Minimum Spanning vs. Principal Trees for Structured Approximations of Multi-Dimensional Datasets
Abstract
1. Introduction
2. Results
2.1. Comparing and Benchmarking Graph-Based Data Approximation Methods Using Data Point Partitioning by Graph Segments
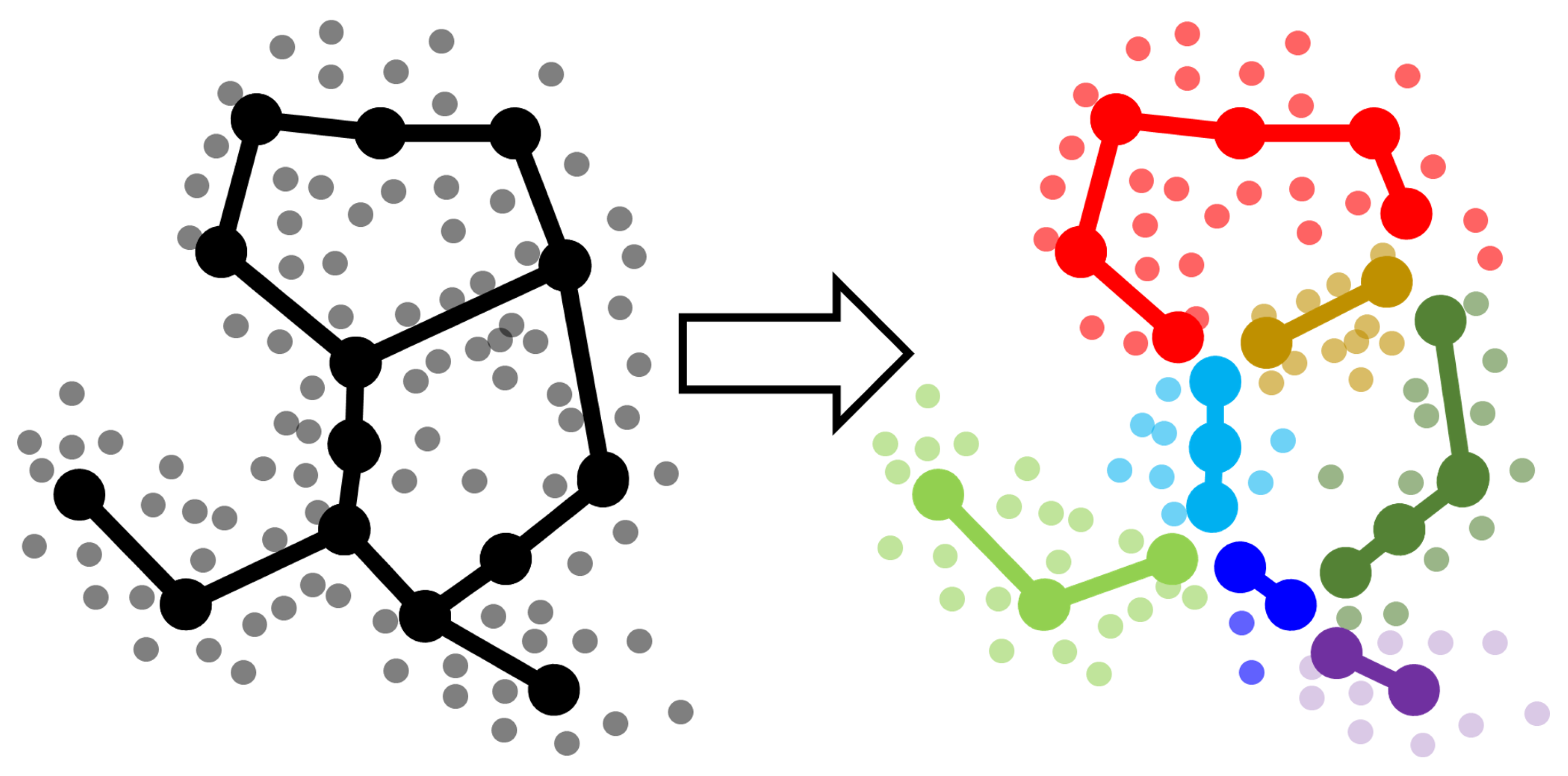
2.1.1. Outlining the Method
- Split each approximating graph into segments. By a segment we mean certain path from one branching point or leaf node to another.
- Create clustering (partitioning) of the dataset by segments. Each data point is associated with the nearest segment of the graph. Thus, in general, if graph is partitioned into N segments at the previous step then the dataset will be partitioned into N clusters.
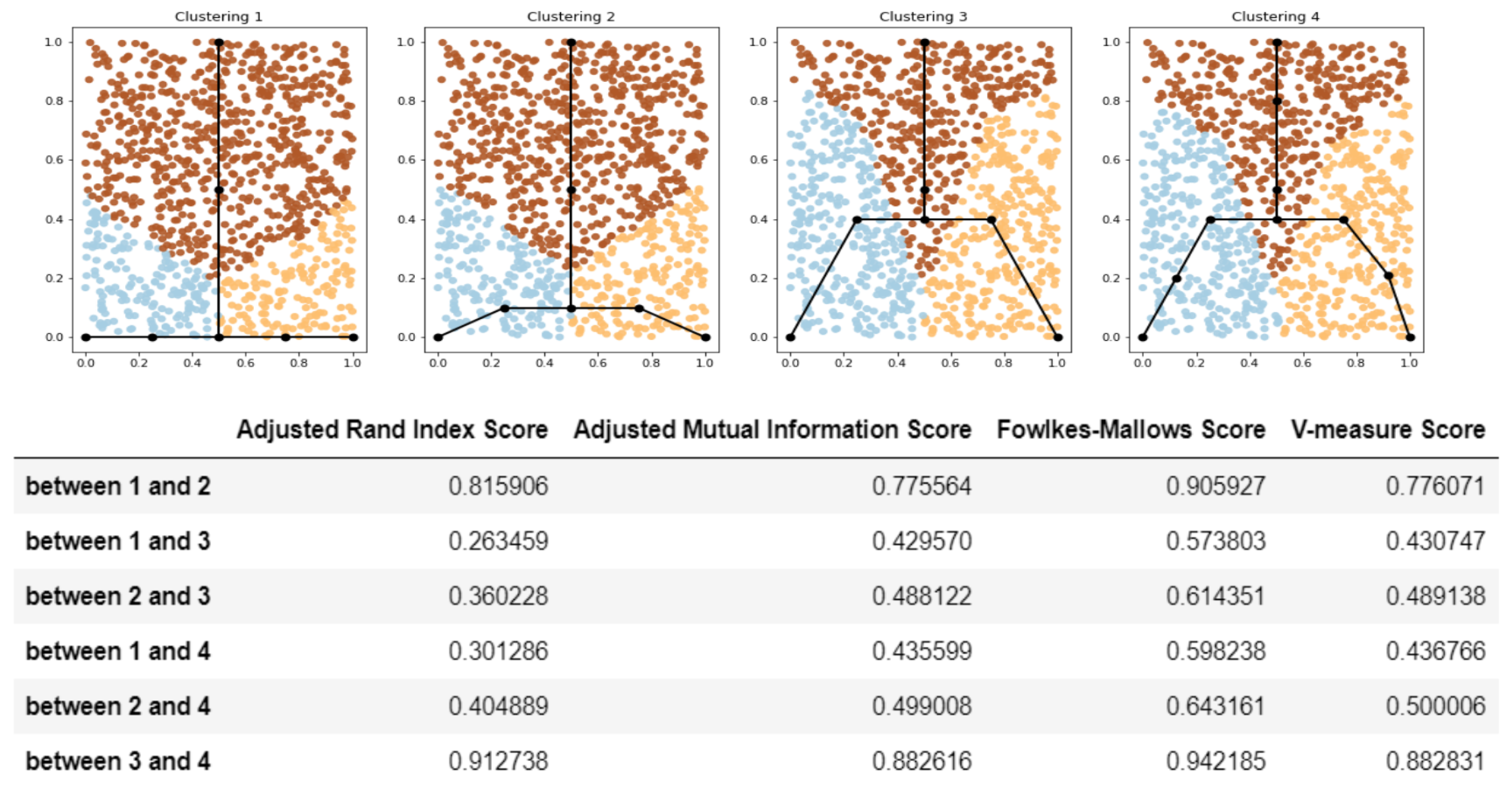
- Compare the clusterings for the dataset using standard metrics (like adjusted Rand index or adjusted mutual information or any other suitable measure). Since the clusterings are produced from the structure of the approximating graphs, the score shows how two GBDA results are similar to each other.
2.1.2. Details on Step 1: Segmenting the Graphs
2.1.3. Details on Step 2: Clustering (Partitioning) Data Points by Graph Segments
2.1.4. Details on Step 3: Compare the Clustering Measures for the Datasets
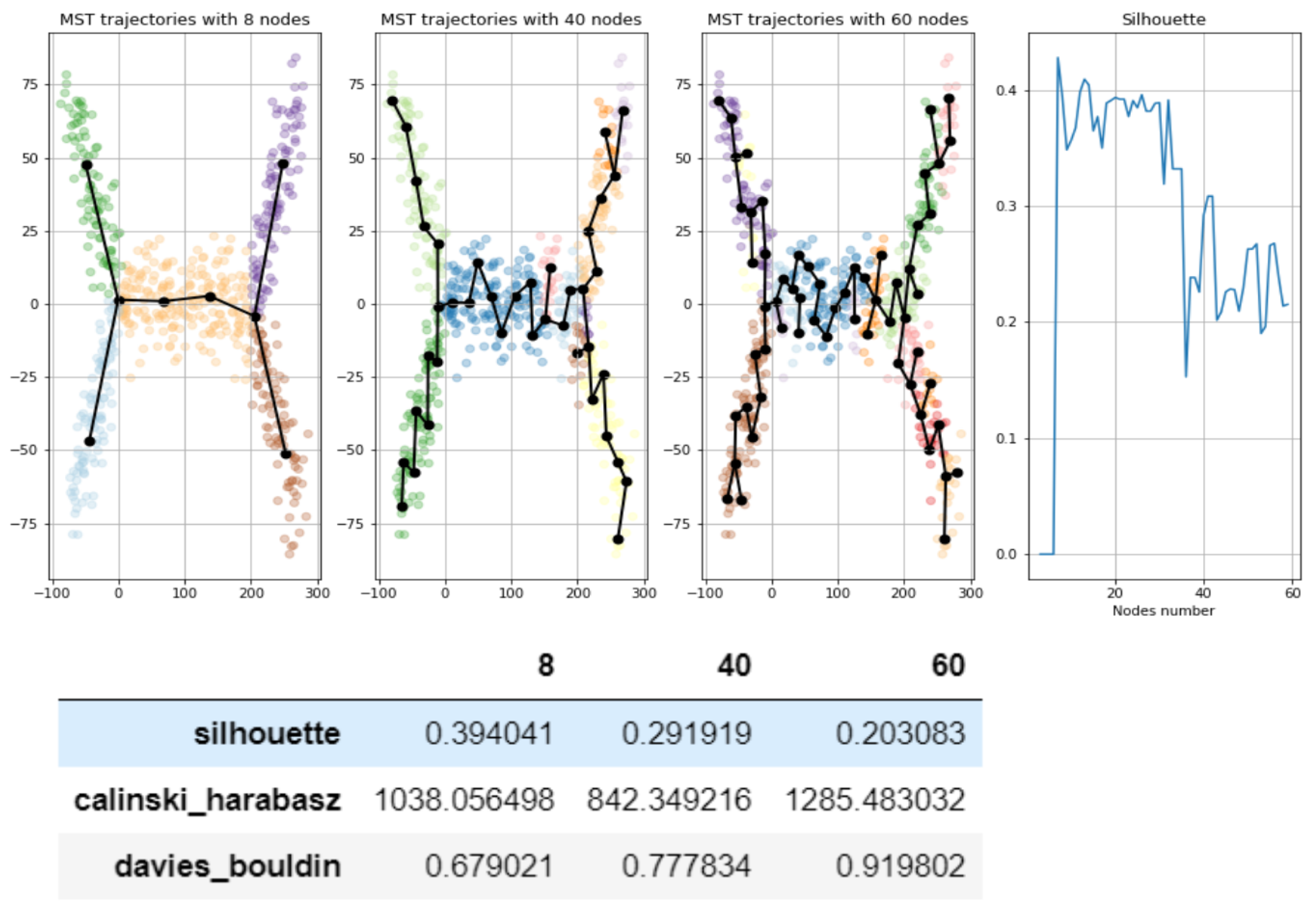
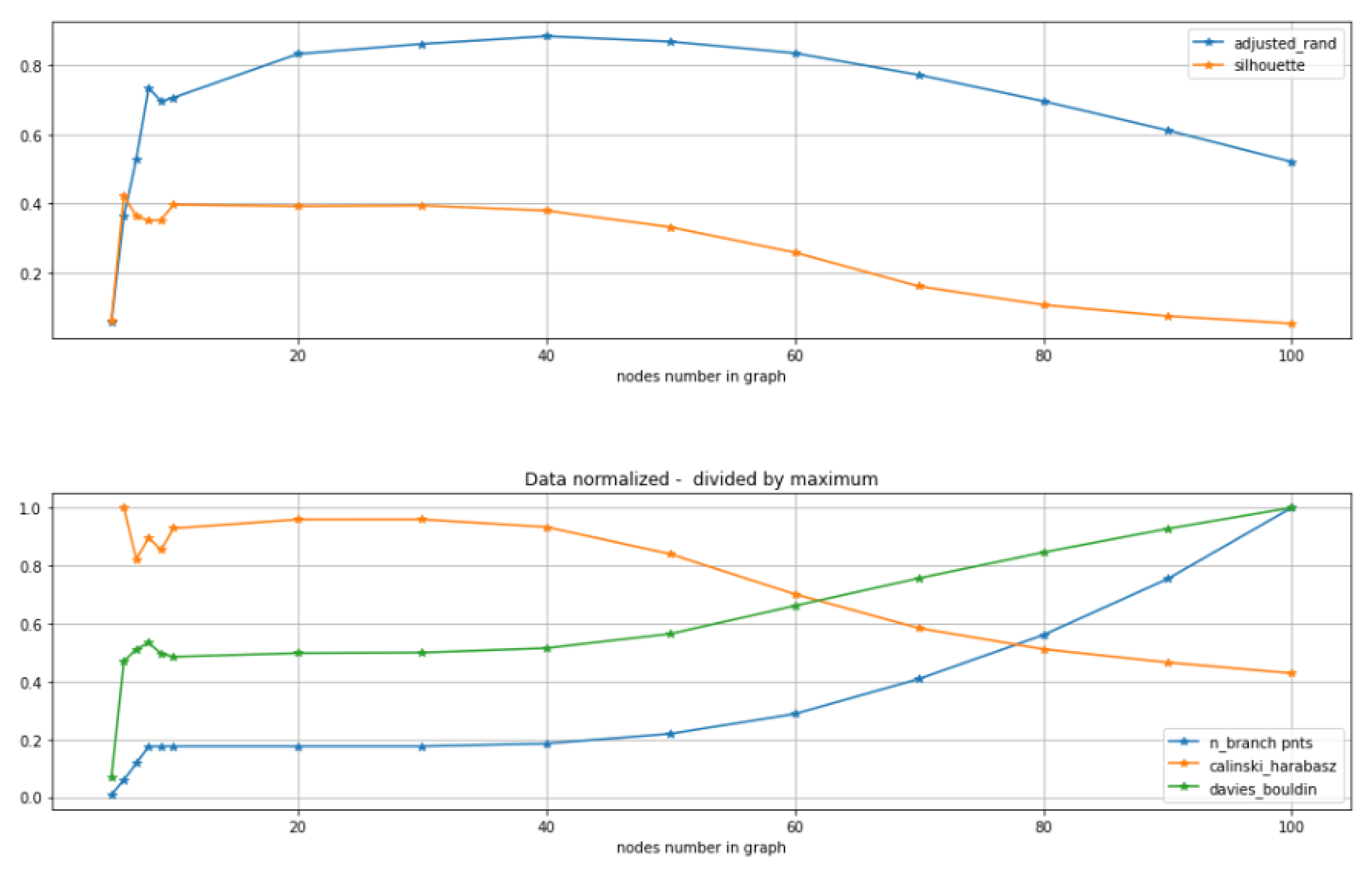
2.2. Unsupervised Scores for Comparing GBDA Methods and Parameter Fine-Tuning
- Split each approximating graph into segments
- Create clustering (partitioning) of the dataset by the segments
- Use standard metrics and tools to calculate how good is the clustering:, e.g., calculate silhouette, Calinski–Harabasz, Davies-Bouldin score etc.
2.3. Comparison of Clustering Based Scores with Other GBDA Metrics
2.4. Comparison of MST-Based Graph-Based Approximations and ElPiGraph
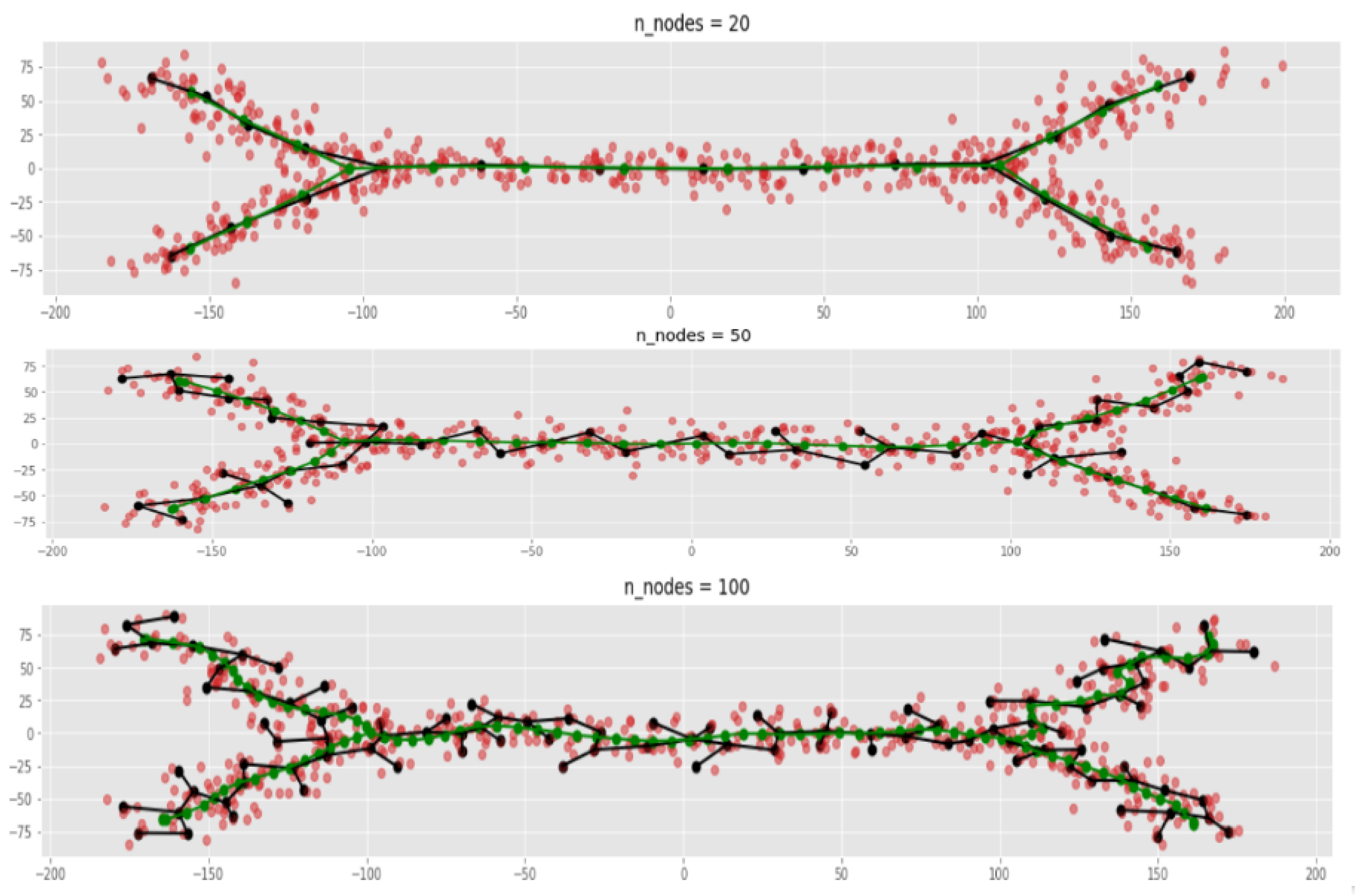
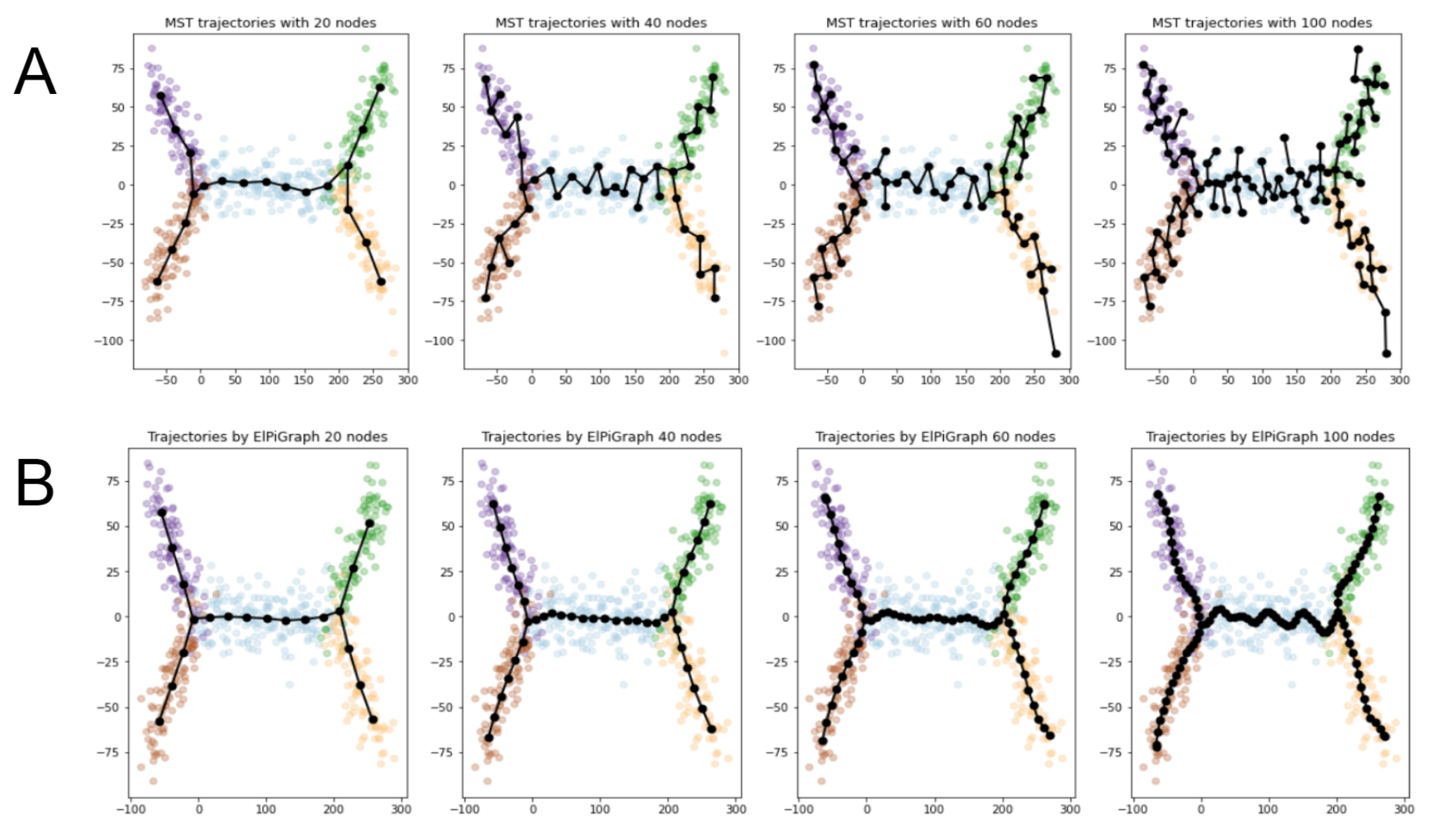
- Both MST and ElPiGraph can achieve similar quality of graph-based data approximation by tuning their main parameter, number of nodes. The quality here is estimated by the scores introduced above and in comparison with ground truth for simulated datasets. However, the advantage of ElPiGraph is in its stability with respect to this parameter. Small modifications do not change the quality dramatically, while for MST even small modifications of the parameter may lead to quite a drastic change of the approximating graph structure and, respectively, the quality of ground truth approximation. That means that in practical situation where ground truth is not available, ElPiGraph has an advantage since one should not be afraid of choosing the parameter incorrectly. Therefore, a reasonably wide range of parameters gives similar results, while for MST approach choosing an incorrect parameter would give a significant loss in approximation quality.
- The embedment of approximating graphs produced by ElPiGraph are much smoother comparing to MST. This observation is consistent with the previous one and with ElPiGraph algorithm which penalizes for non-smoothness and hence it produces graphs which would not produce too much branching points, the problem which underlies the MST instability.
- We analyse a possible combination of MST+ElPiGraph methods, where the initial graph approximation is obtained by MST and then ElPiGraph algorithm starts from this initialization. One of our conclusions is that ElPiGraph “forgets” the initial conditions quite fast. Nevertheless the combination of methods may have certain advantages.
- We consider a real-life single cell RNA sequencing dataset and demonstrate that our approach for choosing the parameters of the trajectory inference method based on unsupervised clustering scores provides reasonable results.
2.4.1. Smoothness of ElpiGraph-Based Graph Approximators Comparing to MST
2.4.2. Comparison between ElPiGraph and MST on Large Number of Different Datasets
2.4.3. Initialization of ElPiGraph with MST-Based Tree
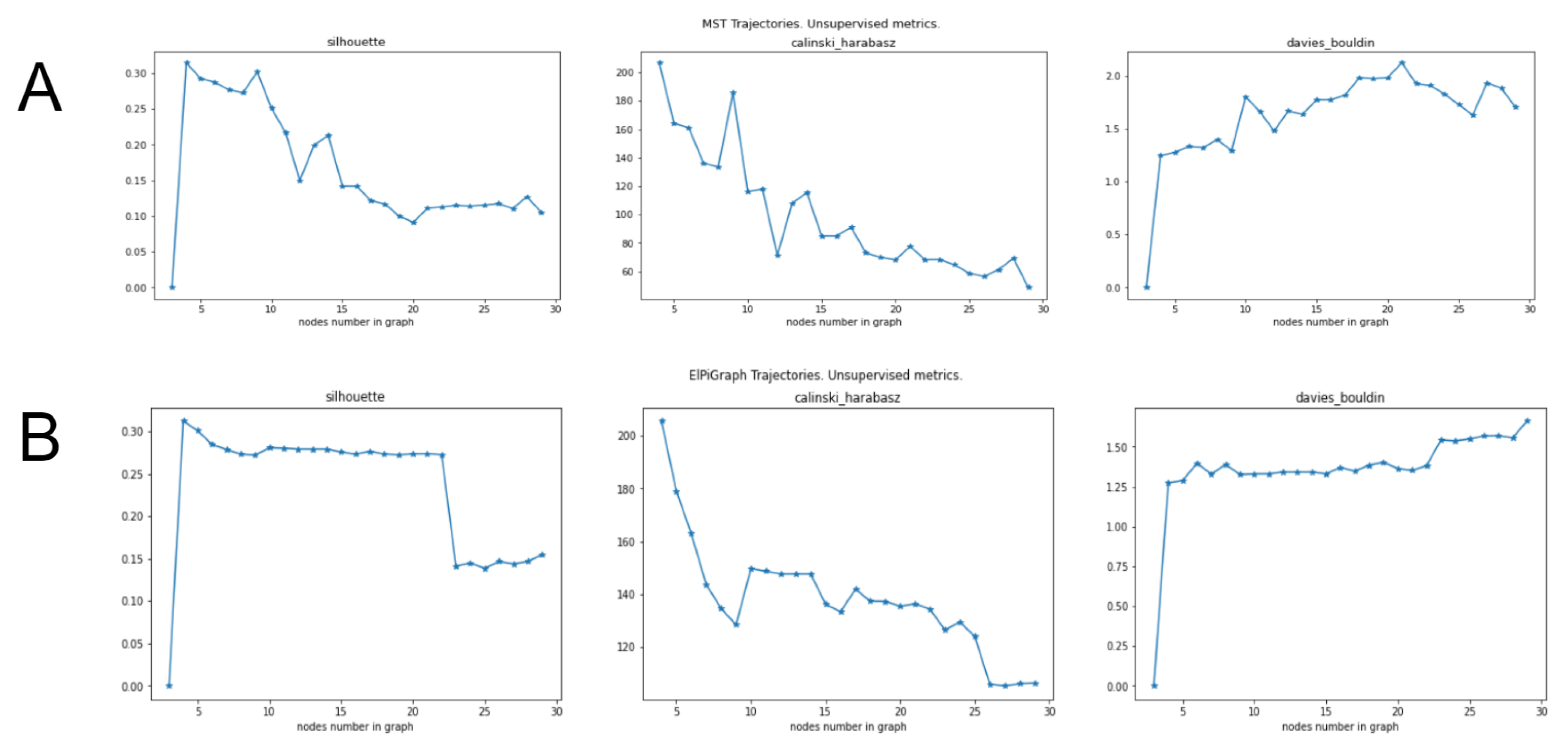
2.5. Tuning Parameters of GBDA Algorithms Using Unsupervised Clustering Quality Scores
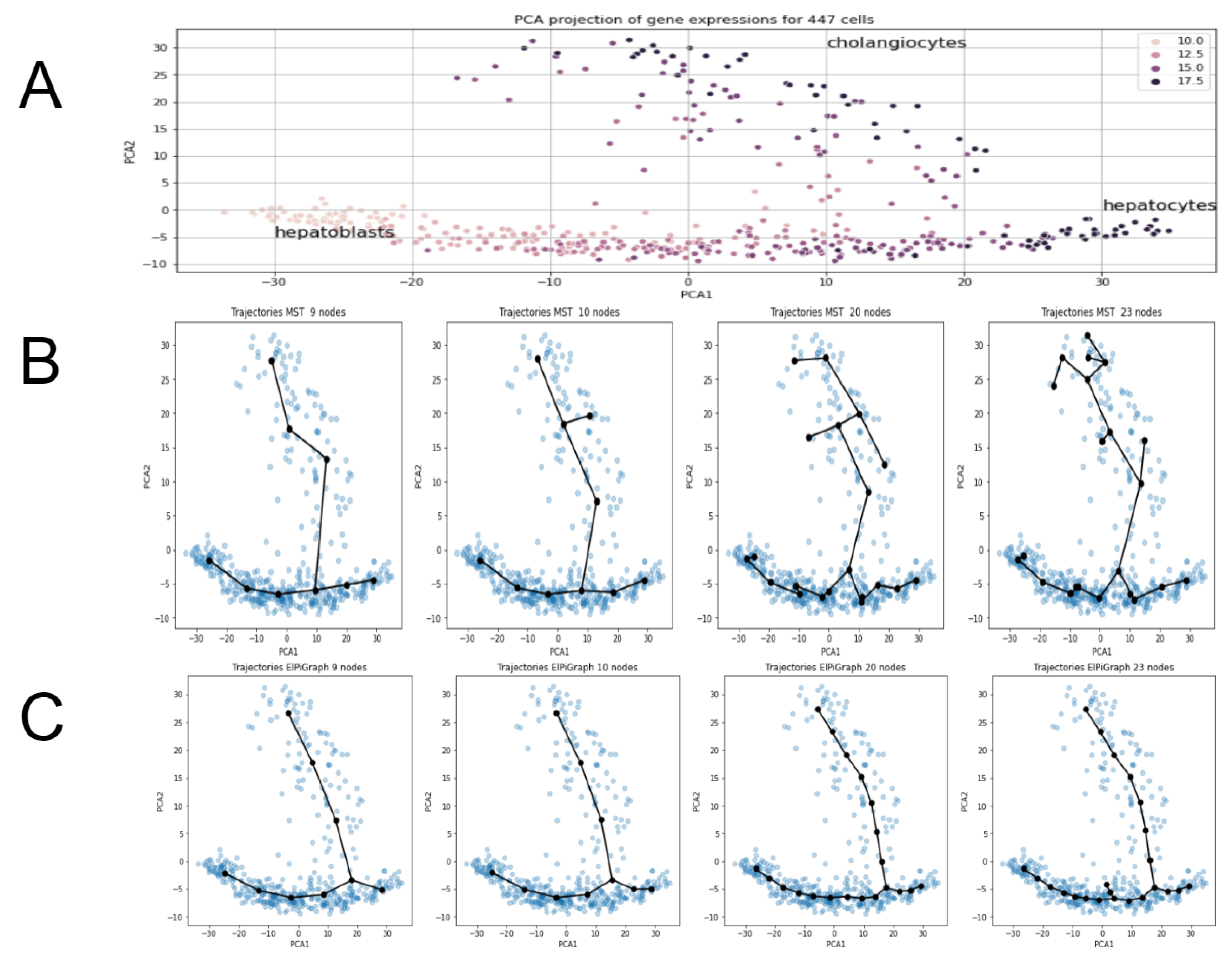
2.6. Single Cell Data Analysis Example
- ElPiGraph demonstrates much higher stability than MST and choice of main parameter (node number) can be made more easily and reliably.
- Unsupervised metrics for trajectories introduced above (based on silhouette, Calinski–Harabas, Davies-Bouldin clustering scores) provides insights for the choice of parameters for trajectory inference methods.
- Heuristic for MST, to take number of nodes equal to approximately square root of number of datapoints fails significantly for the present example. It would give 21 nodes, however trajectories of MST with 21 nodes show significant “overbranching” (see figures below). Reasonable node numbers for MST is 9 or a little below, but 10 and above leads to biologically incorrect branching.
3. Discussion
4. Materials and Methods
4.1. Minimal Spanning Tree-Based Data Approximation Method
- Split the dataset to certain pieces (for example, by applying K-means clustering for some K).
- Construct kNN (k-nearest neighbour) graph with nodes at the centers of clusters.
- Compute an MST (minimal spanning tree) of the kNN graph computed as described above.
4.2. Method of Elastic Principal Graphs (ElPiGraph)
4.3. Benchmark Implementation
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| PG | Principal graph |
| SOM | Self-Organizing Map |
| kNN | k-nearest neighbours |
| GBDA | Graph-based data approximations |
| TI | Trajectory inference |
| MST | Minimal spanning tree |
| PAGA | Partition-based graph abstraction |
| ElPiGraph | Elastic Principal Graph |
References
- Kohonen, T. Self-organized formation of topologically correct feature maps. Biol. Cybern. 1982, 43. [Google Scholar] [CrossRef]
- Gorban, A.N.; Zinovyev, A.Y. Method of elastic maps and its applications in data visualization and data modeling. Int. J. Comput. Anticipatory Syst. CHAOS 2001, 12, 353–369. [Google Scholar]
- Gorban, A.N.; Zinovyev, A. Principal manifolds and graphs in practice: From molecular biology to dynamical systems. Int. J. Neural Syst. 2010, 20, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Hastie, T.; Stuetzle, W. Principal Curves. J. Am. Stat. Assoc. 1989, 84. [Google Scholar] [CrossRef]
- Kégl, B.; Krzyzak, A.; Linder, T.; Zeger, K. A polygonal line algorithm for constructing principal curves. In Proceedings of the Advances in Neural Information Processing Systems, Denver, CO, USA, 1–5 December 1998. [Google Scholar]
- Gorban, A.; Kégl, B.; Wunch, D.; Zinovyev, A. (Eds.) Principal Manifolds for Data Visualisation and Dimension Reduction; Springer: Berlin, Germany, 2008; p. 340. [Google Scholar] [CrossRef]
- Amsaleg, L.; Chelly, O.; Furon, T.; Girard, S.; Houle, M.E.; Kawarabayashi, K.I.; Nett, M. Estimating local intrinsic dimensionality. In Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Sydney, Australia, 10–13 August 2015. [Google Scholar] [CrossRef]
- Albergante, L.; Bac, J.; Zinovyev, A. Estimating the effective dimension of large biological datasets using Fisher separability analysis. In Proceedings of the International Joint Conference on Neural Networks, Budapest, Hungary, 14–19 July 2019. [Google Scholar] [CrossRef]
- Bac, J.; Zinovyev, A. Local intrinsic dimensionality estimators based on concentration of measure. arXiv 2020, arXiv:2001.11739. [Google Scholar]
- Gorban, A.N.; Sumner, N.R.; Zinovyev, A.Y. Topological grammars for data approximation. Appl. Math. Lett. 2007, 20, 382–386. [Google Scholar] [CrossRef]
- Kruskal, J.B. On the shortest spanning subtree of a graph and the traveling salesman problem. Proc. Am. Math. Soc. 1956, 7, 48–50. [Google Scholar] [CrossRef]
- Gorban, A.N.; Zinovyev, A.Y. Principal Graphs and Manifolds. arXiv 2008, arXiv:0809.0490. [Google Scholar]
- Mao, Q.; Yang, L.; Wang, L.; Goodison, S.; Sun, Y. SimplePPT: A simple principal tree algorithm. In Proceedings of the 2015 SIAM International Conference on Data Mining, Vancouver, BC, Canada, 30 April–2 May 2015; pp. 792–800. [Google Scholar]
- Mao, Q.; Wang, L.; Tsang, I.W.; Sun, Y. Principal Graph and Structure Learning Based on Reversed Graph Embedding. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2227–2241. [Google Scholar] [CrossRef]
- Lähnemann, D.; Köster, J.; Szczurek, E.; McCarthy, D.J.; Hicks, S.C.; Robinson, M.D.; Vallejos, C.A.; Campbell, K.R.; Beerenwinkel, N.; Mahfouz, A.; et al. Eleven grand challenges in single-cell data science. Genome Biol. 2020, 21, 1–35. [Google Scholar] [CrossRef]
- Saelens, W.; Cannoodt, R.; Todorov, H.; Saeys, Y. A comparison of single-cell trajectory inference methods. Nat. Biotechnol. 2019, 37, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Aynaud, M.M.; Mirabeau, O.; Gruel, N.; Grossetête, S.; Boeva, V.; Durand, S.; Surdez, D.; Saulnier, O.; Zaïdi, S.; Gribkova, S.; et al. Transcriptional Programs Define Intratumoral Heterogeneity of Ewing Sarcoma at Single-Cell Resolution. Cell Rep. 2020, 30, 1767–1779. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Tan, Y.; Cahan, P. Understanding development and stem cells using single cell-based analyses of gene expression. Development 2017, 144, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Wolf, F.A.; Hamey, F.K.; Plass, M.; Solana, J.; Dahlin, J.S.; Göttgens, B.; Rajewsky, N.; Simon, L.; Theis, F.J. PAGA: Graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol. 2019, 20, 1–9. [Google Scholar] [CrossRef]
- Chen, H.; Albergante, L.; Hsu, J.Y.; Lareau, C.A.; Lo Bosco, G.; Guan, J.; Zhou, S.; Gorban, A.N.; Bauer, D.E.; Aryee, M.J.; et al. Single-cell trajectories reconstruction, exploration and mapping of omics data with STREAM. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef]
- Bac, J.; Zinovyev, A. Lizard Brain: Tackling Locally Low-Dimensional Yet Globally Complex Organization of Multi-Dimensional Datasets. Front. Neurorob. 2020, 13, 110. [Google Scholar] [CrossRef]
- Rousseeuw, P.J. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987, 20, 53–65. [Google Scholar] [CrossRef]
- Rand, W.M. Objective criteria for the evaluation of clustering methods. J. Am. Stat. Assoc. 1971, 66, 846–850. [Google Scholar] [CrossRef]
- Meilǎ, M. Comparing clusterings-an information based distance. J. Multivariate Anal. 2007, 98, 873–895. [Google Scholar] [CrossRef]
- Fowlkes, E.B.; Mallows, C.L. A method for comparing two hierarchical clusterings. J. Am. Stat. Assoc. 1983, 78, 553–569. [Google Scholar] [CrossRef]
- Rosenberg, A.; Hirschberg, J. V-Measure: A conditional entropy-based external cluster evaluation measure. In Proceedings of the 2007 Joint Conference on Empirical Methods in Natural Language Processing and Computational Natural Language Learning, Prague, Czech Republic, 28–30 June 2007. [Google Scholar]
- Shin, J.; Berg, D.A.; Zhu, Y.; Shin, J.Y.; Song, J.; Bonaguidi, M.A.; Enikolopov, G.; Nauen, D.W.; Christian, K.M.; Ming, G.L.; et al. Single-Cell RNA-Seq with Waterfall Reveals Molecular Cascades underlying Adult Neurogenesis. Cell Stem Cell 2015, 17, 360–372. [Google Scholar] [CrossRef] [PubMed]
- Ji, Z.; Ji, H. TSCAN: Pseudo-time reconstruction and evaluation in single-cell RNA-seq analysis. Nucleic Acids Res. 2016, 44, e117. [Google Scholar] [CrossRef] [PubMed]
- Street, K.; Risso, D.; Fletcher, R.B.; Das, D.; Ngai, J.; Yosef, N.; Purdom, E.; Dudoit, S. Slingshot: Cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genom. 2018, 19, 477. [Google Scholar] [CrossRef] [PubMed]
- Parra, R.G.; Papadopoulos, N.; Ahumada-Arranz, L.; Kholtei, J.E.; Mottelson, N.; Horokhovsky, Y.; Treutlein, B.; Soeding, J. Reconstructing complex lineage trees from scRNA-seq data using MERLoT. Nucleic Acids Res. 2019, 47, 8961–8974. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wang, W.H.; Qiu, W.L.; Guo, Z.; Bi, E.; Xu, C.R. A single-cell transcriptomic analysis reveals precise pathways and regulatory mechanisms underlying hepatoblast differentiation. Hepatology 2017, 66, 1387–1401. [Google Scholar] [CrossRef]
- Albergante, L.; Mirkes, E.; Bac, J.; Chen, H.; Martin, A.; Faure, L.; Barillot, E.; Pinello, L.; Gorban, A.; Zinovyev, A. Robust and scalable learning of complex intrinsic dataset geometry via ElPiGraph. Entropy 2020, 22, 296. [Google Scholar] [CrossRef]
- Gorban, A.N.; Mirkes, E.; Zinovyev, A.Y. Robust principal graphs for data approximation. Arch. Data Sci. 2017, 2, 1:16. [Google Scholar]
- Golovenkin, S.E.; Bac, J.; Chervov, A.; Mirkes, E.M.; Orlova, Y.V.; Barillot, E.; Gorban, A.N.; Zinovyev, A. Trajectories, bifurcations and pseudotime in large clinical datasets: Applications to myocardial infarction and diabetes data. GigaScience 2020, in press. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chervov, A.; Bac, J.; Zinovyev, A. Minimum Spanning vs. Principal Trees for Structured Approximations of Multi-Dimensional Datasets. Entropy 2020, 22, 1274. https://doi.org/10.3390/e22111274
Chervov A, Bac J, Zinovyev A. Minimum Spanning vs. Principal Trees for Structured Approximations of Multi-Dimensional Datasets. Entropy. 2020; 22(11):1274. https://doi.org/10.3390/e22111274
Chicago/Turabian StyleChervov, Alexander, Jonathan Bac, and Andrei Zinovyev. 2020. "Minimum Spanning vs. Principal Trees for Structured Approximations of Multi-Dimensional Datasets" Entropy 22, no. 11: 1274. https://doi.org/10.3390/e22111274
APA StyleChervov, A., Bac, J., & Zinovyev, A. (2020). Minimum Spanning vs. Principal Trees for Structured Approximations of Multi-Dimensional Datasets. Entropy, 22(11), 1274. https://doi.org/10.3390/e22111274





