Dynamic Effects in Nucleation of Receptor Clusters
Abstract
:1. Introduction
2. Materials and Methods
2.1. Background
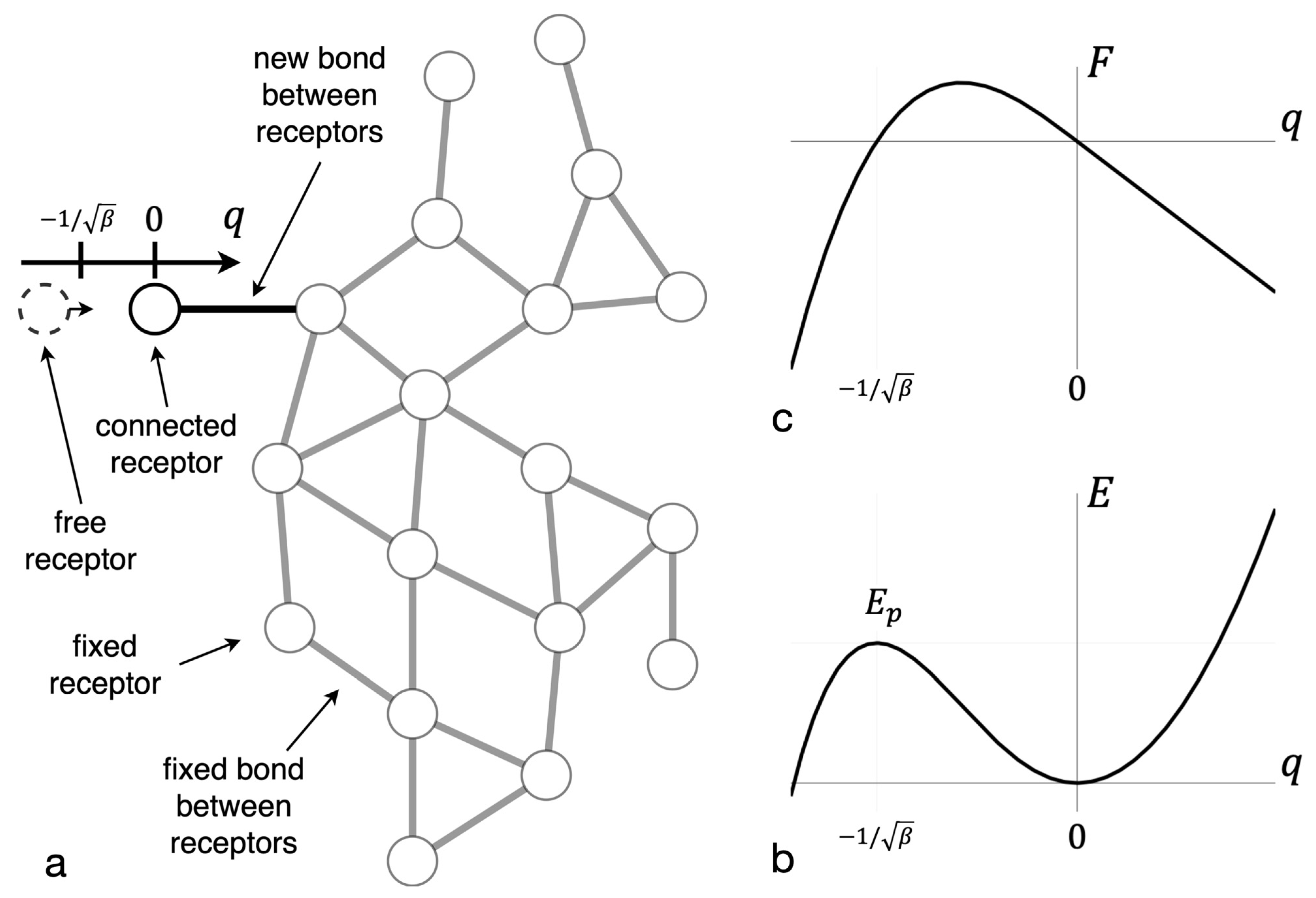
2.2. Mathematical Model
3. Results
3.1. The Potential Barrier of New Bond Formation in a Cluster
3.2. The Kinetic Barrier of Bond Formation
3.3. Von Neumann Entropy Approximation
3.4. Critical Size of Receptor Cluster
3.5. Heterogenous Nucleation Efficiency
3.6. Homologous Series in T Cell Activation by Oligomeric MHC
4. Discussion
- The formation of supercritical clusters by the mechanism of heterogeneous nucleation;
- The growth of these clusters to a productive state that initiates cell’s intrinsic signaling pathways;
- Signal transmission along the signaling pathway to the cell nucleus, where gene expression takes place.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix B
Appendix C
Appendix D
References
- Volmer, M.; Weber, A. Keimbildung in übersättigten Gebilden. Zeitschrift für Physikalische Chemie 1926, 119, 277–301. [Google Scholar] [CrossRef]
- Becker, R.; Döring, W. Kinetische behandlung der keimbildung in übersättigten dämpfen. Ann. Phys. 1935, 416, 719–752. [Google Scholar] [CrossRef]
- Schmelzer, J.W.P. Comments on curvature dependent surface tension and nucleation theory. Nucleation Theory Appl. 1999, 268–289. [Google Scholar]
- Slezov, V.V.; Schmelzer, J. Kinetics of formation of a phase with an arbitrary stoichiometric composition in a multicomponent solid solution. Phys. Rev. E 2002, 65, 31506. [Google Scholar] [CrossRef] [PubMed]
- Schmelzer, J.; Röpke, G.; Priezzhev, V.B. Nucleation Theory and Applications; Wiley-VCH Verlag: Weinheim, Germany, 2005; Volume 76. [Google Scholar] [CrossRef]
- Abyzov, A.S.; Schmelzer, J.W.P.; Fokin, V.M.; Zanotto, E.D. Crystallization of supercooled liquids: Self-consistency correction of the steady-state nucleation rate. Entropy 2020, 22, 558. [Google Scholar] [CrossRef] [PubMed]
- Kelton, K.; Greer, A.L. Nucleation in Condensed Matter: Applications in Materials and Biology; Elsevier: Amsterdam, The Netherlands, 2010. [Google Scholar]
- Frolov, V.A.J.; Chizmadzhev, Y.A.; Cohen, F.S.; Zimmerberg, J. “Entropic traps” in the kinetics of phase separation in multicomponent membranes stabilize nanodomains. Biophys. J. 2006, 91, 189–205. [Google Scholar] [CrossRef] [Green Version]
- Prikhodko, I.V.; Guria, G.T. The role of heterogeneous nucleation in the activation of blood cells. Phys. Solid State 2020, 62, 17–23. [Google Scholar] [CrossRef]
- Prigogine, I. Time, structure, and fluctuations. Science 1978, 201, 777–785. [Google Scholar] [CrossRef] [Green Version]
- Manz, N.; Hamik, C.T.; Steinbock, O. Tracking waves and vortex nucleation in excitable systems with anomalous dispersion. Phys. Rev. Lett. 2004, 92, 248301. [Google Scholar] [CrossRef] [Green Version]
- De Simone, A.; Evanitsky, M.N.; Hayden, L.; Cox, B.D.; Wang, J.; Tornini, V.A.; Ou, J.; Chao, A.; Poss, K.D.; Di Talia, S. Control of osteoblast regeneration by a train of Erk activity waves. Nature 2021, 590, 129–133. [Google Scholar] [CrossRef]
- De Gennes, P.-G. Wetting: Statics and dynamics. Rev. Mod. Phys. 1985, 57, 827. [Google Scholar] [CrossRef]
- Lushnikov, A.A. Gelation in coagulating systems. Phys. D Nonlinear Phenom. 2006, 222, 37–53. [Google Scholar] [CrossRef]
- Alberti, S. Phase separation in biology. Curr. Biol. 2017, 27, R1097–R1102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boeynaems, S.; Alberti, S.; Fawzi, N.L.; Mittag, T.; Polymenidou, M.; Rousseau, F.; Schymkowitz, J.; Shorter, J.; Wolozin, B.; Van Den Bosch, L.; et al. Protein phase separation: A new phase in cell biology. Trends Cell Biol. 2018, 28, 420–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cebecauer, M.; Spitaler, M.; Sergé, A.; Magee, A.I. Signalling complexes and clusters: Functional advantages and methodological hurdles. J. Cell Sci. 2010, 123, 309–320. [Google Scholar] [CrossRef] [Green Version]
- Gaud, G.; Lesourne, R.; Love, P.E. Regulatory mechanisms in T cell receptor signalling. Nat. Rev. Immunol. 2018, 18, 485–497. [Google Scholar] [CrossRef]
- Belardi, B.; Son, S.; Felce, J.H.; Dustin, M.L.; Fletcher, D.A. Cell-cell interfaces as specialized compartments directing cell function. Nat. Rev. Mol. Cell Biol. 2020, 21, 750–764. [Google Scholar] [CrossRef]
- Manz, B.N.; Jackson, B.L.; Petit, R.S.; Dustin, M.L.; Groves, J. T-cell triggering thresholds are modulated by the number of antigen within individual T-cell receptor clusters. Proc. Natl. Acad. Sci. USA 2011, 108, 9089–9094. [Google Scholar] [CrossRef] [Green Version]
- Himanen, J.P.; Yermekbayeva, L.; Janes, P.W.; Walker, J.R.; Xu, K.; Atapattu, L.; Rajashankar, K.R.; Mensinga, A.; Lackmann, M.; Nikolov, D.B.; et al. Architecture of Eph receptor clusters. Proc. Natl. Acad. Sci. USA 2010, 107, 10860–10865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Liu, L.; Zhang, J.; Chen, J.; Ye, J.; Shukla, S.; Qiao, J.; Zhan, X.; Chen, H.; Wu, C.J.; et al. Investigation of antigen-specific T-cell receptor clusters in human cancers. Clin. Cancer Res. 2020, 26, 1359–1371. [Google Scholar] [CrossRef] [PubMed]
- De Nardo, D. Toll-like receptors: Activation, signalling and transcriptional modulation. Cytokine 2015, 74, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Rozbesky, D.; Jones, E.Y. Cell guidance ligands, receptors and complexes-orchestrating signalling in time and space. Curr. Opin. Struct. Biol. 2020, 61, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.; Balagopalan, L.; Nguyen, T.; McIntire, K.M.; Samelson, L.E. TCR microclusters form spatially segregated domains and sequentially assemble in calcium-dependent kinetic steps. Nat. Commun. 2019, 10, 277. [Google Scholar] [CrossRef] [Green Version]
- Pettmann, J.; Santos, A.M.; Dushek, O.; Davis, S.J. Membrane ultrastructure and T cell activation. Front. Immunol. 2018, 9, 2152. [Google Scholar] [CrossRef]
- Lin, J.J.Y.; Low-Nam, S.T.; Alfieri, K.N.; McAffee, D.B.; Fay, N.C.; Groves, J.T. Mapping the stochastic sequence of individual ligand-receptor binding events to cellular activation: T cells act on the rare events. Sci. Signal. 2019, 12, eaat8715. [Google Scholar] [CrossRef]
- Trendel, N.C.; Dushek, O. Mathematical Modelling of T Cell Activation. In Mathematical, Computational and Experimental T Cell Immunology; Springer: Berlin/Heidelberg, Germany, 2021; pp. 223–240. [Google Scholar] [CrossRef]
- Pageon, S.V.; Tabarin, T.; Yamamoto, Y.; Ma, Y.; Bridgeman, J.S.; Cohnen, A.; Benzing, C.; Gao, Y.; Crowther, M.D.; Tungatt, K.; et al. Functional role of T-cell receptor nanoclusters in signal initiation and antigen discrimination. Proc. Natl. Acad. Sci. USA 2016, 113, E5454–E5463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Depoil, D.; Fleire, S.; Treanor, B.L.; Weber, M.; Harwood, N.E.; Marchbank, K.L.; Tybulewicz, V.L.J.; Batista, F.D. CD19 is essential for B cell activation by promoting B cell receptor-antigen microcluster formation in response to membrane-bound ligand. Nat. Immunol. 2008, 9, 63–72. [Google Scholar] [CrossRef]
- Manne, B.K.; Badolia, R.; Dangelmaier, C.A.; Kunapuli, S.P. C-type lectin like receptor 2 (CLEC-2) signals independently of lipid raft microdomains in platelets. Biochem. Pharmacol. 2015, 93, 163–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cebecauer, M.; Amaro, M.; Jurkiewicz, P.; Sarmento, M.J.; Sachl, R.; Cwiklik, L.; Hof, M. Membrane lipid nanodomains. Chem. Rev. 2018, 118, 11259–11297. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Pandzic, E.; Nicovich, P.R.; Yamamoto, Y.; Kwiatek, J.; Pageon, S.V.; Benda, A.; Rossy, J.; Gaus, K. An intermolecular FRET sensor detects the dynamics of T cell receptor clustering. Nat. Commun. 2017, 8, 15100. [Google Scholar] [CrossRef] [Green Version]
- Mbiribindi, B.; Mukherjee, S.; Wellington, D.; Das, J.; Khakoo, S.I. Spatial clustering of receptors and signaling molecules regulates NK cell response to peptide repertoire changes. Front. Immunol. 2019, 10, 605. [Google Scholar] [CrossRef] [Green Version]
- Hancock, J.T. Cell Signalling; Oxford University Press: Oxford, UK, 2017. [Google Scholar]
- Santra, T.; Rukhlenko, O.; Zhernovkov, V.; Kholodenko, B.N. Reconstructing static and dynamic models of signaling pathways using Modular Response Analysis. Curr. Opin. Syst. Biol. 2018, 9, 11–21. [Google Scholar] [CrossRef]
- Sapega, T.S.; Guria, G.T. Stability analysis of the PI3K-Akt-mTOR signaling pathway. Biophysics 2020, 65, 259–267. [Google Scholar] [CrossRef]
- Vlasov, A.A. The Theory of Many Particles; GITTL: Moscow, Russia, 1950. (In Russian) [Google Scholar]
- Sinai, Y.G. Theory of Phase Transitions: Rigorous Results; Elsevier: Amsterdam, The Netherlands, 2014. [Google Scholar]
- Van Kampen, N.G. Stochastic Processes in Physics and Chemistry; Elsevier: Amsterdam, The Netherlands, 1992; Volume 1. [Google Scholar]
- Fermi, E.; Pasta, J.; Ulam, S. Study of Nonlinear Problems; Technical Report No. LA-1940; Los Alamos Scientific Laboratory of the University of California: Los Alamos, NM, USA, 1955; p. 21. [Google Scholar]
- Chirikov, B.V. A universal instability of many-dimensional oscillator systems. Phys. Rep. 1979, 52, 263–379. [Google Scholar] [CrossRef]
- Dhar, A. Heat transport in low-dimensional systems. Adv. Phys. 2008, 57, 457–537. [Google Scholar] [CrossRef] [Green Version]
- Izrailev, F.M.; Chirikov, B.V. Statistical properties of a nonlinear string. Sov. Phys. Doklady 1966, 11, 30. (In Russian) [Google Scholar]
- Kuznetsov, A.P.; Kuznetsov, S.P.; Shchegoleva, N.A.; Stankevich, N.V. Dynamics of coupled generators of quasiperiodic oscillations: Different types of synchronization and other phenomena. Phys. D Nonlinear Phenom. 2019, 398, 1–12. [Google Scholar] [CrossRef]
- Eyring, H. The activated complex in chemical reactions. J. Chem. Phys. 1935, 3, 107–115. [Google Scholar] [CrossRef]
- Shlapintokh, V.; Zamaraev, K.I.; Purmal, A.P. Chemical Thermodynamics; MIPT: Moscow, Russia, 1975. (In Russian) [Google Scholar]
- Steinfeld, J.I.; Francisco, J.S.; Hase, W.L. Chemical Kinetics and Dynamics; Prentice Hall: Upper Saddle River, NJ, USA, 1999. [Google Scholar]
- Bogoliubov, N. Problems of Dynamic Theory in Statistical Physics; OGIZ: Moscow, Russia, 1946. (In Russian) [Google Scholar]
- Kuznetsov, S.P. Dynamical Chaos; FizMatLit: Moscow, Russia, 2001. (In Russian) [Google Scholar]
- Dmitriev, A.; Efremova, E.; Maximov, N.; Panas, A. Chaos Generation; LitRes: Moscow, Russia, 2021. (In Russian) [Google Scholar]
- Giannakis, D.; Majda, A.J. Nonlinear Laplacian spectral analysis for time series with intermittency and low-frequency variability. Proc. Natl. Acad. Sci. USA 2012, 109, 2222–2227. [Google Scholar] [CrossRef] [Green Version]
- Raigorodskii, A.M. Linear Algebraic Method in Combinatorics; Moscow Center for Continuous Mathematical Education: Moscow, Russia, 2007. [Google Scholar]
- Barabási, A.-L. Linked: The New Science of Networks; Perseus Book Group: New York, NY, USA, 2003. [Google Scholar] [CrossRef]
- Anand, K.; Bianconi, G.; Severini, S. Shannon and von Neumann entropy of random networks with heterogeneous expected degree. Phys. Rev. E 2011, 83, 36109. [Google Scholar] [CrossRef] [Green Version]
- Mowshowitz, A.; Dehmer, M. Entropy and the complexity of graphs revisited. Entropy 2012, 14, 559–570. [Google Scholar] [CrossRef]
- Chirkov, B.; Zaslavsky, G. On the mechanism of fermi acceleration in the one-dimensional case. Sov. Phys. Doklady 1964, 159, 98–110. (In Russian) [Google Scholar]
- Moelwyn-Hughes, E.A. A Short Course of Physical Chemistry; American Elsevier Publishing Company: New York, NY, USA, 1967. [Google Scholar]
- Nikitin, E. Theory of Slow Atomic Collisions; Springer Science & Business Media: Berlin, Germany, 2012. [Google Scholar]
- Iwasaki, A.; Medzhitov, R. Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 2004, 5, 987–995. [Google Scholar] [CrossRef]
- Shomuradova, A.S.; Vagida, M.S.; Sheetikov, S.A.; Zornikova, K.V.; Kiryukhin, D.; Titov, A.; Peshkova, I.O.; Khmelevskaya, A.; Dianov, D.V.; Malasheva, M.; et al. SARS-CoV-2 epitopes are recognized by a public and diverse repertoire of human T cell receptors. Immunity 2020, 53, 1245–1257. [Google Scholar] [CrossRef] [PubMed]
- Gantmakher, F.R. Lectures in Analytical Mechanics; Mir Publishers: Moscow, Russia, 1970. [Google Scholar]
- Lurie, A.I. Analytical Mechanics; Springer Science & Business Media: Berlin, Germany, 2013. [Google Scholar]
- Minello, G.; Rossi, L.; Torsello, A. On the von Neumann entropy of graphs. J. Complex Netw. 2019, 7, 491–514. [Google Scholar] [CrossRef]
- Cochran, J.R.; Cameron, T.O.; Stern, L.J. The relationship of MHC-peptide binding and T cell activation probed using chemically defined MHC class II oligomers. Immunity 2000, 12, 241–250. [Google Scholar] [CrossRef] [Green Version]
- Karthika, S.; Radhakrishnan, T.K.; Kalaichelvi, P. A review of classical and nonclassical nucleation theories. Cryst. Growth Des. 2016, 16, 6663–6681. [Google Scholar] [CrossRef]
- Butenin, N.V.; Neimark, Y.I.; Fufaev, N.A. Introduction to the Theory of Nonlinear Oscillations; Nauka: Moscow, Russia, 1987. (In Russian) [Google Scholar]
- Neishtadt, A.I. The separation of motions in systems with rapidly rotating phase. J. Appl. Math. Mech. 1984, 48, 133–139. [Google Scholar] [CrossRef]
- Neimark, J.I. Stochastic and Chaotic Oscillations; Springer Science & Business Media: Berlin, Germany, 1992. [Google Scholar]
- Waddington, C.H. The epigenotype. Endeavour 1942, 1, 18–20. [Google Scholar] [CrossRef] [Green Version]
- Thom, R. Stabilité structurelle et morphogenèse. Poetics 1974, 3, 7–19. [Google Scholar] [CrossRef]
- Franceschelli, S. Morphogenesis, Structural Stability and Epigenetic Landscape. In Morphogenesis; Springer: Berlin/Heidelberg, Germany, 2011; pp. 283–293. [Google Scholar] [CrossRef]
- Kolmogorov, A.N. The local structure of turbulence in incompressible viscous fluid for very large Reynolds numbers. Cr Acad. Sci. URSS 1941, 30, 301–305. [Google Scholar]
- Frisch, U. Turbulence: The Legacy of AN Kolmogorov; Cambridge university press: Cambridge, UK, 1995. [Google Scholar]
- Ruelle, D.; Takens, F. On the nature of turbulence. Les Rencontres Physiciens-Mathématiciens Strasbourg-RCP25 1971, 12, 1–44. [Google Scholar]
- Campi, G.; Varma, R.; Dustin, M.L. Actin and agonist MHC-peptide complex-dependent T cell receptor microclusters as scaffolds for signaling. J. Exp. Med. 2005, 202, 1031–1036. [Google Scholar] [CrossRef]
- Katrukha, E.A.; Mikhaylova, M.; van Brakel, H.X.; Van Bergen en Henegouwen, P.M.; Akhmanova, A.; Hoogenraad, C.C.; Kapitein, L.C. Probing cytoskeletal modulation of passive and active intracellular dynamics using nanobody-functionalized quantum dots. Nat. Commun. 2017, 8, 14772. [Google Scholar] [CrossRef] [Green Version]
- Vavilov, N. The law of homologous series in variation. J. Genet. 1922, 12, 47–89. [Google Scholar] [CrossRef]
- Zlobina, K.E.; Guria, G.T. Platelet activation risk index as a prognostic thrombosis indicator. Sci. Rep. 2016, 6, 30508. [Google Scholar] [CrossRef] [Green Version]
- Pushin, D.M.; Salikhova, T.Y.; Zlobina, K.E.; Guria, G.T. Platelet activation via dynamic conformational changes of von Willebrand factor under shear. PLoS ONE 2020, 15, e0234501. [Google Scholar] [CrossRef] [PubMed]
- Stockschlaeder, M.; Schneppenheim, R.; Budde, U. Update on von Willebrand factor multimers: Focus on high-molecular-weight multimers and their role in hemostasis. Blood Coagul. Fibrinolysis 2014, 25, 206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vangenechten, I.; Gadisseur, A. Improving diagnosis of von Willebrand disease: Reference ranges for von Willebrand factor multimer distribution. Res. Pract. Thromb. Haemost. 2020, 4, 1024–1034. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prikhodko, I.V.; Guria, G.T. Dynamic Effects in Nucleation of Receptor Clusters. Entropy 2021, 23, 1245. https://doi.org/10.3390/e23101245
Prikhodko IV, Guria GT. Dynamic Effects in Nucleation of Receptor Clusters. Entropy. 2021; 23(10):1245. https://doi.org/10.3390/e23101245
Chicago/Turabian StylePrikhodko, Ivan V., and Georgy Th. Guria. 2021. "Dynamic Effects in Nucleation of Receptor Clusters" Entropy 23, no. 10: 1245. https://doi.org/10.3390/e23101245
APA StylePrikhodko, I. V., & Guria, G. T. (2021). Dynamic Effects in Nucleation of Receptor Clusters. Entropy, 23(10), 1245. https://doi.org/10.3390/e23101245





