Photon Dissipation as the Origin of Information Encoding in RNA and DNA
Abstract
:1. Introduction
2. Foundations
3. Salient Properties of Amino Acid, Nucleic Acids, and Their Complexes
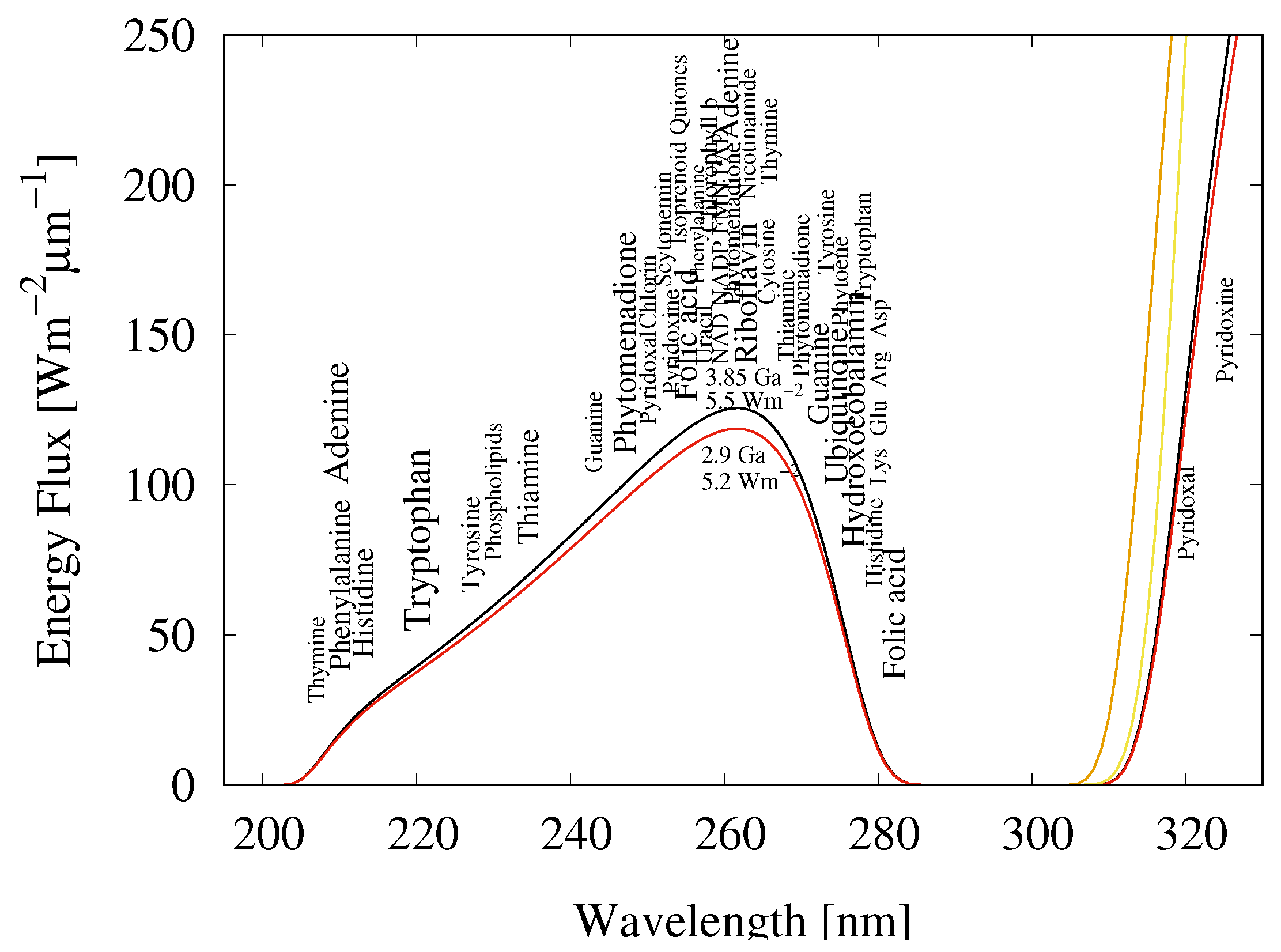
3.1. Absorption and Dissipation
3.2. Amino Acid Affinity to Codons (Or Anti-Codons) of DNA and RNA
4. Ultraviolet and Temperature Assisted Replication (Uvtar)
5. Accumulation of Information through the Dissipation-Replication Mechanism
5.1. Surface Entrapment: Amphipathic Molecules
5.2. Charge Neutralization
5.3. Antenna Molecules
5.4. Intercalation
5.5. Catalysis
6. Amino Acids Which Promote the Dissipation-Replication Relation Have Greatest Affinity to Their Codons/Anticodons
7. Discussion and Conclusions
“I am particularly struck by the difficulty of getting [the genetic code] started unless there is some basis in the specificity of interaction between nucleic acids and amino acids or polypeptides to build upon.”
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| CIT | Clasical Irreversible Thermodynamics |
| DNA | Deoxyribonucleic Acid |
| DRT | Direct-RNA-Template theory for the code’s origin |
| EET | Excitation Energy Transfer |
| Ga | Giga (1000 million) years ago |
| NADH | reduced form of Nicotinamide Adenine Dinucleotide |
| NMR | Nuclear Magnetic Resonance |
| RNA | Ribonucleic Acid |
| RNPs | Ribonucleoprotein Particles |
| UV | Ultraviolet |
| UVB | Ultraviolet B 280–315 nm |
| UVC | Ultraviolet C 100–280 nm |
| UVTAR | enzymeless Ultraviolet and Temperature Assisted Replication mechanism |
References
- Woese, C.R. The Genetic Code: The Molecular Basis for Genetic Expression; Harper & Row: New York, NY, USA, 1967. [Google Scholar]
- Yarus, M. Amino Acids as RNA Ligands: A Direct-RNA-Template Theory for the Codes Origin. J. Mol. Evol. 1998, 47, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Yarus, M.; Caporaso, J.G.; Knight, R. Origins The Genetic Code: The Escaped Triplet Theory. Annu. Rev. Biochem. 2005, 74, 179–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crick, F. The origin of the genetic code. J. Mol. Biol. 1968, 38, 367–379. [Google Scholar] [CrossRef]
- Koonin, E.V. Frozen Accident Pushing 50: Stereochemistry, Expansion, and Chance in the Evolution of the Genetic Code. Life 2017, 7, 22. [Google Scholar] [CrossRef]
- Wong, J.T.F. Co-Evolution Theory of Genetic Code. Proc. Natl. Acad. Sci. USA 1975, 72, 1909–1912. [Google Scholar] [CrossRef] [Green Version]
- Michaelian, K. Thermodynamic origin of life. arXiv 2009, arXiv:0907.0042. [Google Scholar] [CrossRef] [Green Version]
- Michaelian, K. Thermodynamic dissipation theory for the origin of life. Earth Syst. Dynam. 2011, 224, 37–51. [Google Scholar] [CrossRef] [Green Version]
- Michaelian, K. Microscopic Dissipative Structuring and Proliferation at the Origin of Life. Heliyon 2017, 3, e00424. [Google Scholar] [CrossRef]
- Michaelian, K. Photochemical Dissipative Structuring, Proliferation and Evolution at the Origin of Life. arXiv 2020, arXiv:2007.00618. [Google Scholar]
- Onsager, L. Reciprocal Relations in Irreversible Processes, I. Phys. Rev. 1931, 37, 405–426. [Google Scholar] [CrossRef]
- Onsager, L. Reciprocal Relations in Irreversible Processes, II. Phys. Rev. 1931, 38, 2265. [Google Scholar] [CrossRef] [Green Version]
- Prigogine, I.; Nicolis, G. Biological order, structure and instabilities. Q. Rev. Biophys. 1971, 4, 107–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glansdorff, P.; Prigogine, I. Thermodynamic Theory of Structure, Stability and Fluctuations; Wiley-Interscience: Hoboken, NJ, USA, 1971. [Google Scholar]
- Gaspard, P. Engineering of Chemical Complexity; chapter Self-Organization at the Nanoscale Scale in Far-From-Equilibrium Surface Reactions and Copolymerizations; World Scientific: Singapore, 2013; pp. 51–77. [Google Scholar]
- Gaspard, P. Fluctuation theorem for nonequilibrium reactions. J. Chem. Phys. 2004, 120, 8898–8905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrieux, D.; Gaspard, P. Fluctuation Theorem for Currents and Schnakenberg Network Theory. J. Stat. Phys. 2007, 127, 107–131. [Google Scholar] [CrossRef] [Green Version]
- Onsager, L.; Machlup, S. Fluctuations and Irreversible Processes. Phys. Rev. 1953, 91, 1505–1512. [Google Scholar] [CrossRef]
- Prigogine, I. Introduction to Thermodynamics Of Irreversible Processes, 1st ed.; John Wiley & Sons: Hoboken, NJ, USA, 1955. [Google Scholar]
- Ferris, J.P.; Chen, C.T. Chemical evolution. XXVI. Photochemistry of methane, nitrogen, and water mixtures as a model for the atmosphere of the primitive earth. J. Am. Chem. Soc. 1975, 97, 2962–2967. [Google Scholar] [CrossRef]
- Michaelian, K. Thermodynamic Dissipation Theory of the Origina and Evolution of Life: Salient characteristics of RNA and DNA and Other Fundamental Molecules Suggest an Origin of Life Driven by UV-C Light; Self-published; CreateSpace: Mexico City, Mexico, 2016; ISBN 9781541317482. [Google Scholar]
- Kleidon, A.; Lorenz, R.D. Non-Equilibrium Thermodynamics and the Production of Entropy: Life, EARTH, and Beyond; chapter Entropy Production by Earth System Processes; Springer: Berlin/Heidelberg, Germany, 2005; pp. 1–20. ISBN 3-540-22495-5. [Google Scholar]
- Kleidon, A. Maximum entropy production and general trends in biospheric evolution. Paleontol. J. 2009, 43, 980–985. [Google Scholar] [CrossRef]
- Sagan, C. Ultraviolet Selection Pressure on the Earliest Organisms. J. Theor. Biol. 1973, 39, 195–200. [Google Scholar] [CrossRef] [Green Version]
- Kasting, J. Earth’s Early Atmosphere. Science 1993, 259, 920–926. [Google Scholar] [CrossRef]
- Michaelian, K.; Simeonov, A. Fundamental molecules of life are pigments which arose and co-evolved as a response to the thermodynamic imperative of dissipating the prevailing solar spectrum. Biogeosciences 2015, 12, 4913–4937. [Google Scholar] [CrossRef] [Green Version]
- Michaelian, K. A non-linear irreversible thermodynamic perspective on organic pigment proliferation and biological evolution. J. Phys. Conf. Ser. 2013, 475, 012010. [Google Scholar] [CrossRef]
- Prigogine, I. Introduction to Thermodynamics Of Irreversible Processes, 3rd ed.; John Wiley & Sons: Hoboken, NJ, USA, 1967. [Google Scholar]
- Michaelian, K. The biosphere: A thermodynamic imperative. In The Biosphere; IntechOpen: London, UK, 2012. [Google Scholar]
- Middleton, C.T.; de la Harpe, K.; Su, C.; Law, Y.K.; Crespo-Hernández, C.E.; Kohler, B. DNA Excited—State dyanmics: from single bases to the double helix. Annu. Rev. Phys. Chem. 2009, 60, 217–239. [Google Scholar] [CrossRef] [Green Version]
- Ferris, J.P.; Orgel, L.E. An Unusual Photochemical Rearrangement in the Synthesis of Adenine from Hydrogen Cyanide. J. Am. Chem. Soc. 1966, 88, 1074. [Google Scholar] [CrossRef]
- Michaelian, K.; Simeonov, A. Thermodynamic explanation of the cosmic ubiquity of organic pigments. Astrobiol. Outreach 2017, 5, 156. [Google Scholar] [CrossRef]
- Crick, F.H.C. The Genetic Code—Yesterday, Today, and Tomorrow. In Cold Spring Harbor Symposia on Quantitative Biology; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1966; Volume 31, pp. 1–9. [Google Scholar]
- Woese, C.R.; Dugre, D.H.; Saxinger, W.C.; Dugre, S.A. On the fundamental nature and evolution of the genetic code. In Cold Spring Harbor Symposia on Quantitative Biology; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1966; Volume 31, pp. 723–736. [Google Scholar]
- Toulmé, J.; Charlier, M.; Heléme, C. Specific Recognition of Single-Stranded Regions in Ultraviolet Irradiated and Heat-Denatured DNA by Tryptophan-Containing Peptides. Proc. Natl. Acad. Sci. USA 1974, 71, 3185–3188. [Google Scholar] [CrossRef] [Green Version]
- Orgel, L.E. A possible Step in the Origin of the Genetic Code. In Cosmochemical Evolution & the Origins of Life Volume II.; D Reidel Publishing Company: Boston, MA, USA, 1974; p. 313. [Google Scholar]
- Lacey, J.C.J.; Weber, A.L. Protein Structure and Evolution; Marcel-Dekker: New York, NY, USA, 1976; p. 213. [Google Scholar]
- Weber, A.L.; Lacey, J.C. Genetic Code Corelations: Amino Acids and Their Anticodon Nucleotides. J. Mol. Evol. 1978, 11, 199–210. [Google Scholar] [CrossRef]
- Hendry, L.B.; Witham, F.H. Gene regulation: The involvement of stereochemical recognition in DNA-small molecule interactions. Perspect. Biol. Med. 1979, 21, 120–130. [Google Scholar] [CrossRef]
- Hendry, L.B.; Bransome, E.D.; Hutson, M.S.; Campbell, L.K. First approximation of a stereochemical rationale for the genetic code based on the topography and physicochemical properties of “cavities” constructed from models of DNA. Proc. Natl. Acad. Sci. USA 1981, 78, 7440–7444. [Google Scholar] [CrossRef] [Green Version]
- Alberti, S. The Origin of Genetic Code and Protein Synthesis. J. Mol. Evol. 1997, 45, 352–358. [Google Scholar] [CrossRef]
- Knight, R.D.; Landweber, L.F. Rhyme or reason: RNA-arginine interations and the genetic code. Chem. Biol. 1998, 5, R215–R220. [Google Scholar] [CrossRef] [Green Version]
- Majerfeld, I.; Puthenvedu, D.; Yarus, M. RNA Affinity for Molecular L-Histidine; Genetic Code Origins. J. Mol. Evol. 2005, 61, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Serganov, A.; Huang, L.; Patel, D.J. Structural insights into amino acid binding and gene control by a lysine riboswitch. Nature 2008, 455, 1263–1267. [Google Scholar] [CrossRef] [Green Version]
- Yarus, M.; Widmann, J.; Knight, R. RNA-Amino Acid Binding: A Stereochemecal Era for the Genetic Code. J. Mol. Evol. 2009, 69, 406–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, D.; Wang, L. Imprints of the genetic code in the ribosome. Proc. Natl. Acad. Sci. USA 2010, 107, 8298–8303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dougherty, D.A. Cation-p Interactions Involving Aromatic Amino Acids. J. Nutr. 2007, 137, 1504S–1508S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamiichi, K.; Danshita, M.; Minamino, N.; Doi, M.; Ishida, T.; Inoue, M. Indole Ring Binds to 7-methylguanine Base by π-π Stacking Interaction. FEBS Lett. 1986, 195, 57–60. [Google Scholar] [CrossRef] [Green Version]
- AD, G.; Heroux, A.; RP, R.; RT, B. Crystal structure of the lysine riboswitch regulatory mRNA element. J. Biol. Chem. 2008, 283, 22347–22351. [Google Scholar]
- Govi, G.; Kumar, N.Y.; Kumar, M.R.; Hosur, R.V.; Roy, K.B.; Miles, H.T. Recognition Schemes for Protein-Nucleic Acid Interactions. J. Biosci. 1985, 8, 645–656. [Google Scholar] [CrossRef]
- Hendry, L.B.; Bransome, E.D. Are there Structural Analogies Between Amino Acids and Nucleic Acids? Orig. Life 1981, 11, 203–221. [Google Scholar] [CrossRef]
- Copley, S.D.; Smith, E.; Morowitz, H.J. A mechanism for the association of amino acids with their codons and the origin of the genetic code. Proc. Natl. Acad. Sci. USA 2005, 102, 4442–4447. [Google Scholar] [CrossRef] [Green Version]
- Fox, S.; Lacey, J.J.; Nakashima, T. Nucleic Acid-Protein Interactions; Ribbon, D.W., Woessner, J.F., Eds.; North-Holland Pub. Co.: Amsterdam, The Netherlands, 1971; p. 113. [Google Scholar]
- Takagi, Y.; Ikeda, Y.; Taira, K. New Aspects in Phosphorus Chemistry IV; Chapter Ribozyme Mechanisms; Springer: Berlin/Heidelberg, Germany, 2004; pp. 213–266. [Google Scholar]
- Lipfert, J.; Norman, J.O.D.G.; Doniach, S.; Lilley, D.M. The complete VS ribozyme in solution studied by small-angle Xray scattering. Structure 2008, 16, 1357–1367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakano, S.i.; Chadalavada, D.M.; Bevilacqua, P.C. General Acid-Base Catalysis in the Mechanism of a Hepatitis Delta Virus Ribozyme. Science 2000, 287, 1493–1497. [Google Scholar] [CrossRef] [PubMed]
- Lowe, D.R.; Tice, M.M. Geologic evidence for Archean atmospheric and climatic evolution: Fluctuating levels of CO2, CH4, and O2 with an overriding tectonic control. Geology 2004, 32, 493–496. [Google Scholar] [CrossRef] [Green Version]
- Michaelian, K.; Santillán, N. DNA Denaturing through UV-C Photon Dissipation: A Possible Route to Archean Non-enzymatic Replication. bioRxiv 2014, 009126. [Google Scholar] [CrossRef] [Green Version]
- Michaelian, K.; Santillán, N. UVC photon-induced denaturing of DNA: A possible dissipative route to Archean enzyme-less replication. Heliyon 2019, 5, e01902. [Google Scholar] [CrossRef] [Green Version]
- Szostak, J.W. The eightfold path to non-enzymatic RNA replication. J. Syst. Chem. 2012, 3, 2. [Google Scholar] [CrossRef] [Green Version]
- Dubey, R.K.; Tripathi, D.N. A study of thermal denaturataion/renaturation in DNA using laser light scattering: A new approach. Indian J. Biochem. Biophys. 2005, 42, 301–307. [Google Scholar]
- Vekshin, N. Photonics of Biopolymers, 1st ed.; KomKniga: Moscow, Russia, 2005. [Google Scholar]
- Horowitz, E.D.; Engelhart, A.E.; Chen, M.C.; Quarles, K.A.; Smit, h.M.W.; Lynn, D.G.; Hud, N.V. Intercalation as a means to suppress cyclization and promote polymerization of base-pairing oligonucleotides in a prebiotic world. Proc. Natl. Acad. Sci. USA 2010, 107, 5288–5293. [Google Scholar] [CrossRef] [Green Version]
- Eickbush, T.H.; Moudrianakis, E.N. A Mechanism for the Entrapment of DNA at Air-Water Interface. Biophys. J. 1977, 18, 275–288. [Google Scholar] [CrossRef] [Green Version]
- Trifonov, E.N. Evolution of the Code and the Earliest Proteins: Reconstruction from Present-Day Sequences. Biophysics 2002, 47, 539–544. [Google Scholar]
- Prasad, S.; Mandal, I.; Singh, S.; Paul, A.; Mandal, B.; Venkatramani, R.; Swaminathan, R. Near UV-Visible electronic absorption originating from charged amino acids in a monomeric protein. Chem. Sci. 2017, 8, 5416–5433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knauth, L.P. Temperature and salinity history of the Precambrian ocean: Implications for the course of microbial evolution. Paleogeographys. Paleoclimatol. Paleoecol. 2005, 219, 53–69. [Google Scholar] [CrossRef]
- Lovelock, J.E. The Ages of Gaia; A Biography of Our Living Earth; W. W. Norton & Company: New York, NY, USA, 1988. [Google Scholar]
- Bensasson, R.V.; Land, E.J.; Truscott, T.G. Flash Photolysis and Pulse Radiolysis: Contributions to the Chemistry of Biology and Medicine; Pergamon Press: Oxford, UK, 1983. [Google Scholar]
- Cadet, J.; Vigni, P. Bioorganic Photochemistry: Photochemistry and the Nucleic Acids; John Wiley & Sons: Hoboken, NJ, USA, 1990; pp. 1–273. [Google Scholar]
- Montenay-Garestier, T.; Helene, C. Molecular Interactions between Tryptophan and Nucleic Acid Components in Frozen Aqueous Solutions. Nature 1968, 217, 844–845. [Google Scholar] [CrossRef] [PubMed]
- Montenay-Garestier, T.; Helene, C. Reflectance and luminescence studies of molecular complex formation between tryptophan and nucleic acid components in frozen aqueous solutions. Biochemistry 1971, 10, 300–309. [Google Scholar] [PubMed]
- Montenay-Garestier, T.; Toulmi, F.; Fidy, J.; Toulmi, J.J.; Le Doan, T.; Helene, C. Structure, Dynamics, Interactions and Evolution of Biological Macromolecules; Reidel: Kufstein, Austria, 1982; pp. 113–128. [Google Scholar]
- Rajeswari, M.R.; Montenay-Garestier, T.; Helene, C. Does tryptophan intercalate in DNA? A comparative study of peptide binding to alternating and non-alternating A.cntdot.T sequences. Biochemistry 1987, 26, 6825–6831. [Google Scholar] [CrossRef] [PubMed]
- Kleinwächter, V.; Koudelka, J. Thermal denaturation of deoxyribonucleic acid-acridine orange complexes. Biochim. Biophys. Acta-(Bba)-Spec. Sect. Nucleic Acids Relat. Subj. 1964, 91, 539–540. [Google Scholar] [CrossRef]
- Gudgin, E.; Lopez-Delgado, R.; Ware, W.R. The tryptophan fluorescence lifetime puzzle. A study of decay times in aqueous solution as a function of pH and buffer composition. Can. J. Chem. 1971, 59, 1037–1045. [Google Scholar] [CrossRef]
- Mondal, M.; Mukherjee, S.; Bhattacharyya, D. Contribution of phenylalanine side chain intercalation to the TATA-box binding protein–DNA interaction: Molecular dynamics and dispersion-corrected density functional theory studies. J. Mol. Model. 2014, 20, 2499. [Google Scholar] [CrossRef]
- Saidel, L.J.; Goldfarb, A.; Waldman, S. The absorption spectra of amino acids in the region two hundred to two hundred and thirty millimicrons. J. Biol. Chem. 1952, 197, 285–291. [Google Scholar]
- Cataldo, F. A study on ethylene and acetylene photoligomerization and photopolymerization. Photochem. Photobiol. 1996, 99, 75–81. [Google Scholar] [CrossRef]
- Michaelian, K.; Rodriguez, O. Prebiotic fatty acid vesicles through photochemical dissipative structuring. Rev. Cuba QuíMica 2019, 31, 354–370. [Google Scholar]
- Botta, O.; Martins, Z.; Emmenegger, C.; Dworkin, J.P.; Glavin, D.P.; Harvey, R.P.; Zenobi, R.; Bada, J.L.; Ehrenfreund, P. Polycyclic aromatic hydrocarbons and amino acids in meteorites and ice samples from LaPaz Icefield, Antarctica. Meteorit. Planet. Sci. 2008, 43, 1465–1480. [Google Scholar] [CrossRef]
- Allamandola, L.; Tielens, A.; Barker, J. Polycyclic aromatic hydrocarbons and the unidentified infrared emission bands: Auto exhaust along the Milky Wayexclamation. Astrophys. J. 1985, 290, L25–L28. [Google Scholar] [CrossRef]
- Rossmann, M.G.; Moras, D.; Olsen, K.W. Chemical and biological evolution of a nucleotide-biding protein. Nature 1974, 250, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Dayhoff, M.; Schwartz, R.; Orcutt, B. A Model of Evolutionary Change in Proteins. Atlas Protein Seq. Struct. 1978, 5, 345–351. [Google Scholar]


| Amino Acid | Abbreviation | Codon | Codon/Anticodon Specificity | Amphipathic | Antenna 260 nm | Intercalating | Catalysis | Charge Neutralizing |
|---|---|---|---|---|---|---|---|---|
| Aliphatic non-polar R group (Hydrophobic) | ||||||||
| Glycine | Gly | GGU GGC GGA GGG | +/ | |||||
| Alanine | Ala | GCU GCC GCA GGG | ||||||
| Proline | Pro | CCU CCC CCA GGG | ||||||
| Valine | Val | GUU GUC GUA GUG | ||||||
| Leucine | Leu | UUA UUG CUU CUC CUA CUG | +/+ /vw | |||||
| Isoleucine | Ile | AUU | s/ | |||||
| AUC | +/+ | yes | ||||||
| AUA | /s | |||||||
| methionine | Met | AUG | /+ | yes | yes | |||
| Aromatic R group (Slightly hydrophobic) | ||||||||
| Phenylalanine | Phe | UUU UUC | /vw /+ /m | yes | yes | |||
| Tyrosine | Tyr | UAU UAC | vw/s +/+ vw/w | yes | yes | yes | ||
| Tryptophan | Trp | UGG | /s /+ | yes | yes | yes | ||
| Polar R group without charge | ||||||||
| Serine | Ser | UCU UCC UCA UCG AGU AGC | ||||||
| Threonine | Thr | ACU ACC ACA ACG | +/ | |||||
| Cysteine | Cys | UGU UGC | ||||||
| Aspargine | Asn | AAU AAC | ||||||
| Glutamine | Gln | CAA CAG | /+ | |||||
| R group positively charged | ||||||||
| Lysine | Lys | AAA AAG | /s +/ | yes | yes * | yes | yes | |
| Histidine | His | CAU CAC | /vw /+ /s | yes * | yes | yes | yes | |
| Arginine | Arg | CGU CGC CGA CGG AGA AGG | vw/vw +/+ /s w/ s/ | yes * | yes | yes | ||
| R group negatively charged | ||||||||
| Aspartic acid | Asp | GAU GAC | /+ | yes * | yes | |||
| Glutamic acid | Glu | GAA GAG | +/ | yes * | yes | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mejía Morales, J.; Michaelian, K. Photon Dissipation as the Origin of Information Encoding in RNA and DNA. Entropy 2020, 22, 940. https://doi.org/10.3390/e22090940
Mejía Morales J, Michaelian K. Photon Dissipation as the Origin of Information Encoding in RNA and DNA. Entropy. 2020; 22(9):940. https://doi.org/10.3390/e22090940
Chicago/Turabian StyleMejía Morales, Julián, and Karo Michaelian. 2020. "Photon Dissipation as the Origin of Information Encoding in RNA and DNA" Entropy 22, no. 9: 940. https://doi.org/10.3390/e22090940






