Augmentation of Dispersion Entropy for Handling Missing and Outlier Samples in Physiological Signal Monitoring
Abstract
:1. Introduction
- The quantification of the effect of missing and outlier samples on the performance of DisEn.
- The introduction of new variations of the DisEn algorithm to improve its performance when applied to time-series with missing and outlier samples.
- The assessment of the performance of the original algorithm and its variations, across different physiological datasets and under separate experimental setups defined by the percentage of missing or outlier samples and the degree to which these samples are grouped together or exist individually.
2. Methods
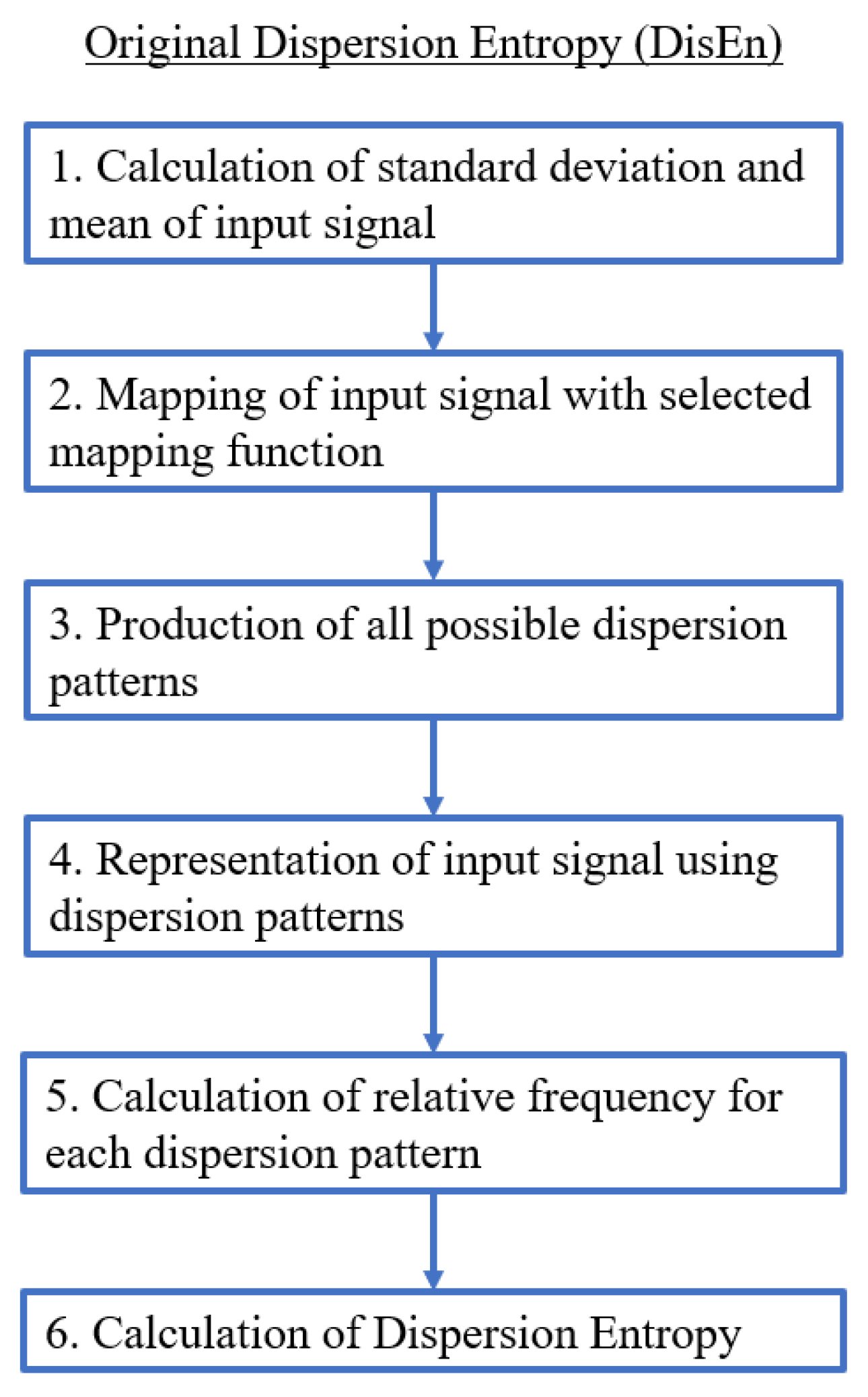
2.1. Dispersion Entropy (DisEn)
- A first and optional step is the mapping of the time-series with a linear or non-linear mapping function. For the formulation of the majority of mapping functions, the mean and standard deviation of the time-series are computed and used.
- A number of classes (c) is then mapped to the resulting signal by being distributed across its amplitude range. Each sample is allocated to the nearest respective class based on its amplitude. As a result, a classified signal is retrieved.
- With the classified signal defined, an embedding dimension (m) and a time delay (d) are set for the creation of multiple time-series, of length m, for each . Each time-series is mapped to its respective dispersion pattern, with the number of potential dispersion patterns being .
- For each dispersion pattern, its relative frequency is obtained and used to calculate the DisEn value of the input time-series based on Shannon’s definition of entropy.
2.2. Dispersion Entropy Variations
2.2.1. Skip Sample Dispersion Entropy (SkipDisEn)
2.2.2. Linearly Interpolated Dispersion Entropy (LinInterDisEn)
2.2.3. Alternative Statistical Metrics Dispersion Entropy (AltMetDisEn)
2.2.4. Dynamic Skip Sample Dispersion Entropy (DynSkipDisEn)
2.3. Experimental Datasets
2.4. Generation of Disrupted Time-Series
- Extraction of ground truth DisEn values. Each original time series containing N points without missing samples is separated in non-overlapping windows of 360 samples each. The choice of window length is made with the aim to test algorithmic performance under the restriction of small sample lengths, which is considered one of the advantages of the original DisEn algorithm [22,33]. The original algorithm DisEn is used to calculate the ground truth DisEn value of each respective window.
- Segmentation of time-series. Copies of the initial time-series are segmented in groups of 1–5 samples, as defined by the grouping factor G. The G factor values used are 1, 2, 3, 4, and 5 samples.
- Marking of missing samples. Based on the percentage factor P, a percentage of segments are uniformly drawn from each time-series and their samples are marked as missing. The P factor values used in this study are 10%, 20%, 30%, 40%, and 50%.
- Production of random variations. Finally, the above process is replicated 10 times for each combination of P and G values producing different random variations for each experimental setup.
- Total number of disrupted time-series. As a result from each initial time series, “disruptedM” versions are produced, and these are used to assess the performance of the DisEn algorithm variations.
2.5. Performance Assessment
- the windows of each time-series,
- the 10 different “disrupted” editions of each time series, and
- the total number of time-series that have been chosen from the respective dataset records.
- Embedding dimension: samples.
- Number of classes: classes.
- Mapping approach: logarithm sigmoid function.
- Time delay: 1 sample.
- Cutoff: 0.7 standard deviation (used only by DynSkipDisEn).
2.6. Statistical Testing
- Kolmogorov-Smirnov Test. Each separate distribution is standardised and compared to a standard normal distribution using a Kolmogorov-Smirnov test.
- Mann-Whitney U Test. Based on the results of the Kolmogorov-Smirnov test, a Mann-Whitney U test is chosen and applied to all distribution pairs produced within the same experimental setup to test whether the distribution of error percentages produced by one DisEn variation is significantly different from the distributions produced from the other variations tested under the same experimental setup.
3. Results
3.1. Summary of Statistical Testing Results
3.2. Experimental Setups for Time-Series with Missing Samples
3.2.1. Performance for RR Time-Series with Missing Samples
- , with a p-value of .
- , with a p-value of .
- , with a p-value of .
- , with a p-value of .
3.2.2. Performance for EEG Time-Series with Missing Samples
3.2.3. Performance for RI Time-Series with Missing Samples
3.3. Experimental Setups for Time-Series with Outlier Samples
3.3.1. Performance on RR Time-Series with Outlier Samples
3.3.2. Performance for EEG Time-Series with Outlier Samples
3.3.3. Performance RI Time-Series with Outlier Samples
3.4. Computation Time
4. Discussion
4.1. Differences in the Effect of Missing versus Outlier Samples
4.2. Effect of Signal’s Spectral Characteristics on the Performance of LinInterDisEn
4.3. Standard Deviations of Performance Measurements
4.4. Effect of Outlier Sample Percentage across Physiological Signals
4.5. Setting the Cutoff Parameter of DynSkipDisEn
4.6. Limitations of Current Study and Future Work
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ApEn | Approximate Entropy |
| SampEn | Sample Entropy |
| PEn | Permutation Entropy |
| FuzzyEn | Fuzzy Entropy |
| EMG | Electromyography |
| DisEn | Dispersion Entropy |
| DisEn | Skip Sample Dispersion Entropy |
| LinInterDisEn | Linearly Interpolated |
| AltMetDisEn | Alternative Statistical Metrics Dispersion Entropy |
| DynSkipDisEn | Dynamic Skip Sample Dispersion Entropy |
| RR | Heart rate interval |
| ECG | Electrocardiogram |
| EEG | Electroencephalogram |
| RI | Respiratory Impedance |
Appendix A















References
- Witt, D.R.; Kellogg, R.A.; Snyder, M.P.; Dunn, J. Windows into human health through wearables data analytics. Curr. Opin. Biomed. Eng. 2019, 9, 28–46. [Google Scholar] [CrossRef]
- Paine, C.W.; Goel, V.V.; Ely, E.; Stave, C.D.; Stemler, S.; Zander, M.; Bonafide, C.P. Systematic Review of Physiologic Monitor Alarm Characteristics and Pragmatic Interventions to Reduce Alarm Frequency: Review of Physiologic Monitor Alarms. J. Hosp. Med. 2016, 11, 136–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azimi, I.; Pahikkala, T.; Rahmani, A.M.; Niela-Vilén, H.; Axelin, A.; Liljeberg, P. Missing data resilient decision-making for healthcare IoT through personalization: A case study on maternal health. Future Gener. Comput. Syst. 2019, 96, 297–308. [Google Scholar] [CrossRef]
- Kumar, R.B.; Goren, N.D.; Stark, D.E.; Wall, D.P.; Longhurst, C.A. Automated integration of continuous glucose monitor data in the electronic health record using consumer technology. J. Am. Med Inform. Assoc. 2016, 23, 532–537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moody, G.B. The PhysioNet/Computing in Cardiology Challenge 2010: Mind the Gap. In Proceedings of the Computing in Cardiology, Belfast, UK, 26–29 September 2010; p. 13. [Google Scholar]
- Shivers, J.P.; Mackowiak, L.; Anhalt, H.; Zisser, H. “Turn it Off!”: Diabetes Device Alarm Fatigue Considerations for the Present and the Future. J. Diabetes Sci. Technol. 2013, 7, 789–794. [Google Scholar] [CrossRef] [Green Version]
- Keller, J.P. Clinical alarm hazards: A “top ten” health technology safety concern. J. Electrocardiol. 2012, 45, 588–591. [Google Scholar] [CrossRef]
- Johnson, K.R.; Hagadorn, J.I.; Sink, D.W. Alarm Safety and Alarm Fatigue. Clin. Perinatol. 2017, 44, 713–728. [Google Scholar] [CrossRef]
- Shannon, C.E. A Mathematical Theory of Communication. Bell Syst. Tech. J. 1948, 27, 623–656. [Google Scholar] [CrossRef]
- Pincus, S.M. Approximate entropy as a measure of system complexity. Proc. Natl. Acad. Sci. USA 1991, 88, 2297–2301. [Google Scholar] [CrossRef] [Green Version]
- Richman, J.S.; Moorman, J.R. Physiological time-series analysis using approximate entropy and sample entropy. Am. J. Physiol.-Heart Circ. Physiol. 2000, 278, H2039–H2049. [Google Scholar] [CrossRef] [Green Version]
- Bandt, C.; Pompe, B. Permutation Entropy: A Natural Complexity Measure for Time Series. Phys. Rev. Lett. 2002, 88, 174102. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Wang, Z.; Xie, H.; Yu, W. Characterization of Surface EMG Signal Based on Fuzzy Entropy. IEEE Trans. Neural Syst. Rehabil. Eng. 2007, 15, 266–272. [Google Scholar] [CrossRef] [PubMed]
- Rostaghi, M.; Azami, H. Dispersion Entropy: A Measure for Time-Series Analysis. IEEE Signal Process. Lett. 2016, 23, 610–614. [Google Scholar] [CrossRef]
- Caldirola, D.; Bellodi, L.; Caumo, A.; Migliarese, G.; Perna, G. Approximate Entropy of Respiratory Patterns in Panic Disorder. Am. J. Psychiatry 2004, 161, 79–87. [Google Scholar] [CrossRef] [Green Version]
- Lake, D.E.; Richman, J.S.; Griffin, M.P.; Moorman, J.R. Sample entropy analysis of neonatal heart rate variability. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2002, 283, R789–R797. [Google Scholar] [CrossRef] [Green Version]
- Olofsen, E.; Sleigh, J.; Dahan, A. Permutation entropy of the electroencephalogram: A measure of anaesthetic drug effect. Br. J. Anaesth. 2008, 101, 810–821. [Google Scholar] [CrossRef] [Green Version]
- Cirugeda-Roldan, E.; Cuesta-Frau, D.; Miro-Martinez, P.; Oltra-Crespo, S. Comparative Study of Entropy Sensitivity to Missing Biosignal Data. Entropy 2014, 16, 5901–5918. [Google Scholar] [CrossRef]
- Dong, X.; Chen, C.; Geng, Q.; Cao, Z.; Chen, X.; Lin, J.; Jin, Y.; Zhang, Z.; Shi, Y.; Zhang, X.D. An Improved Method of Handling Missing Values in the Analysis of Sample Entropy for Continuous Monitoring of Physiological Signals. Entropy 2019, 21, 274. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Gonzalez, M.; Fernandez-Chimeno, M.; Ramos-Castro, J. Errors in the Estimation of Approximate Entropy and Other Recurrence-Plot-Derived Indices Due to the Finite Resolution of RR Time Series. IEEE Trans. Biomed. Eng. 2009, 56, 345–351. [Google Scholar] [CrossRef]
- Molina-Picó, A.; Cuesta-Frau, D.; Aboy, M.; Crespo, C.; Miró-Martínez, P.; Oltra-Crespo, S. Comparative study of approximate entropy and sample entropy robustness to spikes. Artif. Intell. Med. 2011, 53, 97–106. [Google Scholar] [CrossRef]
- Azami, H.; Escudero, J. Amplitude- and Fluctuation-Based Dispersion Entropy. Entropy 2018, 20, 210. [Google Scholar] [CrossRef] [Green Version]
- Rostaghi, M.; Ashory, M.R.; Azami, H. Application of dispersion entropy to status characterization of rotary machines. J. Sound Vib. 2019, 438, 291–308. [Google Scholar] [CrossRef]
- Kim, K.K.; Baek, H.J.; Lim, Y.G.; Park, K.S. Effect of missing RR-interval data on nonlinear heart rate variability analysis. Comput. Methods Programs Biomed. 2012, 106, 210–218. [Google Scholar] [CrossRef] [PubMed]
- Rousseeuw, P.J.; Croux, C. Alternatives to the Median Absolute Deviation. J. Am. Stat. Assoc. 1993, 88, 1273–1283. [Google Scholar] [CrossRef]
- Leys, C.; Ley, C.; Klein, O.; Bernard, P.; Licata, L. Detecting outliers: Do not use standard deviation around the mean, use absolute deviation around the median. J. Exp. Soc. Psychol. 2013, 49, 764–766. [Google Scholar] [CrossRef] [Green Version]
- Pontet, J.; Contreras, P.; Curbelo, A.; Medina, J.; Noveri, S.; Bentancourt, S.; Migliaro, E.R. Heart rate variability as early marker of multiple organ dysfunction syndrome in septic patients. J. Crit. Care 2003, 18, 156–163. [Google Scholar] [CrossRef]
- Augustyniak, P. Wearable wireless heart rate monitor for continuous long-term variability studies. J. Electrocardiol. 2011, 44, 195–200. [Google Scholar] [CrossRef]
- Iyengar, N.; Peng, C.K.; Morin, R.; Goldberger, A.L.; Lipsitz, L.A. Age-related alterations in the fractal scaling of cardiac interbeat interval dynamics. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 1996, 271, R1078–R1084. [Google Scholar] [CrossRef] [Green Version]
- Goldberger, A.L.; Amaral, L.A.N.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.K.; Stanley, H.E. PhysioBank, PhysioToolkit, and PhysioNet: Components of a New Research Resource for Complex Physiologic Signals. Circulation 2000, 101, e215–e220. [Google Scholar] [CrossRef] [Green Version]
- Shoeb, A.H. Application of Machine Learning to Epileptic Seizure Onset Detection and Treatment. Ph.D. Thesis, Harvard University–MIT Division of Health Sciences and Technology, Boston, MA, USA, 2009. [Google Scholar]
- Pimentel, M.A.F.; Johnson, A.E.W.; Charlton, P.H.; Birrenkott, D.; Watkinson, P.J.; Tarassenko, L.; Clifton, D.A. Toward a Robust Estimation of Respiratory Rate From Pulse Oximeters. IEEE Trans. Biomed. Eng. 2017, 64, 1914–1923. [Google Scholar] [CrossRef]
- Kafantaris, E.; Piper, I.; Lo, T.Y.M.; Escudero, J. Application of Dispersion Entropy to Healthy and Pathological Heartbeat ECG Segments. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; IEEE: Berlin, Germany, 2019; pp. 2269–2272. [Google Scholar] [CrossRef]
- Rajendra Acharya, U.; Paul Joseph, K.; Kannathal, N.; Lim, C.M.; Suri, J.S. Heart rate variability: A review. Med. Biol. Eng. Comput. 2006, 44, 1031–1051. [Google Scholar] [CrossRef] [PubMed]
- Kaczka, D.W.; Barnas, G.M.; Suki, B.; Lutchen, K.R. Assessment of time-domain analyses for estimation of low-frequency respiratory mechanical properties and impedance spectra. Ann. Biomed. Eng. 1995, 23, 135–151. [Google Scholar] [CrossRef] [PubMed]
- Diong, B.; Nazeran, H.; Nava, P.; Goldman, M. Modeling Human Respiratory Impedance. IEEE Eng. Med. Biol. Mag. 2007, 26, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Dressler, O.; Schneider, G.; Stockmanns, G.; Kochs, E. Awareness and the EEG power spectrum: Analysis of frequencies. Br. J. Anaesth. 2004, 93, 806–809. [Google Scholar] [CrossRef] [Green Version]










| RR | EEG | RI | |
|---|---|---|---|
| DisEn | 1.6 | 1.5 | 1.9 |
| SkipDisEn | 1.5 | 1.4 | 1.9 |
| AltMetDisEn | 1.7 | 1.8 | 1.9 |
| LinInterDisEn | 1.6 | 1.7 | 2 |
| DynSkipDisEn | 1.5 | 1.4 | 1.5 |
| RR | EEG | RI | |
|---|---|---|---|
| DisEn | 2.1 | 2.3 | 2.3 |
| SkipDisEn | 2.4 | 2.5 | 2.2 |
| AltMetDisEn | 2.6 | 2.8 | 2.7 |
| LinInterDisEn | 3.1 | 3.3 | 3.2 |
| DynSkipDisEn | 2.5 | 2.5 | 2.6 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kafantaris, E.; Piper, I.; Lo, T.-Y.M.; Escudero, J. Augmentation of Dispersion Entropy for Handling Missing and Outlier Samples in Physiological Signal Monitoring. Entropy 2020, 22, 319. https://doi.org/10.3390/e22030319
Kafantaris E, Piper I, Lo T-YM, Escudero J. Augmentation of Dispersion Entropy for Handling Missing and Outlier Samples in Physiological Signal Monitoring. Entropy. 2020; 22(3):319. https://doi.org/10.3390/e22030319
Chicago/Turabian StyleKafantaris, Evangelos, Ian Piper, Tsz-Yan Milly Lo, and Javier Escudero. 2020. "Augmentation of Dispersion Entropy for Handling Missing and Outlier Samples in Physiological Signal Monitoring" Entropy 22, no. 3: 319. https://doi.org/10.3390/e22030319
APA StyleKafantaris, E., Piper, I., Lo, T.-Y. M., & Escudero, J. (2020). Augmentation of Dispersion Entropy for Handling Missing and Outlier Samples in Physiological Signal Monitoring. Entropy, 22(3), 319. https://doi.org/10.3390/e22030319






