Gene Network Analysis of Alzheimer’s Disease Based on Network and Statistical Methods
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Preparation
Data Sources
2.2. Methods
2.2.1. Prediction of Essential Genes based on Modular Network
Network Module Partition Method
Entropy
Prediction of Functional Gene Modules
Screening of Essential Genes
2.2.2. Integrated Algorithm for Predicting Essential Genes
Acquisition of Priori Probability of Genes
Binary Label Assignment
Obtain Feature Vectors according to the Integrated Network and Binary Labels
Estimate Parameters and Calculate the Posterior Probability
Get Decision Score
3. Results and Discussion
3.1. Network Construction
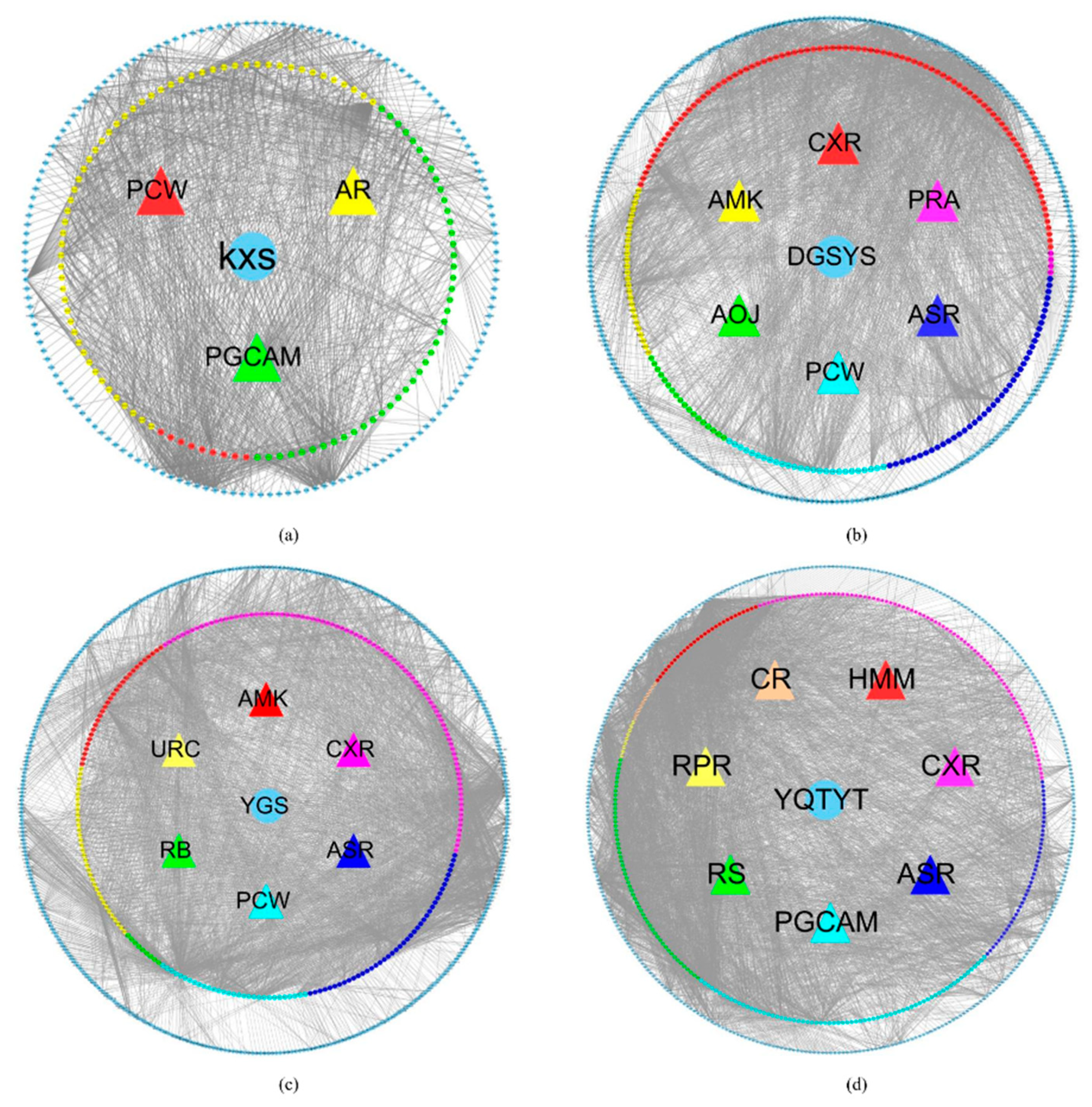
3.1.1. Herb-Active Compound-Target Network
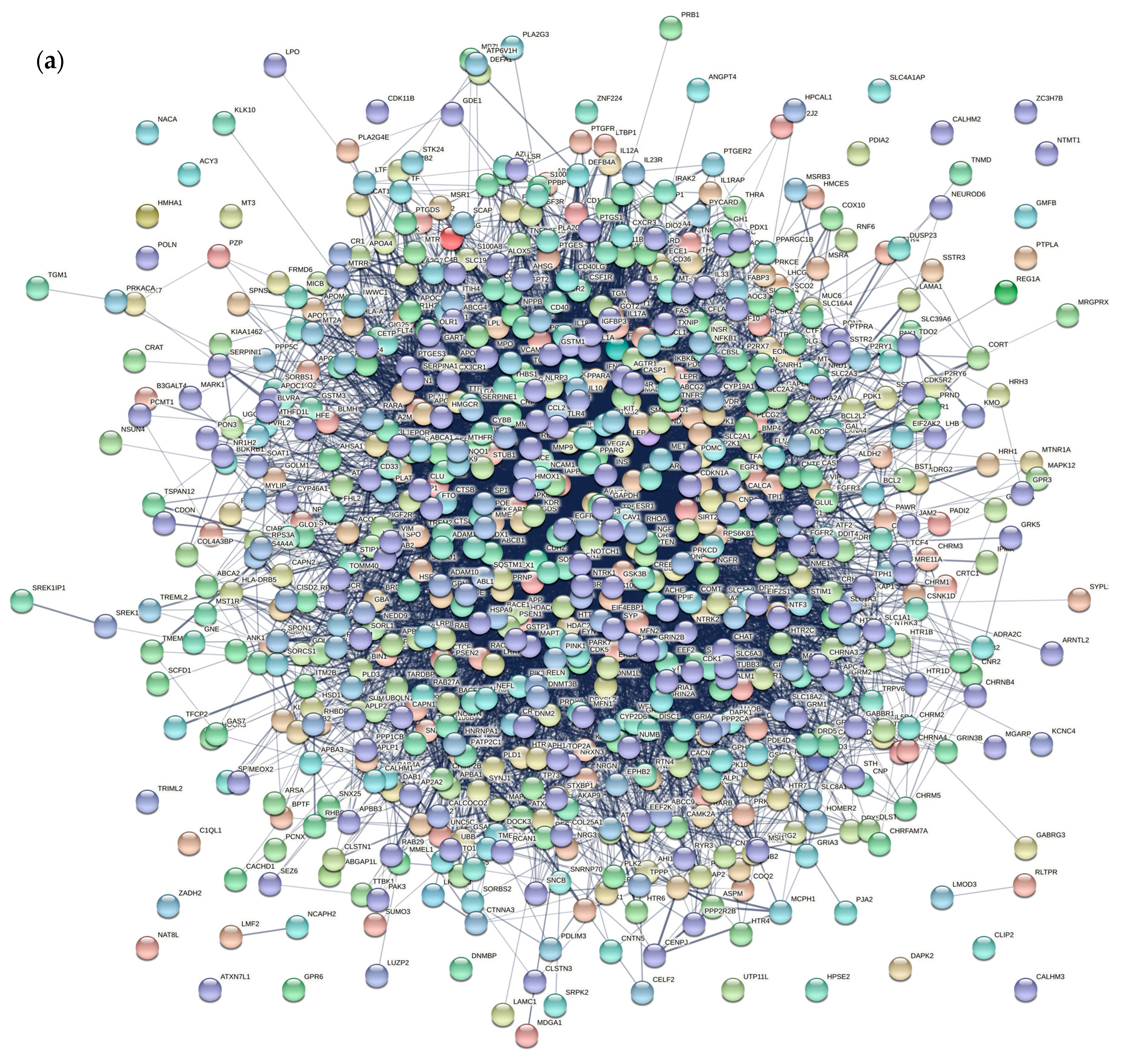
3.1.2. AD Gene Network Construction
3.2. Prediction of Essential Genes based on Modular Network
3.2.1. Module Partition
3.2.2. Calculation of Association Indices
3.2.3. Prediction of Essential Genes
3.3. Integrated Algorithm for Predicting Essential Genes
3.4. Screening of Essential Genes of AD
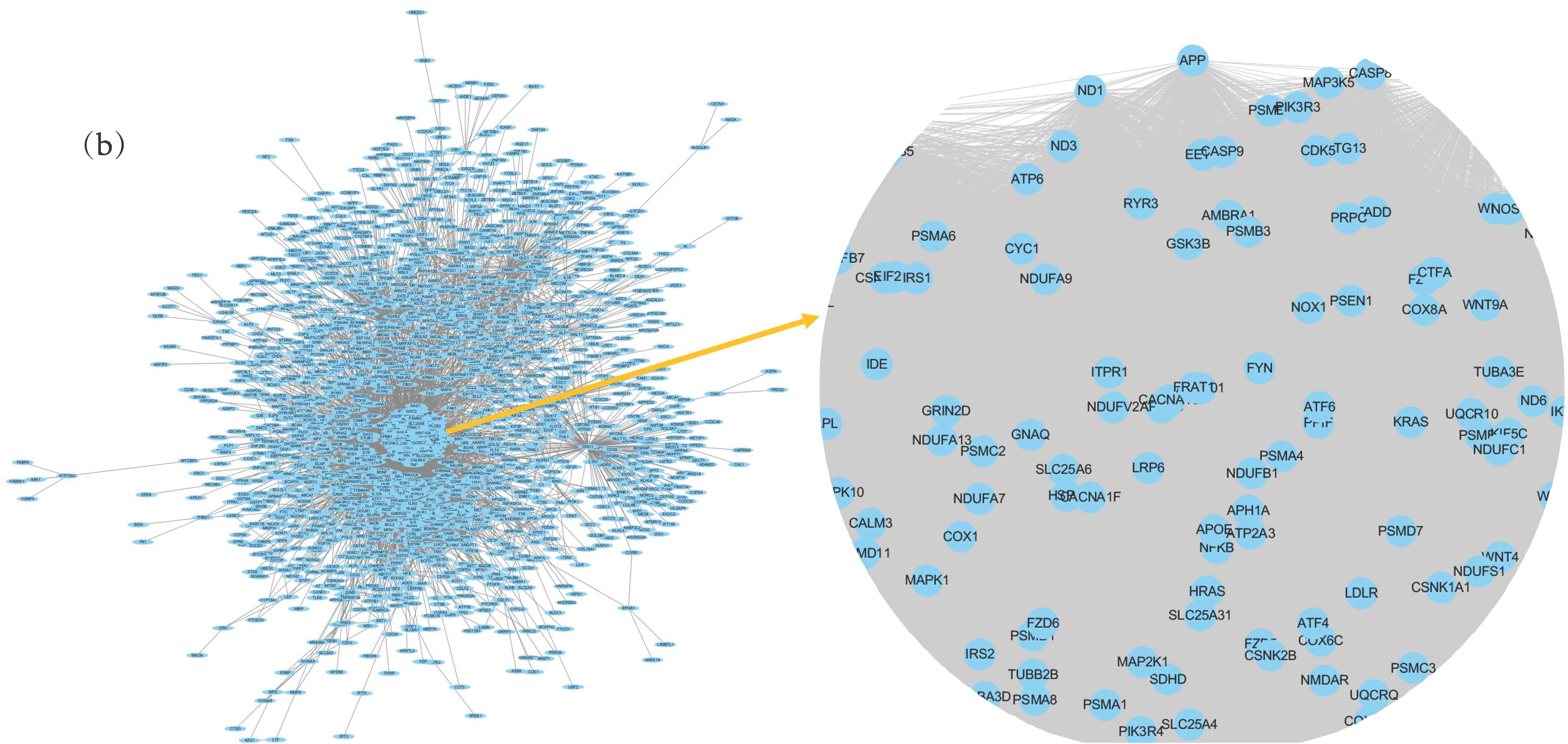
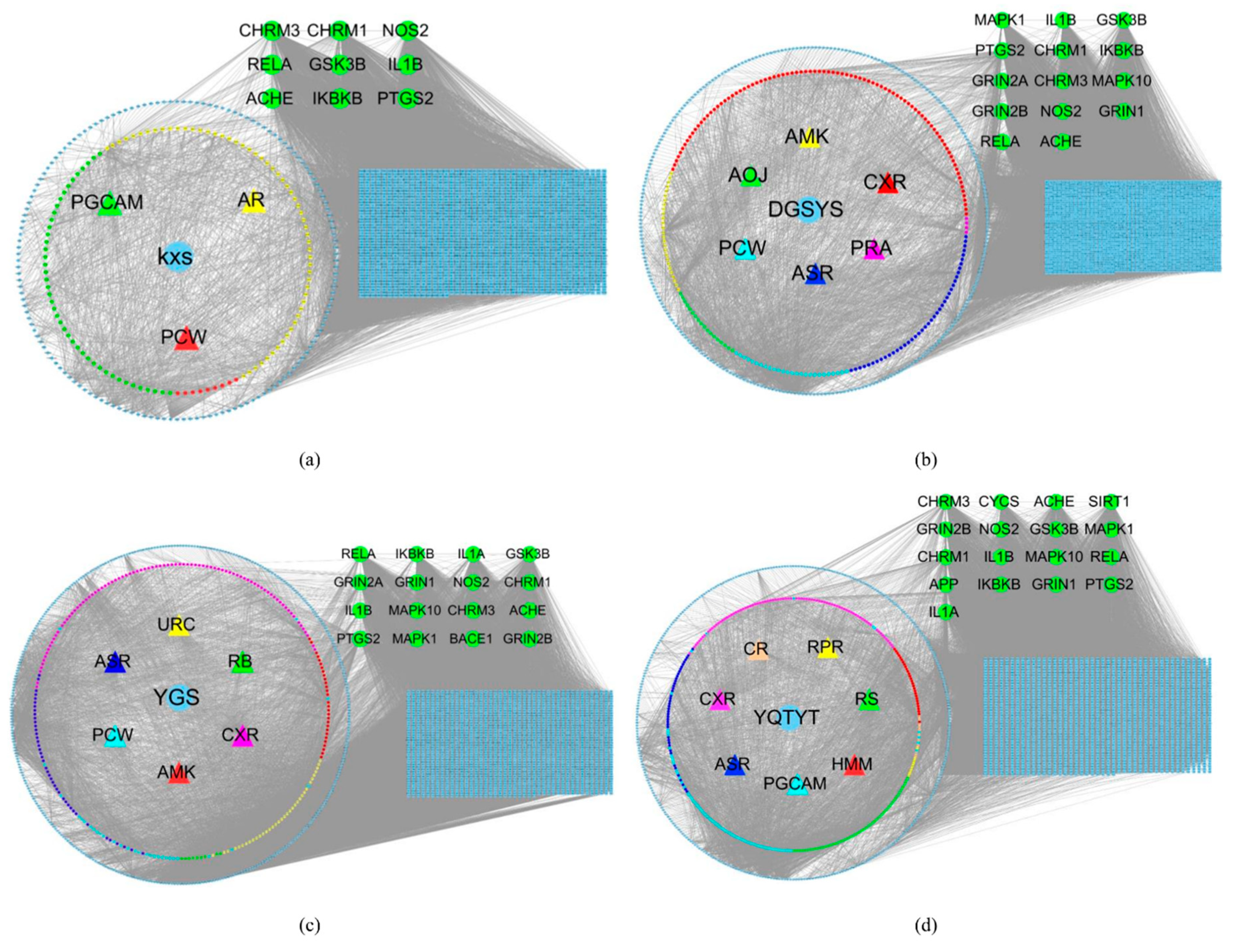
3.5. Herb- Active Compounds-Target Genes-Essential Genes Network
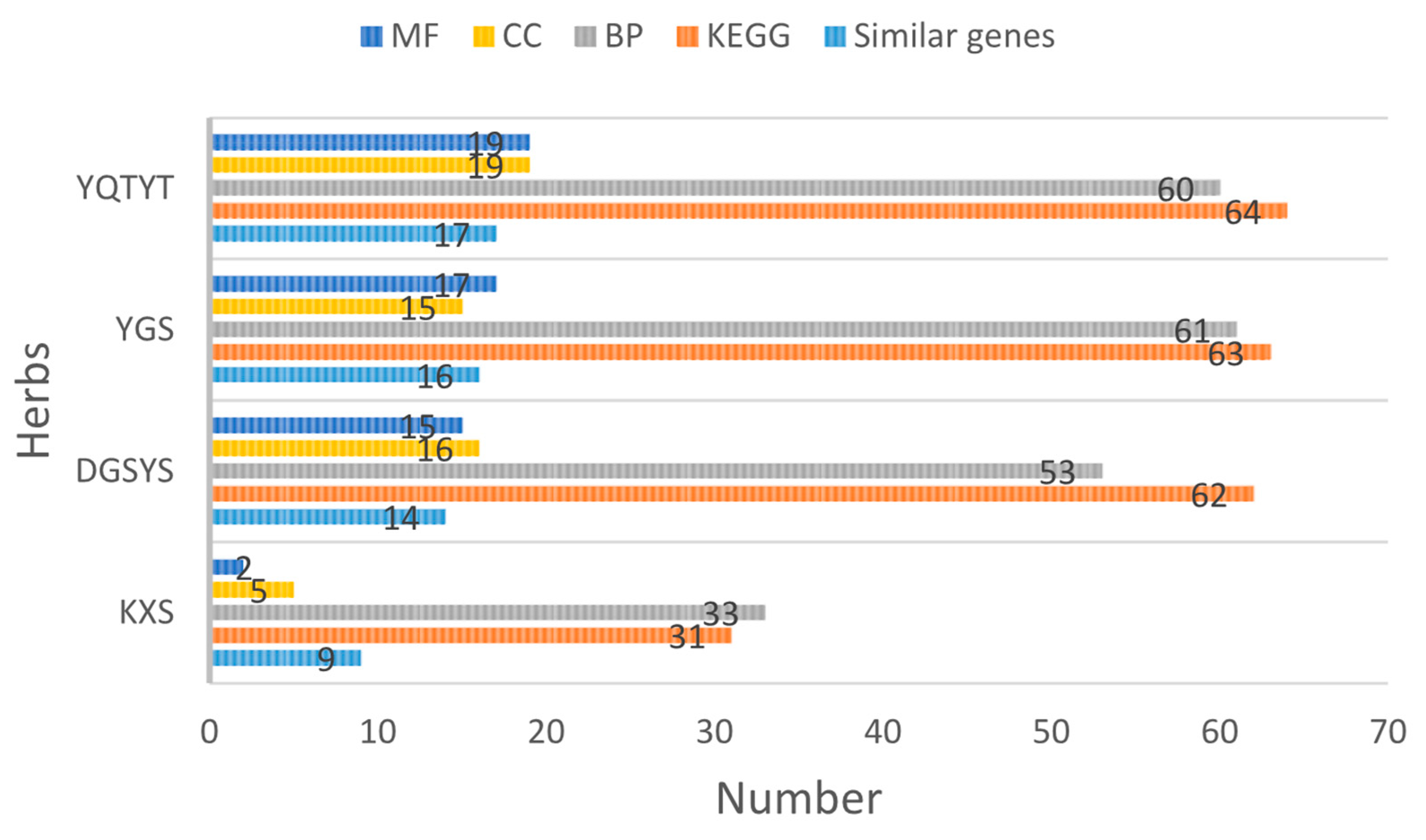
3.6. Enrichment Analysis of Herb Compound Target
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s disease |
| MCL | Markov Clustering |
| MCODE | Molecular Complex Detection |
| Glay | Community Clustering |
| PPI | protein-protein interaction |
| PCC | Pearson Correlation Coefficient |
| TCMSP | Traditional Chinese Medicine Systems Pharmacology |
| NCBI | National Center for Biotechnology Information |
| OMIM | Online Mendelian Inheritance in Man |
| TTD | Therapeutic Target Database |
| IntAct | IntAct Molecular Interaction Database |
| GEO | Gene Expression Omnibus |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| CORUM | Comprehensive Resource of Mammalian protein complexes |
| KXS | Kaixinsan (herb) |
| DGSYS | Dangguishaoyaosan (herb) |
| YGS | Yigansan (herb) |
| YQTYT | Yiqitongyutang (herb) |
| PGCAM | Panax Ginseng C. A. Mey. (compound of KXS and YQTYT) |
| AR | Acoritataninowii Rhizoma (compound of KXS) |
| PCW | Poria Cocos(Schw.) Wolf. (compound of KXS, DGSYS and YGS) |
| ASR | Angelicae Sinensis Radix (compound of DGSYS, YGS and YQTYT) |
| PRA | Paeoniae Radix Alba (compound of DGSYS) |
| CXR | Chuanxiong Rhizoma (compound of DGSYS, YGS and YQTYT) |
| AMK | Atractylodes Macrocephala Koidz. (compound of DGSYS and YGS) |
| AOJ | Alisma Orientale (Sam.) Juz. (compound of DGSYS) |
| RB | Radix Bupleuri (compound of YGS) |
| URC | Uncariae Ramulus Cumuncis (compound of YGS) |
| RS | Radix Salviae (compound of YQTYT) |
| CR | Codonopsis Radix (compound of YQTYT) |
| RPR | Radix Paeoniae Rubra (compound of YQTYT) |
| HMM | Hedysarum Multijugum Maxim. (compound of YQTYT) |
| ROC | Receiver Operating Characteristic |
| AUC | Area Under Curve |
| TPR | True Positive Rate |
| FPR | False Positive Rate |
References
- Yao, X.; Li, X.; Zhou, J.; Wang, Q.; Liu, G.Z.; Zhou, Y.Y. Experimental Research Progress on Traditional Chinese Medicine in Treatment of Alzheimer’s Disease by Regulating and Controlling Calcium Ions in SteadyState. Chin. Arch. Tradit. Chin. Med. 2018, 36, 49–52. [Google Scholar] [CrossRef]
- Hopkins, A.L. Network pharmacology. Nat. Biotechnol. 2007, 25, 1110–1111. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, A.L. Network pharmacology: The next paradigm in drug discovery. Nat. Chem. Biol. 2008, 4, 682–690. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, G.; Palmieri, F. Network traffic classification using deep convolutional recurrent autoencoder neural networks for spatial—Temporal features extraction. J. Netw. Comput. Appl. 2021, 173, 102890. [Google Scholar] [CrossRef]
- D’Angelo, G.; Palmieri, F. Knowledge elicitation based on genetic programming for non destructive testing of critical aerospace systems. Future Gener. Comput. Syst. 2020, 102, 633–642. [Google Scholar] [CrossRef]
- Zhang, W.; Sun, F.; Jiang, R. Integrating multiple protein-protein interaction networks to prioritize disease genes: A Bayesian regression approach. BMC Bioinform. 2011, 12, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, B.L.; Wang, J.X.; Li, M.; Wu, F.X. Identifying disease genes by integrating multiple data sources. BMC Med. Genom. 2014, 7 (Suppl. 2), S2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, B.L.; Li, M.; Wang, J.X.; Wu, F.X. Disease gene identification by using graph kernels and Markov random fields. Sci China Life Sci. 2014, 57, 1054–1063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, B.L.; Li, M.; Wang, J.X.; Shang, X.Q.; Wu, F.X. A Fast and high performance multiple data integration algorithm for identifying human disease genes. BMC Med. Genom. 2015, 8, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, P.G.; Gao, L.; Han, S.S. Identification of overlapping and non-overlapping community structure by fuzzy clustering in complex networks. Inf. Sci. 2010, 181, 1060–1071. [Google Scholar] [CrossRef]
- Bader, G.D.; Hogue, C.W. An automated method for finding molecular complexes in large protein interaction networks. BioMed Cent. 2003, 4, 1–27. [Google Scholar] [CrossRef] [Green Version]
- Lancichinetti, A.; Fortunato, S.; Kertesz, J. Detecting the overlapping and hierarchical community structure in complex networks. New J. Phys. 2009, 11, 33015. [Google Scholar] [CrossRef]
- Xia, J.; Zhang, R.C.; Cheng, S.J. Discussion on Treatment of Senile Dementia with Traditional Chinese Medicine. J. Sichuan Tradit. Chin. Med. 2008, 36, 40–42. [Google Scholar]
- Qiu, X.F.; Yuan, D.P.; Wang, P.; Zhang, L.T.; Hu, Y.G. The Basic Pathogenesis of Alzheimer’s Disease (AD) Being Deficiency of Kidney and Debility of Marrow Blockage of Brain Collateral. J. Henan Univ. Chin. Med. 2006, 21, 11–13. [Google Scholar] [CrossRef]
- Yildirim, M.A.; Goh, K.I.; Cusick, M.E.; Barabasi, A.L.; Vidal, M. Drug-target network. Nat. Biotechnol. 2007, 25, 1119–1126. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.H. New paradigm for drug discovery based on network pharmacology. Chin. J. New Drugs Clin. Rem. 2009, 28, 721–726. [Google Scholar]
- Liu, C.; Zhang, A.; Wang, X.J. Recent research on Kaixin San. Acta Chin. Med. Pharmacol. 2014, 42, 164–165. [Google Scholar] [CrossRef]
- Xie, J. Formulating Rules of Senile Dementiabased on Statistical Analysis. Ph.D. Thesis, Nanjing University of Traditional Chinese Medicine, Nanjing, China, 2009. [Google Scholar]
- Ru, J.L.; Li, P.; Wang, J.A.; Zhou, W.; Li, C.; Huang, P.; Li, P.D.; Guo, Z.H.; TAO, W.H.; Yang, Y.F.; et al. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform. 2014, 6, 13. [Google Scholar] [CrossRef] [Green Version]
- Federhen, S. The NCBI Taxonomy database. Nucleic Acids Res. 2012, 40, D136–D143. [Google Scholar] [CrossRef] [Green Version]
- McKusick, V.A. Mendelian Inheritance in Man and its online version, OMIM. Am. J. Hum. Genet. 2007, 80, 588–604. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Ji, Z.L.; Chen, Y.Z. TTD: Therapeutic Target Database. Nucleic Acids Res. 2002, 30, 412–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kerrien, S.; Alam-Faruque, Y.; Aranda, B.; Bancarz, I.; Bridge, A.; Derow, C.; Dimmer, E.; Feuermann, M.; Friedrichsen, A.; Huntley, R.; et al. IntAct—Open source resource for molecular interaction data. Nucleic Acids Res. 2007, 35, D561–D565. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991–D9955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef] [Green Version]
- Ruepp, A.; Waegele, B.; Lechner, M.; Brauner, B.; Dunger-Kaltenbach, I.; Fobo, G.; Frishman, G.; Montrone, C.; Mewes, H. CORUM: The comprehensive resource of mammalian protein complexes—2009. Nucleic Acids Res. 2010, 38, D497–D501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.Q.; Lei, H.; Shi, Y.T. Measurement Analysis and Application in Network Science; Chemical Industry Press: Beijing, China, 2019; ISBN 978-7-122-33221-9. [Google Scholar]
- Chen, Y.Y. A Multidimensional Comparison of Pharmacological Mechanisms of Different Compound Treatments on Cerebral Ischemia Models. Ph.D. Thesis, China Academy of Chinese Medical Sciences, Beijing, China, 2013. [Google Scholar]
- Gu, H.; Chen, Y.Y.; Wang, P.Q.; Wang, Z. Comparison of different methods of module division by entropyand functional similarity of gene network and its modules forcoronary heart disease. Chin. J. Pharm. Toxicol. 2018, 32, 377–384. [Google Scholar]
- Liu, Q.; Gu, H.; Liu, J.; Chen, Y.Y.; Li, B.; Wang, Z. Module Partition and Biological Mechanism Analysis of Genetic Network of Urinary Tract Infection Based on Entropy. Genom. Appl. Biol. 2018, 37, 4676–4681. [Google Scholar]
- Bass, J.I.F.; Diallo, A.; Nelson, J.; Soto, J.M.; Myers, C.L.; Walhout, A.J.M. Using networks to measure similarity between genes: Association index selection. Nat. Methods Tech. Life Sci. Chem. 2013, 10, 1169–1176. [Google Scholar] [CrossRef] [Green Version]
- Zhu, W.H.; Qiao, Z.H.; Chen, Y.J.; Zeng, P.G.; Cao, S.J.; Zhou, C.; Peng, S.Y.; Zou, Y.M. Module partition and analysis of gene network of Alzheimer’s disease based on graph entropy. Pure and Applied Mathematics. Be published in 2023.
- Lin, C.Y.; Chin, C.H.; Wu, H.H.; Chen, S.H.; Ho, C.W.; Ko, M.T. Hubba: Hub objects analyzer—A framework of interactome hubs identification for network biology. Nucleic Acids Res. 2008, 36, W438–W443. [Google Scholar] [CrossRef]
- Cao, S.J.; Yu, L.; Mao, j.Y.; Wang, Q.; Ruan, J.S. Uncovering the Molecular Mechanism of Actions between Pharmaceuticals and Proteins on the AD Network. PLoS ONE 2017, 10, e0144387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
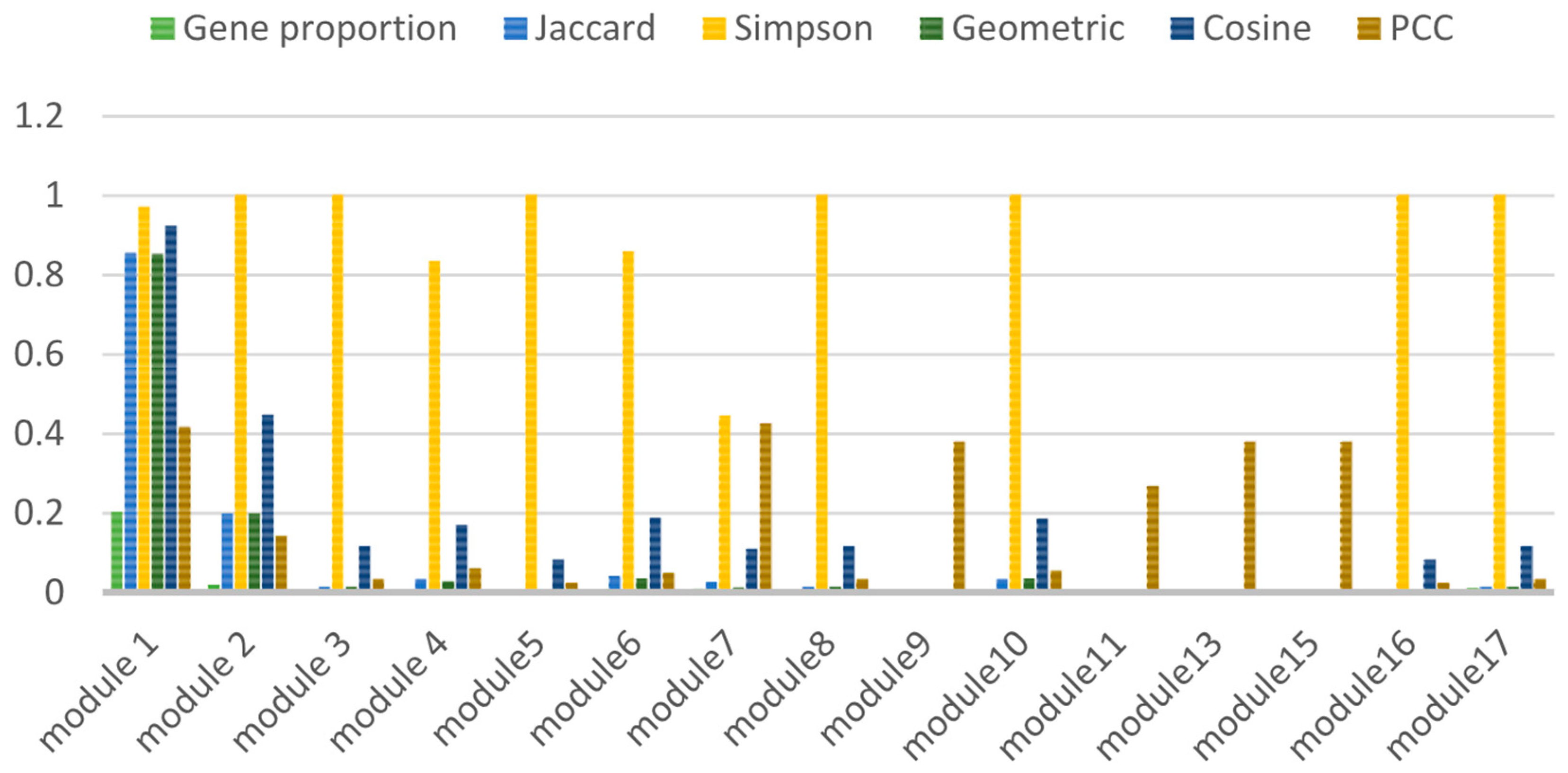

| Correlation Index | Formula | Meaning |
|---|---|---|
| Jaccard | The range of values is [0, 1], and the closer it is to 1, the stronger the correlation. | |
| Simpson | ||
| Geometric | ||
| Cosine | ||
| PCC |
| Division Methods | Number of Modules | Entropy Value |
|---|---|---|
| MCODE | 18 | 6.05 |
| MCL | 89 | 6.19 |
| Glay | 17 | 6.20 |
| Cluster one | 89 | 6.22 |
| Module | The Number of Genes | Module Score | Module | The Number of Genes | Module Score |
|---|---|---|---|---|---|
| 1 | 400 | 400.000 | 10 | 6 | 3.200 |
| 2 | 38 | 6.585 | 11 | 3 | 3.000 |
| 3 | 7 | 5.667 | 12 | 3 | 3.000 |
| 4 | 5 | 4.500 | 13 | 3 | 3.000 |
| 5 | 5 | 4.000 | 14 | 3 | 3.000 |
| 6 | 10 | 3.778 | 15 | 3 | 3.000 |
| 7 | 15 | 3.571 | 16 | 3 | 3.000 |
| 8 | 4 | 3337 | 17 | 20 | 2.947 |
| 9 | 7 | 3.333 | 18 | 4 | 2.667 |
| Module | The Number of Pathways | Intersection | Union | Gene Proportion | Module | The Number of Pathways | Intersection | Union | Gene Proportion |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 132 | 128 | 150 | 20.04% | 9 | 2 | 0 | 146 | 0.35% |
| 2 | 29 | 29 | 146 | 1.90% | 10 | 5 | 5 | 148 | 0.30% |
| 3 | 2 | 2 | 146 | 0.35% | 11 | 1 | 0 | 146 | 0.15% |
| 4 | 6 | 5 | 147 | 0.25% | 13 | 2 | 0 | 147 | 0.15% |
| 5 | 1 | 1 | 146 | 0.25% | 15 | 2 | 0 | 148 | 0.15% |
| 6 | 7 | 6 | 147 | 0.50% | 16 | 1 | 1 | 146 | 0.15% |
| 7 | 9 | 4 | 147 | 0.75% | 17 | 2 | 2 | 146 | 1.00% |
| 8 | 2 | 2 | 151 | 0.20% |
| Gene | Repetitions | Gene | Repetitions | Gene | Repetitions | Gene | Repetitions |
|---|---|---|---|---|---|---|---|
| ABCA1 | 10 | NFKB | 10 | WNT9A | 10 | WNT3 | 9 |
| ACHE | 10 | NMDAR | 10 | WNT9B | 10 | WNT4 | 9 |
| CASP6 | 10 | PKC | 10 | XBP1 | 10 | APBB1 | 8 |
| CHAT | 10 | PP2A | 10 | ADAM10 | 9 | APH1B | 7 |
| CTFA | 10 | PRPC | 10 | APP | 9 | APOE | 7 |
| CYLD | 10 | PSD95 | 10 | BACE1 | 9 | BECN1 | 7 |
| DAG1 | 10 | SIRT1 | 10 | CHRM5 | 9 | CALM1 | 7 |
| DR6 | 10 | SPS | 10 | GRIN1 | 9 | CAPN2 | 7 |
| EETS | 10 | UCHL1 | 10 | IDE | 9 | CDK5 | 7 |
| EPHB2 | 10 | UQCRB | 10 | LRP1 | 9 | CHRM1 | 7 |
| FYN | 10 | VLDLR | 10 | MAPT | 9 | CHRM3 | 7 |
| GRIN3A | 10 | WNT1 | 10 | PSEN1 | 9 | CYCS | 7 |
| HPETE | 10 | WNT3A | 10 | PSEN2 | 9 | DVL2 | 7 |
| HSPG | 10 | WNT5A | 10 | RELA | 9 | GNAQ | 7 |
| IKKA | 10 | WNT5B | 10 | TNF | 9 | GRIN2A | 7 |
| IKKB | 10 | WNT6 | 10 | WNT10B | 9 | GRIN2B | 7 |
| INSP3R | 10 | WNT7A | 10 | WNT11 | 9 | GRM5 | 7 |
| LDLR | 10 | WNT7B | 10 | WNT16 | 9 | GSK3B | 7 |
| LILRB2 | 10 | WNT8A | 10 | WNT2 | 9 | HRAS | 7 |
| MAPK | 10 | WNT8B | 10 | WNT2B | 9 | IKBKB | 7 |
| Gene | Posterior Probability | Score | Gene | Posterior Probability | Score |
|---|---|---|---|---|---|
| APP | 0.9998 | 1 | GRIN1 | 0.9927 | 0.992481 |
| ADAM10 | 0.9991 | 0.999499 | CDK5R1 | 0.9926 | 0.99198 |
| MAPK1 | 0.9989 | 0.998997 | CDK5 | 0.9919 | 0.991479 |
| MAPT | 0.9986 | 0.998496 | MAP2K1 | 0.9918 | 0.990977 |
| RELA | 0.9956 | 0.997995 | AKT2 | 0.9915 | 0.990476 |
| ACHE | 0.9955 | 0.997494 | MTOR | 0.9915 | 0.990476 |
| MAPK10 | 0.9952 | 0.996992 | GRIN2C | 0.9913 | 0.989474 |
| APOE | 0.995 | 0.996491 | SIRT1 | 0.9913 | 0.989474 |
| KIF5A | 0.9945 | 0.99599 | CALM1 | 0.9912 | 0.988471 |
| NFKB1 | 0.9944 | 0.995489 | CACNA1D | 0.9911 | 0.98797 |
| GRIN2A | 0.994 | 0.994987 | ITPR1 | 0.9911 | 0.98797 |
| GNAQ | 0.9937 | 0.994486 | ATP2A2 | 0.991 | 0.986967 |
| HRAS | 0.9935 | 0.993985 | CASP7 | 0.991 | 0.986967 |
| GRIN2B | 0.9933 | 0.993484 | DVL1 | 0.991 | 0.986967 |
| APBB1 | 0.9929 | 0.992982 | INS | 0.991 | 0.986967 |
| Gene | Gene | Gene | Gene | Gene |
|---|---|---|---|---|
| ACHE | DVL2 | ITPR1 | NOX1 | WNT11 |
| ADAM10 | EPHB2 | KLC1 | NOX4 | WNT16 |
| APBB1 | GNAQ | LILRB2 | NRAS | WNT2 |
| APH1B | GRIN1 | LRP1 | PPP3R1 | WNT2B |
| APOE | GRIN2A | MAP2K1 | PSEN1 | WNT3 |
| APP | GRIN2B | MAP2K2 | PTGS2 | WNT3A |
| BACE1 | GSK3B | MAPK1 | RELA | WNT4 |
| CALM1 | HRAS | MAPK10 | SIRT1 | WNT5A |
| CDK5 | IDE | MAPK3 | UCHL1 | WNT5B |
| CHRM1 | IKBKB | MAPK9 | UQCRB | WNT6 |
| CHRM3 | IL1A | MAPT | WNT1 | WNT7A |
| CYCS | IL1B | NOS2 | WNT10B | XBP1 |
| Herb | Similar Genes |
|---|---|
| KXS | ACHE, CHRM1, CHRM3, GSK3B, IKBKB, IL1B, NOS2, PTGS2, RELA |
| DGYSY | ACHE, CHRM1, CHRM3, GRIN1, GRIN2A, GRIN2B, GSK3B, IKBKB, IL1B, MAPK1, MAPK10, NOS2, PTGS2, RELA |
| YGS | ACHE, BACE1, CHRM1, CHRM3, GRIN1, GRIN2A, GRIN2B, GSK3B, IKBKB, IL1A, IL1B, MAPK1, MAPK10, NOS2, PTGS2, RELA |
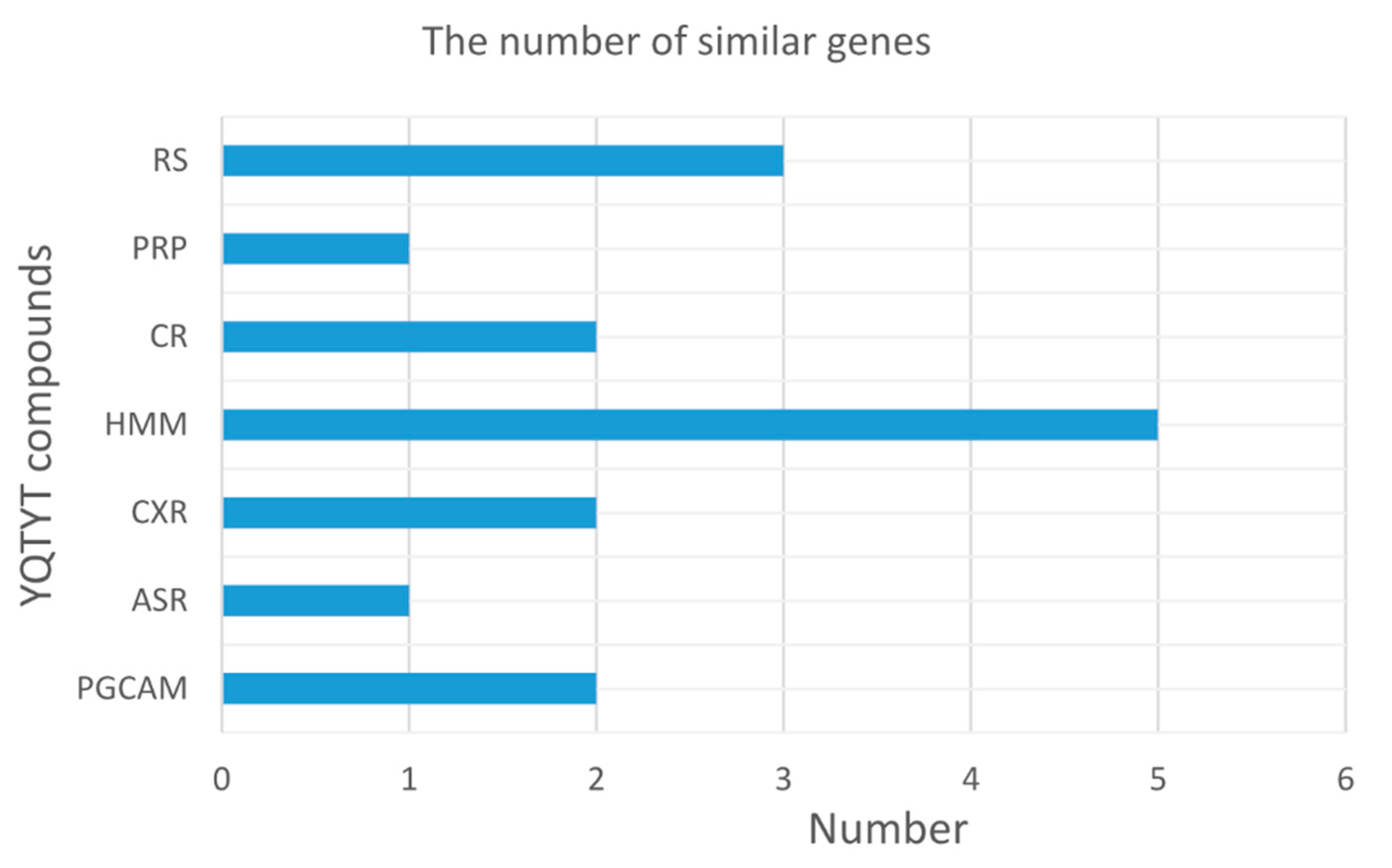
| YQTYT | ACHE, APP, CHRM1, CHRM3, CYCS, GRIN1, GRIN2B, GSK3B, IKBKB, IL1A, IL1B, MAPK1, MAPK10, NOS2, PTGS2, RELA, SIRT1, |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, C.; Guo, H.; Cao, S. Gene Network Analysis of Alzheimer’s Disease Based on Network and Statistical Methods. Entropy 2021, 23, 1365. https://doi.org/10.3390/e23101365
Zhou C, Guo H, Cao S. Gene Network Analysis of Alzheimer’s Disease Based on Network and Statistical Methods. Entropy. 2021; 23(10):1365. https://doi.org/10.3390/e23101365
Chicago/Turabian StyleZhou, Chen, Haiyan Guo, and Shujuan Cao. 2021. "Gene Network Analysis of Alzheimer’s Disease Based on Network and Statistical Methods" Entropy 23, no. 10: 1365. https://doi.org/10.3390/e23101365
APA StyleZhou, C., Guo, H., & Cao, S. (2021). Gene Network Analysis of Alzheimer’s Disease Based on Network and Statistical Methods. Entropy, 23(10), 1365. https://doi.org/10.3390/e23101365






