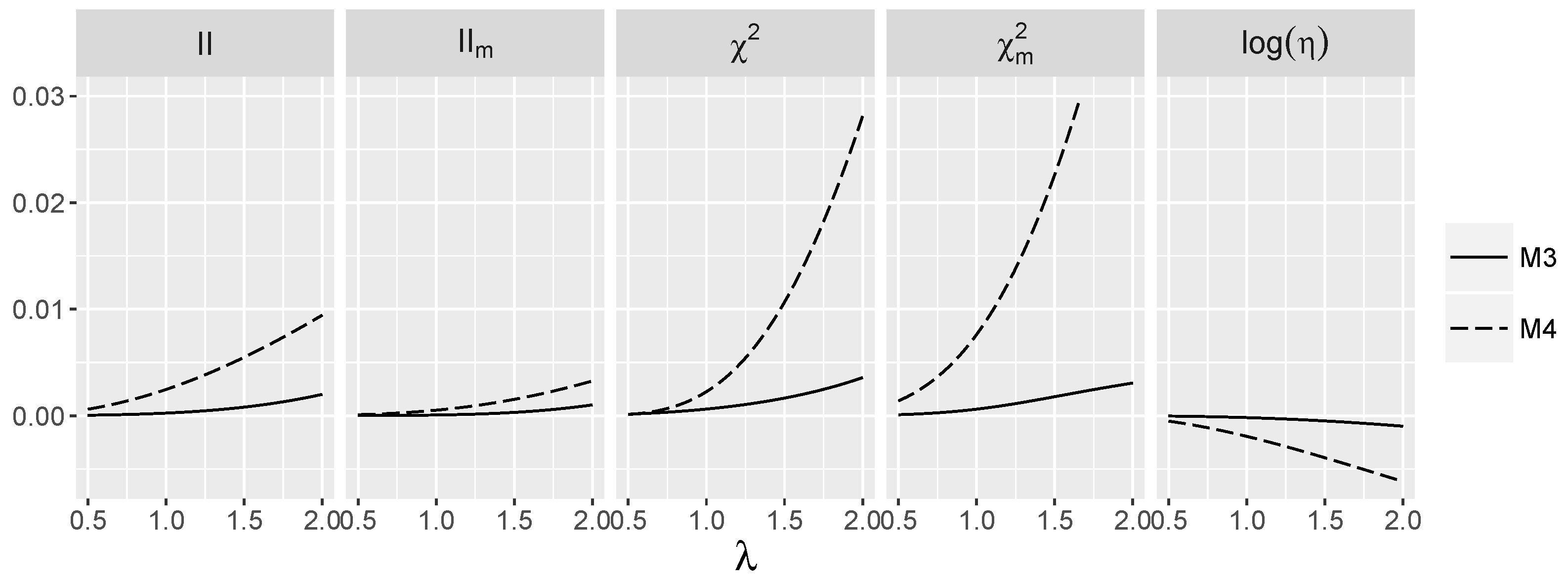
3.4. Behavior of Interaction Indices When and Are Independent
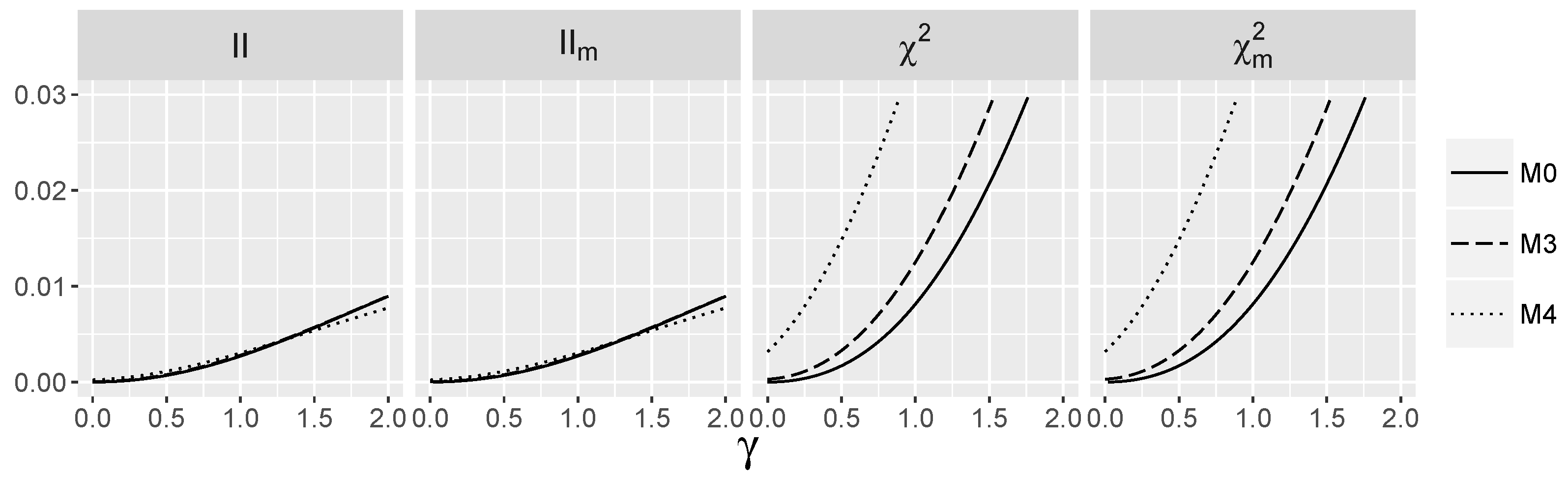
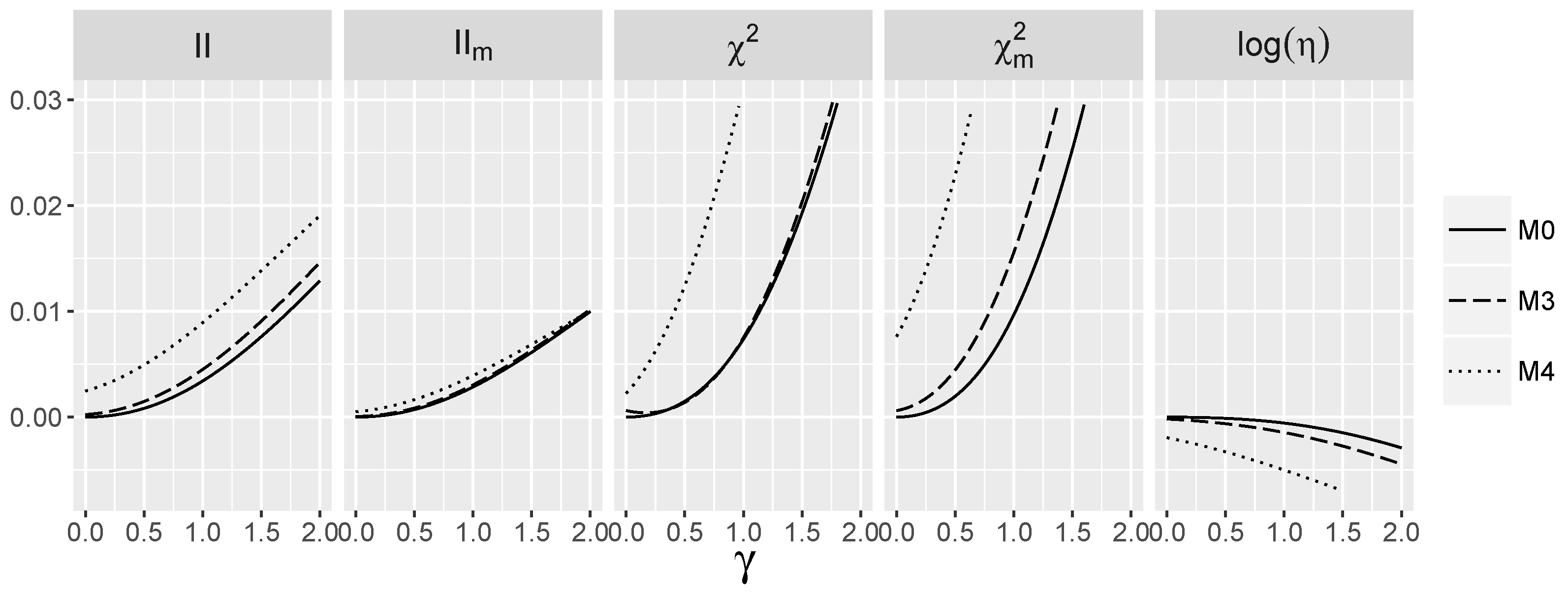
3.5. Behavior of Interaction Indices When and Are Dependent
4. Tests for Predictive Interaction
- , the number of observations in controls () and cases (), set equal in our experiments and , the total number of observations. Values of and were considered.
- MAF, the minor allele frequency for and . We set for both loci.
- copula, the function that determines the cumulative distribution of based on its marginal distributions.
- , the prevalence mapping, which in our experiments was either additive logistic or logistic with nonzero interaction.
4.1. Behavior of Interaction Tests When and Are Independent
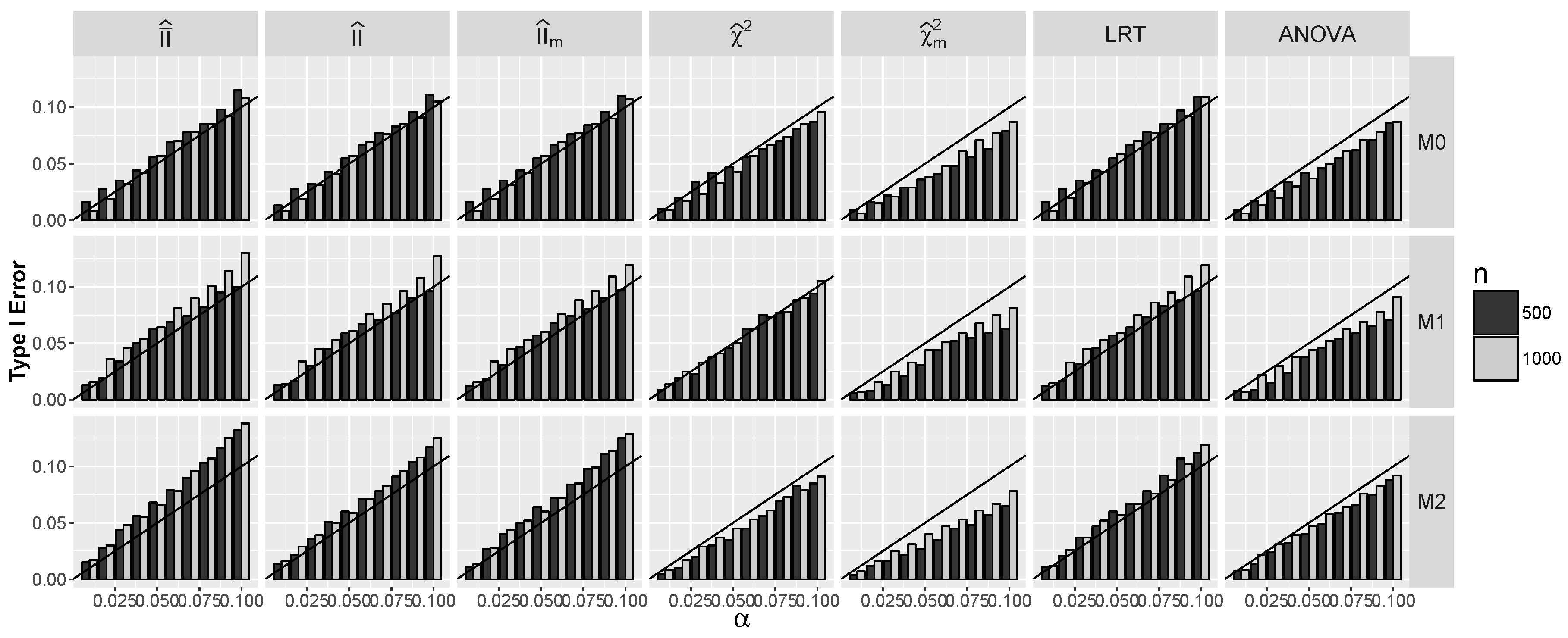
4.1.1. Type I Errors for Models –
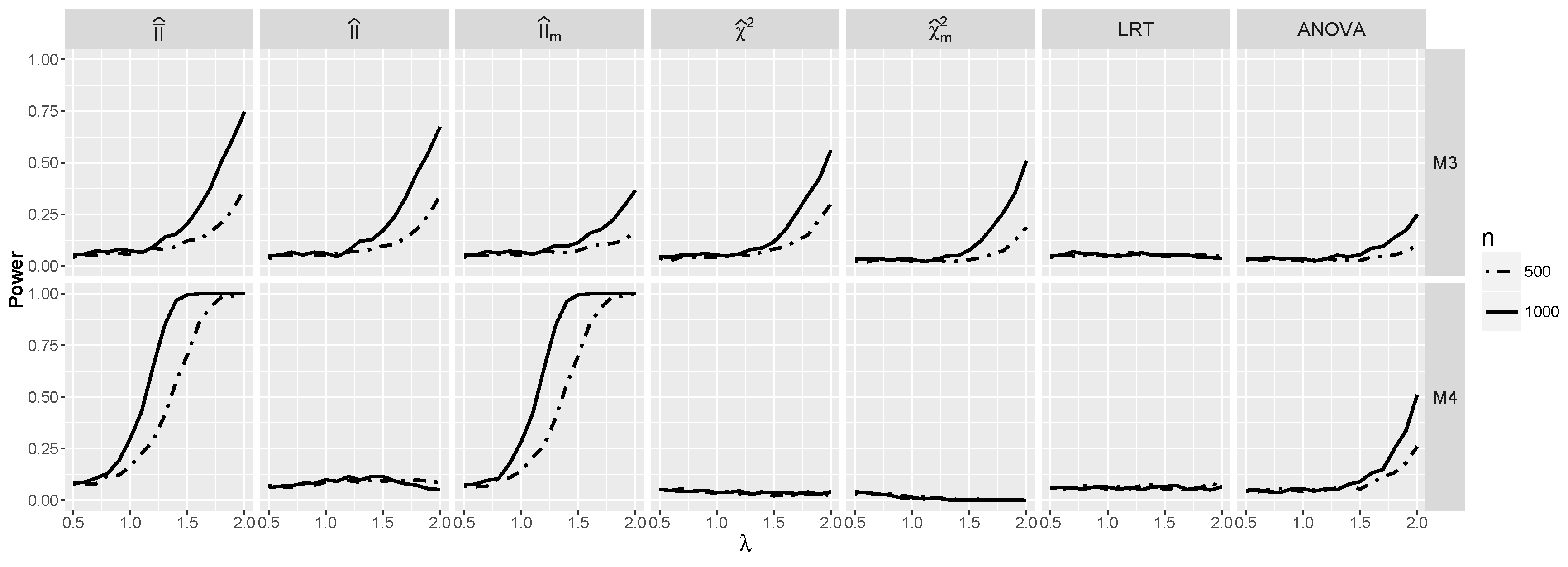
4.1.2. Power for Additive Logistic Models
4.1.3. Power for the Logistic Model with Interactions
4.2. Behavior of the Interaction Tests When and Are Dependent
4.2.1. Type I Errors for Model
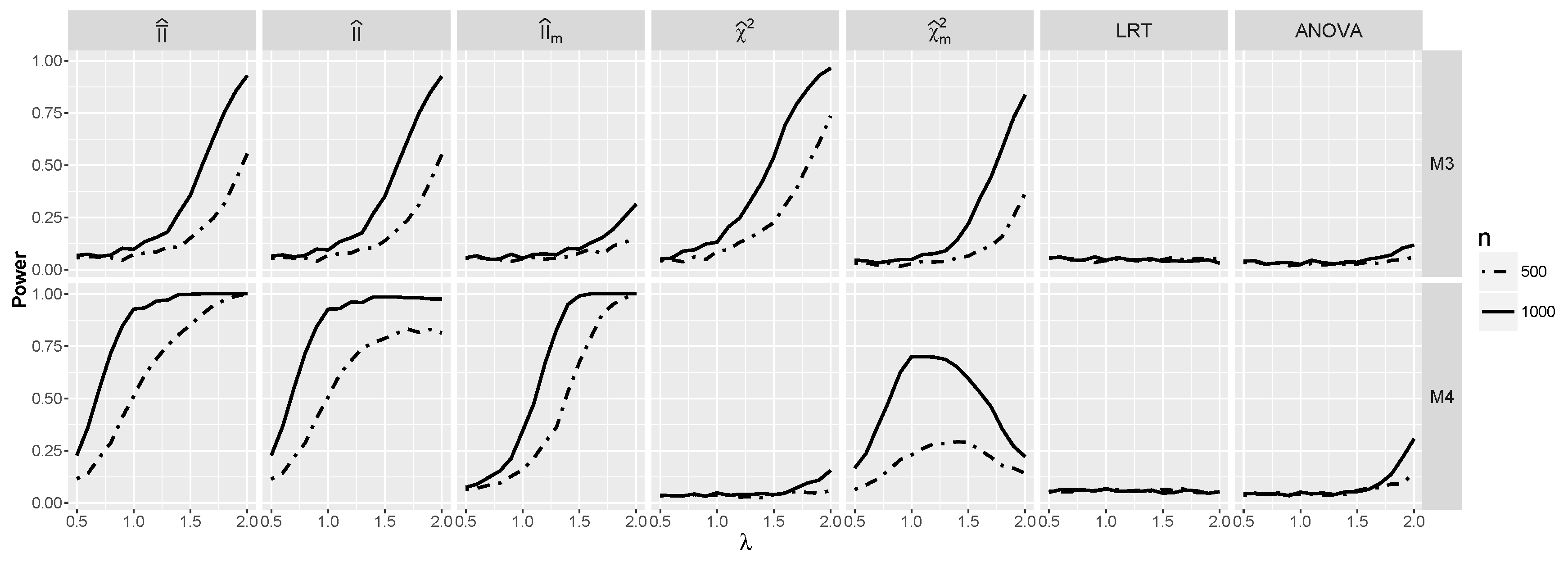
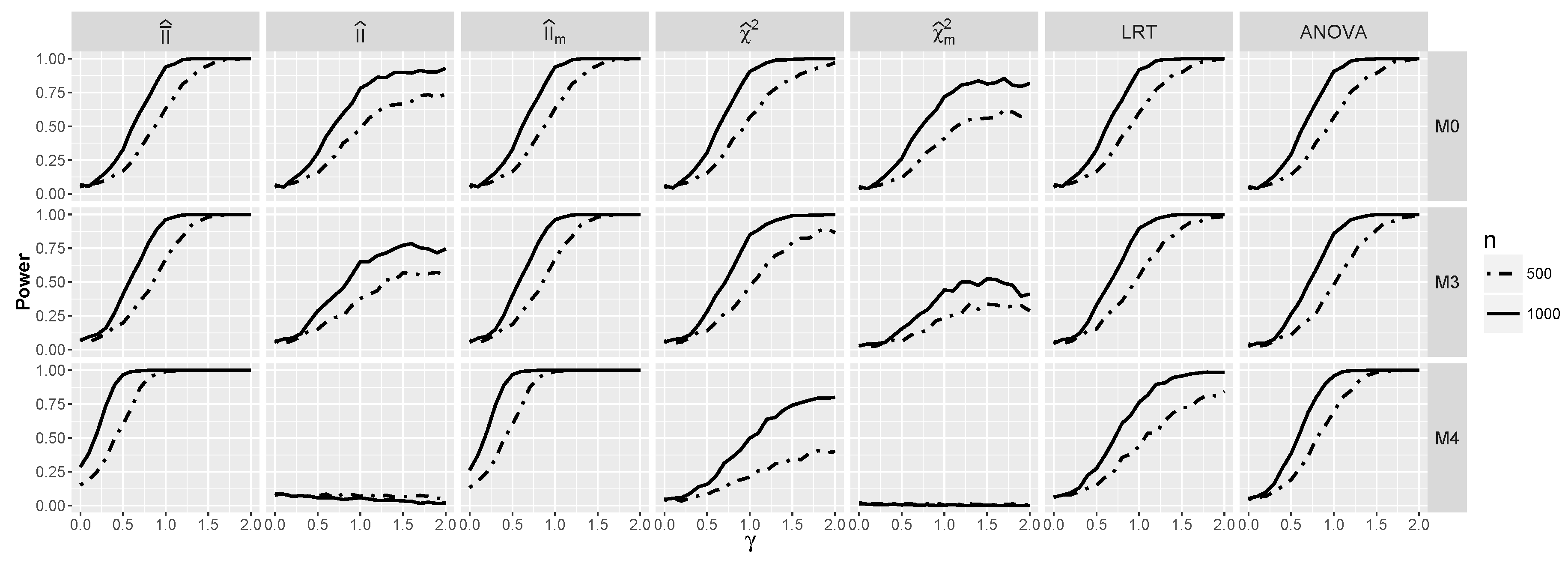
4.2.2. Power for Additive Logistic Models When and Are Dependent
4.2.3. The Powers for Logistic Models with Interaction When and Are Dependent
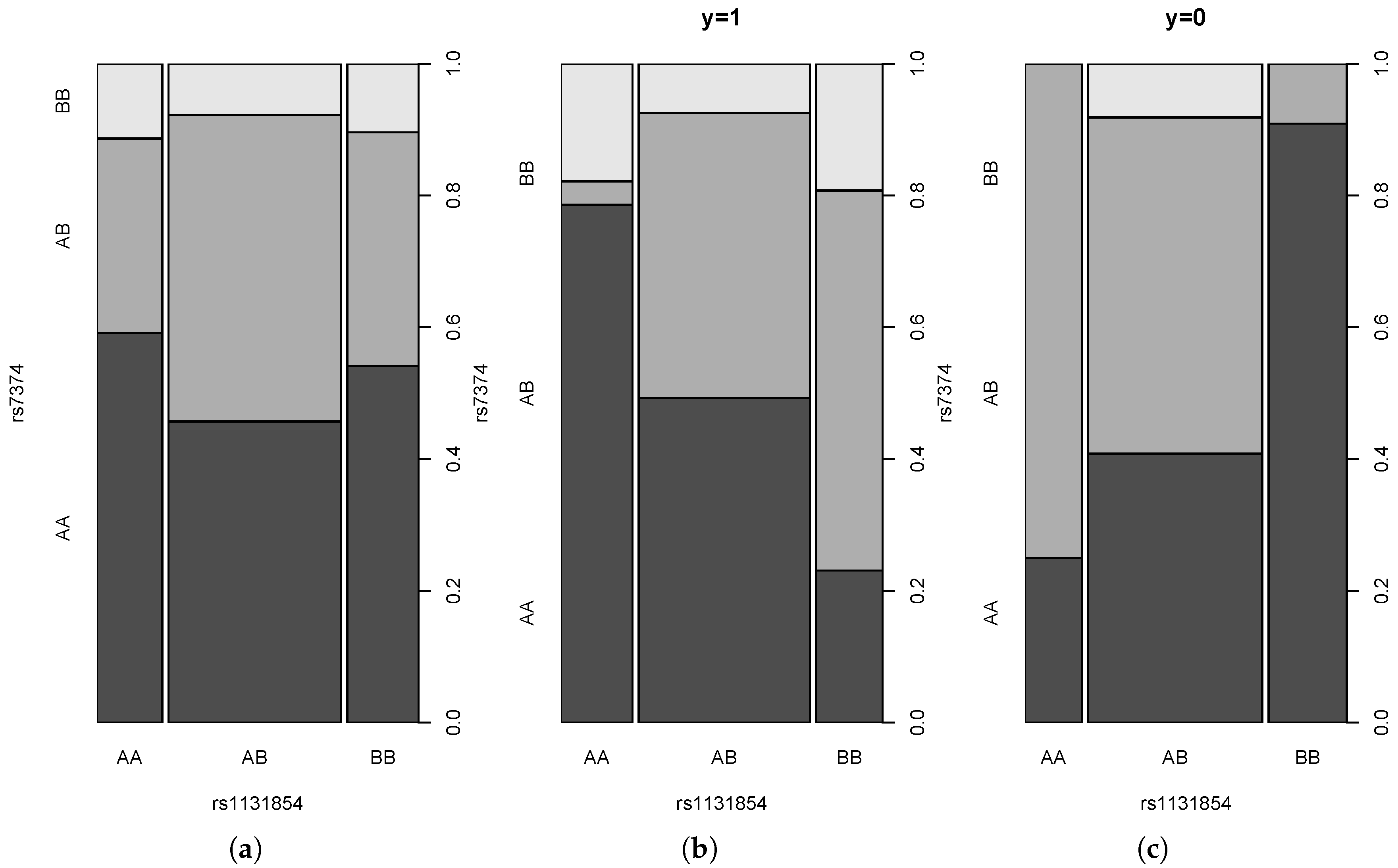
4.3. Real Data Example
5. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix A
Appendix A.1. Distribution of X1,X2
| 1 | 2 | 3 | ∑ | |
|---|---|---|---|---|
| Independent and | ||||
| 1 | ||||
| 2 | ||||
| 3 | ||||
| ∑ | 1 | |||
| Frank Copula with | ||||
| 1 | ||||
| 2 | ||||
| 3 | ||||
| ∑ | 1 | |||
Appendix A.2. Prevalence Mapping with the Logistic Regression Model
References
- Cordell, H.J. Epistasis: What it means, what it doesn’t mean, and statistical methods to detect it in humans. Hum. Mol. Genet. 2002, 11, 2463–2468. [Google Scholar] [CrossRef]
- Ritchie, M.; Hahn, L.; Roodi, N.; Bailey, L.; Dupont, W.; Parl, F.; Moore, J. Multifactor dimensionality reduction reveals high-order interactions in genome-wide association studies in sporadic breast cancer. Am. J. Hum. Genet. 2001, 69, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, R. Bayesian inference of epistatic interactions in case-control studies. Nat. Genet. 2007, 39, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; He, Z.; Wan, X.; Yang, Q.; Xue, H.; Yu, W. SNP Harvester: A filtering-based approach for detecting epistatic interactions in genome-wide association studies. Bioinformatics 2009, 25, 504–511. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xu, H.; Chen, S. Genome-wide interaction based association analysis identified multiple new susceptibility loci for common diseases. PLoS Genet. 2011, 7, e1001338. [Google Scholar] [CrossRef] [PubMed]
- Goudey, B.; Rawlinson, D.; Wang, Q.; Shi, F.; Ferra, H.; Campbell, R.M.; Stern, L.; Inouye, M.T.; Ong, C.S.; Kowalczyk, A. GWIS—Model-free, fast and exhaustive search for epistatic interactions in case-control GWAS. BMC Genom. 2013, 14 (Suppl. 3), S10. [Google Scholar] [CrossRef] [PubMed]
- Chanda, P.; Sucheston, L.; Zhang, A.; Brazeau, D.; Freudenheim, J.; Ambrosone, C.; Ramanathan, M. AMBIENCE: A novel approach and efficient algorithm for identifying informative genetic and environmental associations with complex phenotypes. Genetics 2008, 180, 1191–1210. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Yang, C.; Yang, Q.; Xue, T.; Fan, X.; Tang, N.; Yu, W. BOOST: A fast approach to detecting gene-gene interactions in genome-wide case-control studies. Am. J. Hum. Genet. 2010, 87, 325–340. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.H.; Hermani, G.; Haley, C. Detecting epistasis in human complex traits. Nat. Rev. Genet. 2014, 15, 722–733. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.; Williams, S. Epistasis. Methods and Protocols; Humana Press: New York, NY, USA, 2015. [Google Scholar]
- Duggal, P.; Gillanders, E.; Holmes, T.; Bailey-Wilson, J. Establishing an adjusted p-value threshold to control the family-wide type 1 error in genome wide association studies. BMC Genom. 2008, 9, 516. [Google Scholar] [CrossRef] [PubMed]
- Culverhouse, R.; Suarez, B.K.; Lin, J.; Reich, T. A perspective on epistasis: Limits of models displaying no main effect. Am. J. Hum. Genet. 2002, 70, 461–471. [Google Scholar] [CrossRef] [PubMed]
- Lüdtke, N.; Panzeri, S.; Brown, M.; Broomhead, D.S.; Knowles, J.; Montemurro, M.A.; Kell, D.B. Information-theoretic sensitivity analysis: A general method for credit assignment in complex networks. J. R. Soc. Interface 2008, 5, 223–235. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.; Marchini, J.; Morris, A.; Cardon, L. Two-stage two-locus models in genome-wide asssociation. PLoS Genet. 2006, 2, 1424–1432. [Google Scholar]
- McGill, W. Multivariate information transmission. Psychometrika 1954, 19, 97–116. [Google Scholar] [CrossRef]
- Fano, F. Transmission of Information: Statistical Theory of Communication; MIT Press: Cambridge, MA, USA, 1961. [Google Scholar]
- Cover, T.; Thomas, J. Elements of Information Theory; Wiley: New York, NY, USA, 2006. [Google Scholar]
- Han, T.S. Multiple mutual informations and multiple interactions in frequency data. Inf. Control 1980, 46, 26–45. [Google Scholar] [CrossRef]
- Kang, G.; Yue, W.; Zhang, J.; Cui, Y.; Zuo, Y.; Zhang, D. An entropy-based approach for testing genetic epistasis underlying complex diseases. J. Theor. Biol. 2008, 250, 362–374. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Jin, L.; Xiong, M. Test for interaction between two unlinked loci. Am. J. Hum. Genet. 2006, 79, 831–845. [Google Scholar] [CrossRef] [PubMed]
- Darroch, J. Interactions in multi-factor contingency tables. J. R. Stat. Soc. Ser. B 1962, 24, 251–263. [Google Scholar]
- Sucheston, L.; Chanda, P.; Zhang, A.; Tritchler, D.; Ramanathan, M. Comparison of information-theoretic to statistical methods for gene-gene interactions in the presence of genetic heterogeneity. BMC Genom. 2010, 11, 487. [Google Scholar] [CrossRef] [PubMed]
- Darroch, J. Multiplicative and additive interaction in contingency tables. Biometrika 1974, 9, 207–214. [Google Scholar] [CrossRef]
- Li, W.; Reich, J. A complete enumeration and classification of two-locus disease models. Hum. Hered. 2000, 17, 334–349. [Google Scholar] [CrossRef]
- Culverhouse, R. The use of the restricted partition method with case-control data. Hum. Hered. 2007, 63, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Rencher, A.C.; Schaalje, G.B. Linear Models in Statistics; Wiley: Hoboken, NJ, USA, 2008. [Google Scholar]
- Tan, A.; Fan, J.; Karakiri, C.; Bibikova, M.; Garcia, E.; Zhou, L.; Barker, D.; Serre, D.; Feldmann, G.; Hruban, R.; et al. Allele-specific expression in the germline of patients with familial pancreatic cancer: An unbiased approach to cancer gene discovery. Cancer Biol. Ther. 2008, 7, 135–144. [Google Scholar] [CrossRef]
- SNPsyn. Available online: http://snpsyn.biolab.si (accessed on 4 January 2017).
- Nelson, R.B. An Introduction to Copulas; Springer: New York, NY, USA, 1999. [Google Scholar]


| Model\Coefficients | ||||||
|---|---|---|---|---|---|---|
| 0 | 0 | 0 | 0 | 0 | 0 | |
| λ | 0 | 0 | 0 | 0 | 0 | |
| λ | λ | 0 | 0 | 0 | 0 | |
| λ | 0 | 0 | λ | 0 | 0 | |
| λ | λ | 0 | λ | λ | 0 |
| 1 | 2 | 3 | |
|---|---|---|---|
| 1 | γ | γ | 0 |
| 2 | γ | γ | 0 |
| 3 | 0 | 0 | 0 |
© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mielniczuk, J.; Rdzanowski, M. Use of Information Measures and Their Approximations to Detect Predictive Gene-Gene Interaction. Entropy 2017, 19, 23. https://doi.org/10.3390/e19010023
Mielniczuk J, Rdzanowski M. Use of Information Measures and Their Approximations to Detect Predictive Gene-Gene Interaction. Entropy. 2017; 19(1):23. https://doi.org/10.3390/e19010023
Chicago/Turabian StyleMielniczuk, Jan, and Marcin Rdzanowski. 2017. "Use of Information Measures and Their Approximations to Detect Predictive Gene-Gene Interaction" Entropy 19, no. 1: 23. https://doi.org/10.3390/e19010023
APA StyleMielniczuk, J., & Rdzanowski, M. (2017). Use of Information Measures and Their Approximations to Detect Predictive Gene-Gene Interaction. Entropy, 19(1), 23. https://doi.org/10.3390/e19010023





