An Interval Iteration Based Multilevel Thresholding Algorithm for Brain MR Image Segmentation
Abstract
:1. Introduction
- (1)
- A hybrid L1 − L0 layer decomposition method is used to achieve the base layer of an original image, which can remove noise and preserve edge information in the segmentation process.
- (2)
- An interval iteration multilevel thresholding method is proposed in this paper. In the grayscale histogram of an original image, iterations are separated by the combination of class means and thresholds, and Otsu single thresholding is iteratively applied to each iteration.
- (3)
- A fusion strategy is adopted to fuse different segmentation results. It takes both spatial and intensity information into account, and makes segmentation more accurate.
2. Interval Iteration Based Multilevel Thresholding
2.1. Otsu Method
2.2. Interval Iteration Based Multilevel Thresholding
2.2.1. The First Iteration
2.2.2. The Second Iteration
2.2.3. The sth Iteration
| Algorithm 1. Interval iteration-based multilevel thresholding (IIMT). |
| Input: original image I, number of thresholds K (), constant ; |
| Output: optimal thresholds T1, T2, …, TK; |
| 1: Otsu multilevel thresholding (maximize Equation (1)), obtain thresholds T1,1, T1,2, …, |
| T1,K, corresponding class means μ1,1, μ1,2, …, μ1,K, μ1,K+1, and divided classes C1, CK+1; |
| 2: Otsu single thresholding in interval [μ1,i, μ1,i + 1] (i = 1, …, K), obtain corresponding |
| threshold T2,i, and class means μ2,2i−1, μ2,2i, update classes C1, CK+1, obtain divided |
| classes C2, …, CK; |
| 3: for i = 1, …, K do |
| 4: s = 3; |
| 5: do |
| 6: {Otsu single thresholding in every interval [μs−1,2i−1, μs−1,2i] (i = 1, …, K), obtain |
| corresponding threshold Ts,i and class means μs,2i−1, μs,2i, update divided classes Ci; |
| 7: s++; |
| 8: } while () |
| 9: ; |
| 10: end for |
3. The Proposed Algorithm
3.1. The Framework
- (1)
- A hybrid L1 − L0 layer decomposition method is performed on the original image to obtain its base layer.
- (2)
- The original image and its base layer are segmented by the IIMT algorithm, and their segmentation results are denoted A and B, respectively.
- (3)
- The segmentation fusion method is applied to A and B to obtain the final segmentation result.
3.2. Hybrid L1 − L0 Layer Decomposition
3.3. Segmentation Fusion
4. Experimental Results and Analysis
4.1. Experimental Protocols
4.2. Evaluation Measure
- (1)
- Uniformity measure
- (2)
- Misclassification error
- (3)
- Hausdorff distance
- (4)
- Jaccard index
4.3. Comparison with Otsu-Based Method
4.4. Experimental Results on Images Containing Noise
4.5. Comprehensive Comparison
- (1)
- Proposed
- (2)
- LLF-DCE
- (3)
- PSO
- (4)
- BF
- (5)
- ABF
- (6)
- NMS
- (7)
- RCGA
4.6. Experimental Results on BRATS Database
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Wang, R.; Cao, S.; Ma, K.; Zheng, Y.; Meng, D. Pairwise learning for medical image segmentation. Med. Image Anal. 2020, 67, 101876. [Google Scholar] [CrossRef] [PubMed]
- Oktay, O.; Ferrante, E.; Kamnitsas, K.; Heinrich, M.; Bai, W.; Caballero, J.; Cook, S.A.; De Marvao, A.; Dawes, T.; O‘Regan, D.P.; et al. Anatomically constrained neural networks (ACNNs), application to cardiac image enhancement and segmentation. IEEE Trans. Med. Imaging 2017, 37, 384–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.; Ji, Z.; Yang, J.; Sun, Q.; Shen, X.; Ren, Z.; Ge, Q. Label Group Diffusion for Image and Image Pair Segmentation. Pattern Recognit. 2020, 112, 107789. [Google Scholar] [CrossRef]
- Mărginean, R.; Andreica, A.; Dioşan, L.; Bálint, Z. Butterfly effect in chaotic image segmentation. Entropy 2020, 22, 1028. [Google Scholar] [CrossRef]
- Monteiro, C.; Campilho, C. Performance Evaluation of Image Segmentation. International Conference Image Analysis and Recognition; Springer: Berlin/Heidelberg, Germany, 2006; pp. 248–259. [Google Scholar]
- Zou, L.; Song, L.T.; Weise, T.; Wang, X.F.; Huang, Q.J.; Deng, R.; Wu, Z.Z. A Survey on Regional Level Set Image Segmentation Models based on the Energy Functional Similarity Measure. Neurocomputing 2020, 452, 606–622. [Google Scholar] [CrossRef]
- Apro, M.; Pal, S.; Dedijer, S. Evaluation of single and multi-threshold entropy-based algorithms for folded substrate analysis. J. Graph. Eng. Des. 2011, 2, 1–9. [Google Scholar]
- Raki, M.; Cabezas, M.; Kushibar, K.; Oliver, A.; Lladó, X. Improving the Detection of Autism Spectrum Disorder by Combining Structural and Functional MRI Information. NeuroImage Clin. 2020, 25, 102181. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.G.; Bi, L.X.; Hui, H.G. Survey on Medical Image Computer Aided Detection and Diagnosis Systems. J. Softw. 2018, 29, 1471–1514. [Google Scholar]
- Tian, J.X.; Liu, G.C.; Gu, S.S. Deep Learning in Medical Image Analysis and Its Challenges. Zidonghua Xuebao/Acta Autom. Sin. 2018, 44, 401–424. [Google Scholar]
- Dekhil, O.; Ali, M.; Haweel, R.; Elnakib, Y.; Ghazal, M.; Hajjdiab, H.; Fraiwan, L.; Shalaby, A.; Soliman, A.; Mahmoud, A.; et al. A Comprehensive Framework for Differentiating Autism Spectrum Disorder from Neurotypicals by Fusing Structural MRI and Resting State Functional MRI. Semin. Pediatric Neurol. 2020, 34, 100805. [Google Scholar] [CrossRef]
- Fu, S.; Mui, K. A survey on image segmentation. Pattern Recognit. 1981, 13, 3–16. [Google Scholar] [CrossRef]
- Wu, B.; Zhou, J.; Ji, X.; Yin, Y.; Shen, X. An ameliorated teaching-learning-based optimization algorithm based study of image segmentation for multilevel thresholding using Kapur’s entropy and Otsu’s between class variance. Inf. Sci. 2020, 533, 72–107. [Google Scholar] [CrossRef]
- Zortea, M.; Flores, E.; Scharcanski, J. A simple weighted thresholding method for the segmentation of pigmented skin lesions in macroscopic images. Pattern Recognit. 2017, 64, 92–104. [Google Scholar] [CrossRef]
- Ghamisi, P.; Couceiro, M.S.; Martins, F.M.L.; Benediktsson, J.A. Multilevel image segmentation based on fractional-order Darwinian particle swarm optimization. IEEE Trans. Geosci. Remote Sens. 2013, 52, 2382–2394. [Google Scholar] [CrossRef] [Green Version]
- Tan, K.S.; Isa, N.A.M. Color image segmentation using histogram thresholding-Fuzzy C-means hybrid approach. Pattern Recognit. 2011, 44, 1–15. [Google Scholar]
- Zhang, X.; Sun, Y.; Liu, H.; Hou, Z.; Zhao, F.; Zhang, C. Improved clustering algorithms for image segmentation based on non-local information and back projection. Inf. Sci. 2021, 550, 129–144. [Google Scholar] [CrossRef]
- Huang, J.; You, X.; Tang, Y.Y.; Du, L.; Yuan, Y. A novel iris segmentation using radial-suppression edge detection. Signal Process. 2009, 89, 2630–2643. [Google Scholar] [CrossRef]
- Rampun, A.; López-Linares, K.; Morrow, P.J.; Scotney, B.W.; Wang, H.; Ocaña, I.G.; Maclair, G.; Zwiggelaar, R.; Ballester, M.A.; Macía, I. Breast pectoral muscle segmentation in mammograms using a modified holistically-nested edge detection network. Med. Image Anal. 2019, 57, 1–17. [Google Scholar] [CrossRef]
- Soltani-Nabipour, J.; Khorshidi, A.; Noorian, B. Lung tumor segmentation using improved region growing algorithm. Nucl. Eng. Technol. 2020, 52, 2313–2319. [Google Scholar] [CrossRef]
- Xu, G.; Li, X.; Lei, B.; Lv, K. Unsupervised color image segmentation with color-alone feature using region growing pulse coupled neural network. Neurocomputing 2018, 306, 1–16. [Google Scholar] [CrossRef]
- Siriapisith, T.; Kusakunniran, W.; Haddawy, P. Pyramid graph cut, Integrating intensity and gradient information for grayscale medical image segmentation. Comput. Biol. Med. 2020, 126, 103997. [Google Scholar] [CrossRef] [PubMed]
- Ortuño-Fisac, J.E.; Vegas-Sánchez-Ferrero, G.; Gómez-Valverde, J.J.; Chen, M.Y.; Santos, A.; McVeigh, E.R.; Ledesma-Carbayo, M.J. Automatic estimation of aortic and mitral valve displacements in dynamic CTA with 4D graph-cuts. Med Image Anal. 2020, 65, 101748. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Shi, Y.; Sun, J.; Gao, Y.; Zhu, J.; Dai, Y. Crossbar-net, A novel convolutional neural network for kidney tumor segmentation in ct images. IEEE Trans. Image Process. 2019, 28, 4060–4074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhandari, A.K.; Kumar, A.; Singh, G.K. Modified artificial bee colony based computationally efficient multilevel thresholding for satellite image segmentation using Kapur’s, Otsu and Tsallis functions. Expert Syst. Appl. 2015, 42, 1573–1601. [Google Scholar] [CrossRef]
- Otsu, N. A threshold selection method from gray-level histograms. Automatica 1975, 11, 23–27. [Google Scholar] [CrossRef] [Green Version]
- Kapur, J.; Sahoo, P.; Wong, A. A new method for gray-level picture thresholding using the entropy of the histogram. Comput. Vis. Graph. Image Process. 1985, 29, 273–285. [Google Scholar] [CrossRef]
- Kittler, J.; Illingworth, J. Minimum error thresholding. Pattern Recognit. 1986, 19, 41–47. [Google Scholar] [CrossRef]
- Lifang, H.; Songwei, H. An efficient krill herd algorithm for color image multilevel thresholding segmentation problem. Appl. Soft Comput. J. 2020, 89, 106063. [Google Scholar]
- Abd Elaziz, M.; Lu, S. Many-objectives Multilevel Thresholding Image Segmentation using Knee Evolutionary Algorithm. Expert Syst. Appl. 2019, 125, 305–316. [Google Scholar] [CrossRef]
- Lei, B.; Fan, J. Image thresholding segmentation method based on minimum square rough entropy. Appl. Soft Comput. 2019, 84, 105687. [Google Scholar] [CrossRef]
- Yan, Z.; Zhang, J.; Yang, Z.; Tang, J. Kapur’s entropy for underwater multilevel thresholding image segmentation based on whale optimization algorithm. IEEE Access 2020, 9, 41294–41319. [Google Scholar] [CrossRef]
- Singh, P. A neutrosophic-entropy based adaptive thresholding segmentation algorithm, A special application in MR images of Parkinson’s disease. Artif. Intell. Med. 2020, 104, 101838. [Google Scholar] [CrossRef] [PubMed]
- Omid, T.; Haifeng, S. An adaptive differential evolution algorithm to optimal multi-level thresholding for MRI brain image segmentation. Expert Syst. Appl. 2019, 138, 112820. [Google Scholar]
- Zhao, D.; Liu, L.; Yu, F.; Heidari, A.A.; Wang, M.; Liang, G.; Muhammad, K.; Chen, H. Chaotic random spare ant colony optimization for multi-threshold image segmentation of 2D Kapur entropy. Knowl.-Based Syst. 2021, 216, 106510. [Google Scholar] [CrossRef]
- Cai, H.; Yang, Z.; Cao, X.; Xia, W.; Xu, X. A new iterative triclass thresholding technique in image segmentation. IEEE Trans. Image Process. 2014, 23, 1038–1046. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Shi, Y.; Sun, J.; Wang, L.; Zhou, L.; Gao, Y.; Shen, D. Interactive medical image segmentation via a point-based interaction. Artif. Intell. Med. 2021, 111, 101998. [Google Scholar] [CrossRef]
- Liang, Z.; Xu, J.; Zhang, D.; Cao, Z.; Zhang, L. A hybrid l1-l0 layer decomposition model for tone mapping. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4758–4766. [Google Scholar]
- Feng, Y.; Shen, X.; Chen, H.; Zhang, X. Segmentation fusion based on neighboring information for MR brain images. Multimed. Tools Appl. 2017, 76, 23139–23161. [Google Scholar] [CrossRef]
- Manikandan, S.; Ramar, K.; Iruthayarajan, M.; Srinivasagan, K.G. Multilevel thresholding for segmentation of medical brain images using real coded genetic algorithm. Measurement 2014, 47, 558–568. [Google Scholar] [CrossRef]
- Liew, L.; Anglin, M.; Banks, W.; Sondag, M.; Ito, K.L.; Kim, H.; Chan, J.; Ito, J.; Jung, C.; Khoshab, N.; et al. A large, open source dataset of stroke anatomical brain images and manual lesion segmentations. Sci. Data 2018, 5, 180011. [Google Scholar] [CrossRef] [Green Version]
- Qian, H.; Guo, X.; Xue, I. Image Understanding Based Global Evaluation Algorithm for Segmentation. Acta Electron. Sin. 2012, 40, 1989–1995. [Google Scholar]
- Sathya, P.; Kayalvizhi, R. Optimal segmentation of brain MRI based on adaptive bacterial foraging algorithm. Neurocomputing 2011, 74, 2299–2313. [Google Scholar] [CrossRef]
- Menze, H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging 2015, 34, 1993–2024. [Google Scholar] [CrossRef] [PubMed]






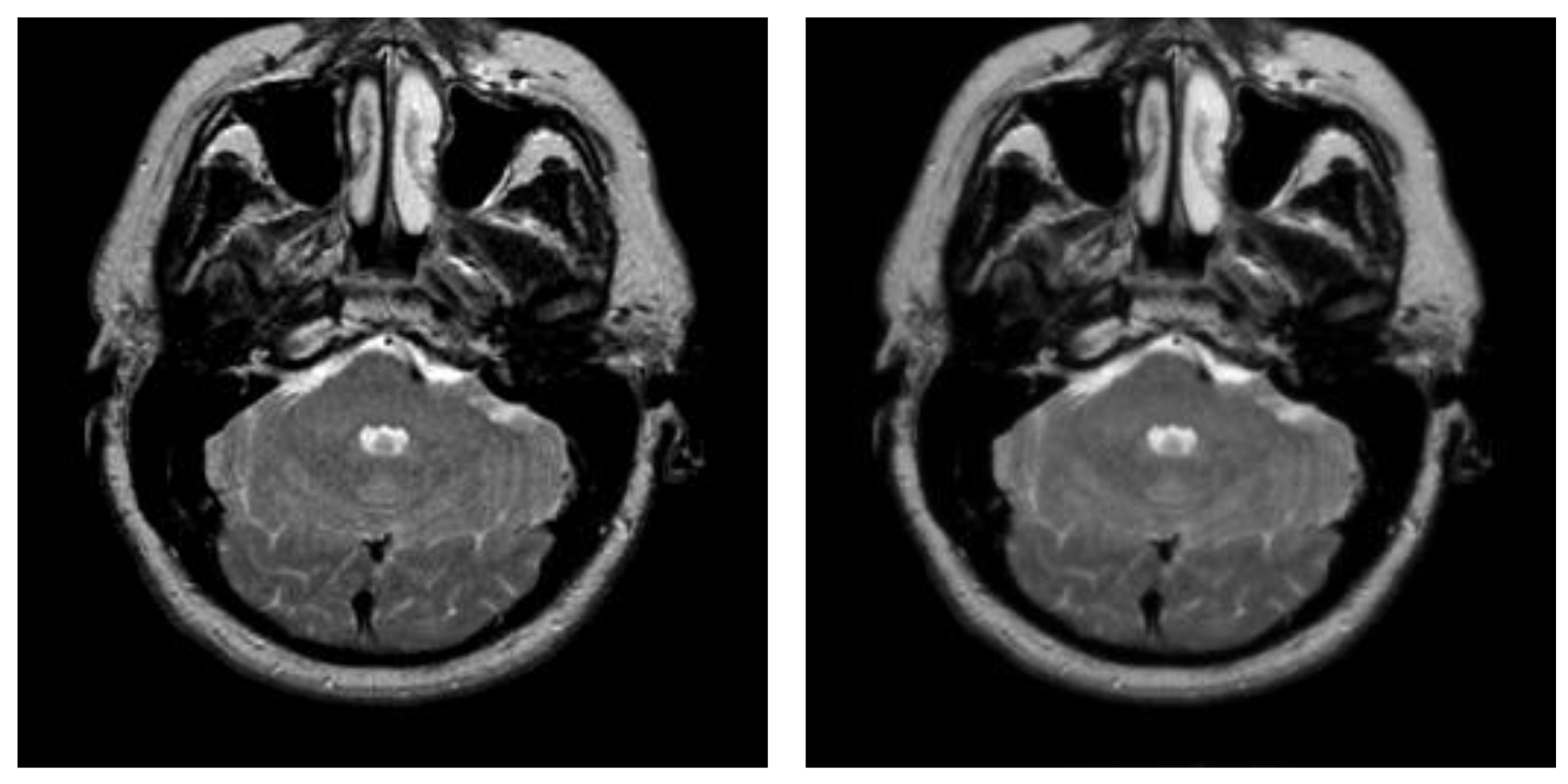












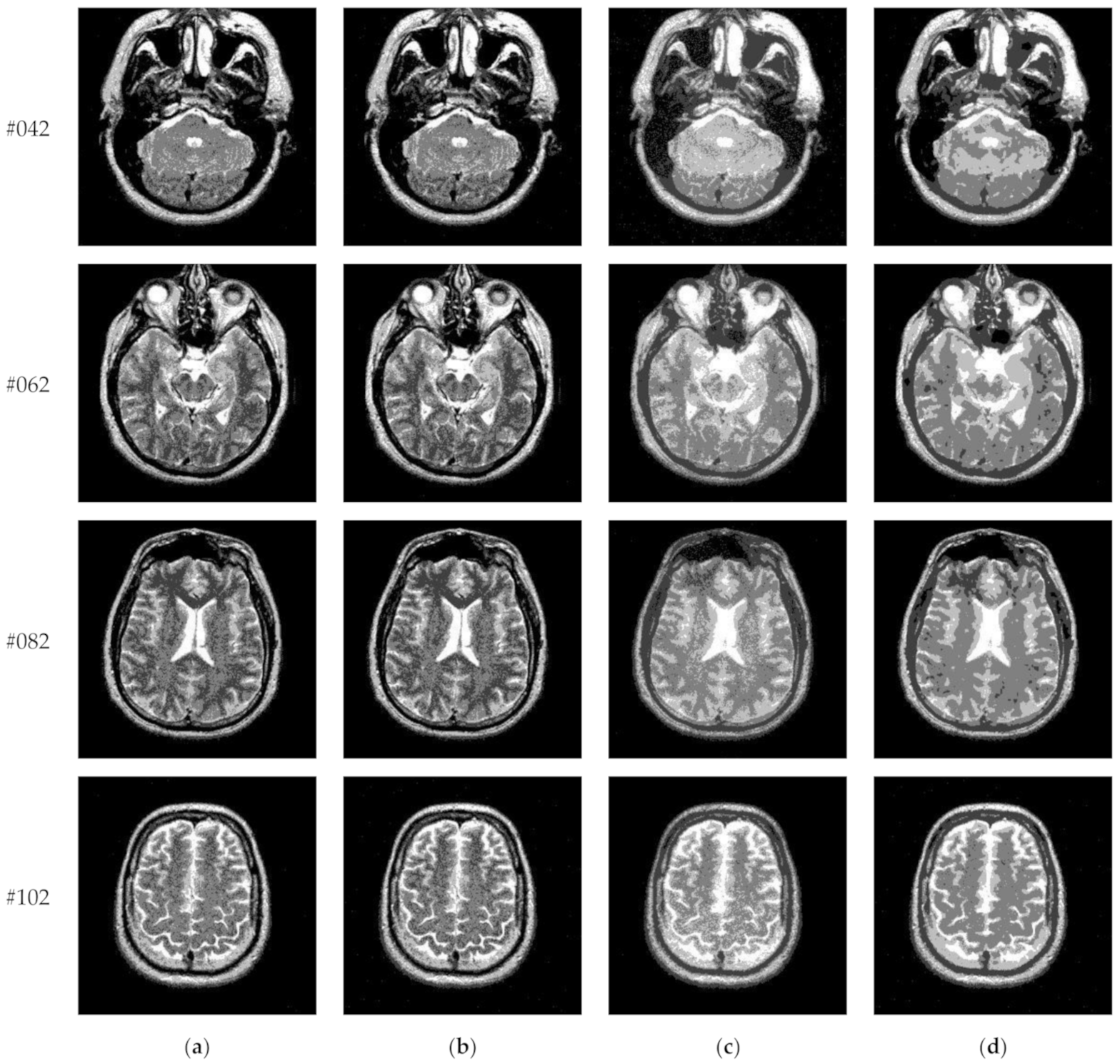








| Parameter Settings | Description |
|---|---|
| δ = 0.01 | Value that stops the iteration for IIMT |
| λ1 = 1 | Weight of base layer for hybrid L1 − L0 layer decomposition |
| λ2 = 0.1λ1 | Weight of detail layer for hybrid L1 − L0 layer decomposition |
| r = 12 | Radius for segmentation fusion |
| K = 1, 2, 3, 4, 5 | Number of the thresholds |
| Test Images | Number of Thresholds (K) | Uniformity Measure (U) | |||
|---|---|---|---|---|---|
| Proposed | HL-IIMT | IIMT | OTSU | ||
| #042 | 1 | 0.9858 | 0.9818 | 0.9773 | 0.9715 |
| 2 | 0.9855 | 0.9805 | 0.9764 | 0.9705 | |
| 3 | 0.9893 | 0.9825 | 0.9759 | 0.9694 | |
| 4 | 0.9893 | 0.9814 | 0.9709 | 0.9608 | |
| 5 | 0.9914 | 0.9831 | 0.9717 | 0.9707 | |
| #082 | 1 | 0.9827 | 0.9796 | 0.9708 | 0.9670 |
| 2 | 0.9836 | 0.9799 | 0.9716 | 0.9687 | |
| 3 | 0.9927 | 0.9823 | 0.9733 | 0.9702 | |
| 4 | 0.9869 | 0.9802 | 0.9749 | 0.9713 | |
| 5 | 0.9938 | 0.9804 | 0.9750 | 0.9714 | |
| Test Images | Number of Thresholds (K) | Uniformity Measure (U) | |||
|---|---|---|---|---|---|
| Proposed | HL-IIMT | IIMT | OTSU | ||
| #022 | 1 | 0.9892 | 0.9786 | 0.9652 | 0.9569 |
| 4 | 0.9895 | 0.9795 | 0.9672 | 0.9608 | |
| #042 | 1 | 0.9817 | 0.9723 | 0.9671 | 0.9646 |
| 4 | 0.9856 | 0.9786 | 0.9685 | 0.9571 | |
| #062 | 1 | 0.9780 | 0.9702 | 0.9519 | 0.9407 |
| 4 | 0.9833 | 0.9728 | 0.9605 | 0.9547 | |
| #082 | 1 | 0.9808 | 0.9719 | 0.9591 | 0.9520 |
| 4 | 0.9856 | 0.9786 | 0.9688 | 0.9572 | |
| #102 | 1 | 0.9869 | 0.9784 | 0.9556 | 0.9503 |
| 4 | 0.9906 | 0.9813 | 0.9685 | 0.9622 | |
| Test Images | K | Optimal Threshold Values | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Proposed | LLF-DCE | PSO | BF | ABF | NMS | RCGA | ||||
| IIMT | HL-IIMT | LLF-Otsu | DCE-Otsu | |||||||
| #022 | 2 | 40, 96 | 34, 103 | 26, 95 | 1, 77 | 97, 184 | 96, 184 | 95, 184 | 96, 184 | 96, 184 |
| 3 | 42, 98, 156 | 22, 69, 125 | 26, 64, 103 | 1, 3, 77 | 69, 138, 207 | 65, 131, 186 | 69, 114, 185 | 58, 116, 185 | 58, 115, 185 | |
| 4 | 20, 48, 86, 126 | 20, 60, 100, 141 | 26, 64, 91, 132 | 1, 3, 5, 79 | 83, 116, 175, 207 | 52, 99, 148, 186 | 58, 113, 174, 208 | 43, 87, 132, 185 | 44, 87, 131, 186 | |
| 5 | 28, 70, 118, 164, 228 | 17, 51, 90, 132, 178 | 14, 26, 64, 91, 132 | 1, 3, 5, 68, 79 | 76, 119, 154, 184, 214 | 44, 90, 127, 170, 208 | 43, 88, 130, 176, 208 | 44, 104, 140, 176, 214 | 44, 86, 127, 174, 208 | |
| #032 | 2 | 50, 112 | 43, 115 | 25, 102 | 1, 77 | 107, 185 | 110, 185 | 110, 185 | 110, 185 | 109, 185 |
| 3 | 28, 70, 120 | 22, 73, 124 | 25, 82, 110 | 1, 3, 79 | 74, 157, 192 | 72, 120, 198 | 81, 134, 187 | 56, 115, 186 | 53, 116, 185 | |
| 4 | 24, 66, 112, 152 | 22, 73, 124, 181 | 25, 82, 94, 151 | 1, 3, 5, 81 | 95, 125, 164, 194 | 63, 119, 173, 208 | 58, 102, 142, 190 | 39, 83, 132, 189 | 39, 84, 131, 189 | |
| 5 | 30, 74, 118, 158, 204 | 15, 47, 81, 112, 148 | 25, 56, 88, 97, 151 | 1, 3, 5, 57, 81 | 80, 112, 139, 186, 213 | 63, 101, 140, 175, 207 | 52, 87, 128, 167, 198 | 29, 75, 124, 173, 207 | 34, 78, 123, 174, 207 | |
| #042 | 2 | 54, 118 | 46, 120 | 29, 111 | 37, 87 | 111, 183 | 114, 184 | 114, 184 | 113, 184 | 114, 183 |
| 3 | 34, 82, 130 | 27, 82, 130 | 29, 69, 132 | 37, 49, 141 | 80, 148, 178 | 70, 136, 188 | 74, 130, 185 | 84, 132, 188 | 84, 132, 187 | |
| 4 | 36, 76, 112, 156 | 21, 67, 105, 149 | 29, 69, 97, 144 | 37, 48, 95, 143 | 81, 125, 164, 197 | 62, 112, 156, 194 | 50, 100, 143, 190 | 29, 76, 128.187 | 30, 75, 127, 188 | |
| 5 | 20, 56, 90, 126, 168 | 18, 60, 94, 128, 170 | 29, 69, 78, 108, 145 | 35, 49, 77, 95, 143 | 82, 115, 142, 184, 214 | 58, 114, 151, 188, 218 | 53, 97, 144, 184, 218 | 31, 76, 126, 178, 217 | 25, 69, 114, 156, 194 | |
| #052 | 2 | 58, 114 | 49, 111 | 30, 103 | 31, 89 | 119, 186 | 117, 186 | 117, 186 | 118, 185 | 118, 185 |
| 3 | 46, 88, 130 | 31, 88, 127 | 30, 75, 111 | 31, 45, 125 | 89, 113, 187 | 102, 156, 206 | 107, 158, 204 | 109, 166, 207 | 109, 165, 203 | |
| 4 | 22, 60, 96, 134 | 19, 65, 99, 135 | 30, 75, 93, 143 | 31, 45, 79, 127 | 79, 111, 141, 208 | 93, 124, 171, 210 | 90, 129, 173, 210 | 94, 132, 175, 210 | 91, 131, 174, 209 | |
| 5 | 22, 58, 88, 120, 156 | 35, 89, 120, 152, 194 | 14, 30, 75, 93, 143 | 31, 45, 79, 100, 127 | 65, 85, 131, 162, 203 | 56, 112, 144, 175, 209 | 56, 95, 133, 167, 203 | 20, 67, 120, 167, 207 | 24, 67, 118, 166, 203 | |
| #062 | 2 | 58, 120 | 51, 118 | 31, 111 | 33, 103 | 109, 186 | 119, 190 | 119, 186 | 121, 187 | 121, 187 |
| 3 | 48, 94, 144 | 42, 97, 142 | 31, 79, 134 | 33, 45, 133 | 112, 167, 187 | 97, 133, 183 | 102, 147, 199 | 101, 148, 195 | 101, 147, 196 | |
| 4 | 42, 84, 120, 164 | 19, 67, 106, 149 | 31, 79, 96, 151 | 33, 45, 81, 135 | 85, 134, 180, 203 | 98, 140, 182, 218 | 93, 135, 175, 212 | 94, 134, 176, 211 | 94, 134, 175, 211 | |
| 5 | 28, 64, 94, 128, 170 | 27, 75, 102, 137, 179 | 17, 31, 79, 96, 151 | 33, 45, 81, 116, 135 | 99, 119, 157, 181, 203 | 73, 104, 139, 184, 213 | 79, 111, 145, 179, 212 | 28, 68, 120, 168, 208 | 20, 65, 113, 158, 200 | |
| #072 | 2 | 60, 120 | 52, 120 | 32, 133 | 33, 111 | 116, 177 | 117, 179 | 117, 179 | 118, 179 | 117, 179 |
| 3 | 54, 100, 156 | 47, 103, 156 | 32, 76, 139 | 33, 45, 139 | 96, 178, 207 | 95, 147, 202 | 99, 150, 190 | 100, 142, 188 | 99, 141, 187 | |
| 4 | 48, 86, 122, 178 | 36, 87, 122, 174 | 32, 76, 93, 155 | 33, 45, 81, 141 | 96, 124, 161, 187 | 94, 129, 173, 214 | 95, 134, 174, 214 | 100, 140, 179, 214 | 99, 140, 179, 213 | |
| 5 | 48, 84, 110, 142, 188 | 17, 62, 94, 128, 179 | 32, 68, 81, 102, 155 | 33, 45, 63, 81, 141 | 72, 112, 151, 178, 197 | 87, 109, 139, 178, 210 | 87, 119, 150, 180, 214 | 10, 64, 120, 172, 211 | 14, 64, 119, 171, 211 | |
| #082 | 2 | 60, 116 | 51, 113 | 32, 110 | 37, 103 | 110, 170 | 112, 169 | 111, 170 | 112, 169 | 111, 169 |
| 3 | 54, 102, 158 | 47, 102, 158 | 32, 83, 143 | 37, 49, 137 | 103, 136, 198 | 114, 155, 210 | 111, 155, 201 | 103, 146, 189 | 103, 146, 190 | |
| 4 | 42, 82, 116, 168 | 20, 70, 105, 158 | 32, 83, 93, 166 | 37, 49, 87, 138 | 100, 129, 167, 188 | 103, 139, 175, 214 | 99, 135, 170, 210 | 98, 134, 169, 210 | 98, 133, 169, 210 | |
| 5 | 52, 88, 118, 154, 210 | 17, 63, 92, 121, 171 | 15, 32, 83, 93, 166 | 37, 49, 87, 99, 139 | 78, 105, 151, 180, 201 | 81, 122, 150, 182, 212 | 84, 113, 146, 178, 214 | 14, 62, 115, 168, 210 | 10, 62, 107, 148, 190 | |
| #092 | 2 | 58, 108 | 55, 115 | 33, 104 | 35, 101 | 109, 175 | 108, 174 | 109, 174 | 109, 173 | 109, 174 |
| 3 | 52, 92, 134 | 46, 97, 135 | 33, 78, 109 | 35, 47, 123 | 115, 134, 178 | 107, 144, 209 | 104, 158, 207 | 106, 158, 206 | 105, 158, 206 | |
| 4 | 40, 78, 106, 144 | 19, 70, 105, 143 | 33, 78, 93, 143 | 35, 47, 81, 125 | 77, 107, 149, 194 | 100, 129, 164, 208 | 102, 138, 171, 212 | 112, 152, 186, 220 | 97, 136, 211, 173 | |
| 5 | 24, 60, 84, 110, 148 | 18, 65, 94, 120, 154 | 33, 66, 83, 104, 143 | 35, 47, 81, 92, 125 | 90, 113, 165, 185, 206 | 85, 114, 147, 175, 212 | 96, 128, 158, 186, 216 | 10, 64, 110, 160, 205 | 5, 62, 109, 159, 205 | |
| #102 | 2 | 56, 108 | 53, 114 | 31, 102 | 33, 99 | 98, 166 | 108, 174 | 108, 174 | 108, 173 | 107, 174 |
| 3 | 50, 92, 136 | 45, 100, 144 | 31, 66, 108 | 33, 47, 127 | 113, 145, 180 | 103, 148, 189 | 98, 146, 189 | 94, 142, 189 | 94, 142, 190 | |
| 4 | 56, 96, 138, 184 | 20, 70, 106, 147 | 31, 66, 94, 143 | 33, 45, 79, 127 | 84, 124, 165, 189 | 79, 122, 164, 200 | 90, 127, 164, 198 | 2, 64, 119, 173 | 1, 63, 120, 174 | |
| 5 | 50, 84, 114, 146, 182 | 19, 67, 97, 125, 158 | 31, 61, 79, 100, 143 | 31, 45, 60, 79, 127 | 99, 128, 147, 194, 218 | 81, 113, 147, 187, 220 | 82, 114, 148, 184, 218 | 9, 62, 106, 147, 190 | 1, 62, 104, 145, 189 | |
| #112 | 2 | 54, 106 | 48, 121 | 25, 96 | 35, 81 | 109, 162 | 105, 165 | 105, 164 | 106, 163 | 106, 163 |
| 3 | 34, 78, 122 | 28, 87, 138 | 25, 78, 106 | 35, 51, 137 | 104, 163, 216 | 79, 134, 180 | 71, 123, 175 | 3, 49, 145 | 1, 70, 142 | |
| 4 | 40, 74, 106, 148 | 25, 79, 119, 164 | 25, 71, 89, 148 | 35, 51, 91, 139 | 63, 130, 153, 206 | 54, 117, 156, 192 | 58, 105, 146, 182 | 4, 63, 132, 178 | 1, 65, 123, 172 | |
| 5 | 28, 66, 100, 144, 194 | 21, 64, 100, 129, 170 | 25, 49, 84, 94, 148 | 35, 51, 91, 93, 141 | 58, 128, 155, 187, 213 | 48, 112, 137, 161, 200 | 47, 108, 142, 171, 197 | 2, 44, 79, 131, 175 | 1, 49, 95, 139, 183 | |
| Test Images | Number of Thresholds (K) | Uniformity Measure (U) | ||||||
|---|---|---|---|---|---|---|---|---|
| Proposed | DCE | PSO | BF | ABF | NMS | RCGA | ||
| #022 | 2 | 0.9879 | 0.9860 | 0.9552 | 0.9569 | 0.9569 | 0.9569 | 0.9569 |
| 3 | 0.9956 | 0.9795 | 0.9672 | 0.9708 | 0.9696 | 0.9769 | 0.9769 | |
| 4 | 0.9912 | 0.9847 | 0.9420 | 0.9765 | 0.9698 | 0.9824 | 0.9824 | |
| 5 | 0.9975 | 0.9837 | 0.9435 | 0.9786 | 0.9785 | 0.9752 | 0.9788 | |
| #032 | 2 | 0.9894 | 0.9844 | 0.9368 | 0.9342 | 0.9342 | 0.9342 | 0.9342 |
| 3 | 0.9910 | 0.9863 | 0.9619 | 0.9716 | 0.9600 | 0.9796 | 0.9801 | |
| 4 | 0.9920 | 0.9855 | 0.9144 | 0.9697 | 0.9766 | 0.9848 | 0.9848 | |
| 5 | 0.9983 | 0.9852 | 0.9422 | 0.9668 | 0.9767 | 0.9851 | 0.9843 | |
| #042 | 2 | 0.9855 | 0.9823 | 0.9271 | 0.9246 | 0.9246 | 0.9246 | 0.9246 |
| 3 | 0.9893 | 0.9826 | 0.9585 | 0.9721 | 0.9689 | 0.9548 | 0.9548 | |
| 4 | 0.9893 | 0.9853 | 0.9465 | 0.9752 | 0.9821 | 0.9865 | 0.9865 | |
| 5 | 0.9914 | 0.9893 | 0.9348 | 0.9724 | 0.9766 | 0.9845 | 0.9877 | |
| #052 | 2 | 0.9882 | 0.9840 | 0.9158 | 0.9128 | 0.9128 | 0.9068 | 0.9128 |
| 3 | 0.9907 | 0.9849 | 0.9523 | 0.9713 | 0.9673 | 0.8800 | 0.9467 | |
| 4 | 0.9892 | 0.9861 | 0.9372 | 0.9764 | 0.9834 | 0.8982 | 0.9856 | |
| 5 | 0.9933 | 0.9875 | 0.9240 | 0.9735 | 0.9782 | 0.9842 | 0.9868 | |
| #062 | 2 | 0.9818 | 0.9802 | 0.9192 | 0.9047 | 0.9049 | 0.9015 | 0.9015 |
| 3 | 0.9906 | 0.9823 | 0.8777 | 0.9135 | 0.9029 | 0.9030 | 0.9030 | |
| 4 | 0.9868 | 0.9805 | 0.9236 | 0.8856 | 0.8988 | 0.8989 | 0.8989 | |
| 5 | 0.9907 | 0.9828 | 0.8505 | 0.9527 | 0.9325 | 0.9835 | 0.9855 | |
| #072 | 2 | 0.9799 | 0.9786 | 0.9068 | 0.9041 | 0.9041 | 0.9041 | 0.9041 |
| 3 | 0.9910 | 0.9821 | 0.9034 | 0.9084 | 0.8985 | 0.8992 | 0.8992 | |
| 4 | 0.9890 | 0.9830 | 0.8809 | 0.8876 | 0.8804 | 0.8666 | 0.8666 | |
| 5 | 0.9917 | 0.9830 | 0.9531 | 0.8881 | 0.8876 | 0.9818 | 0.9825 | |
| #082 | 2 | 0.9836 | 0.9791 | 0.9120 | 0.9091 | 0.9091 | 0.9091 | 0.9091 |
| 3 | 0.9927 | 0.9837 | 0.8852 | 0.8621 | 0.8661 | 0.8849 | 0.8849 | |
| 4 | 0.9869 | 0.9830 | 0.8619 | 0.8479 | 0.8622 | 0.8695 | 0.8695 | |
| 5 | 0.9938 | 0.9860 | 0.9372 | 0.9188 | 0.9105 | 0.9854 | 0.9857 | |
| #092 | 2 | 0.9893 | 0.9887 | 0.9131 | 0.9156 | 0.9131 | 0.9131 | 0.9131 |
| 3 | 0.9948 | 0.9890 | 0.8607 | 0.8751 | 0.8827 | 0.8786 | 0.8786 | |
| 4 | 0.9904 | 0.9865 | 0.9490 | 0.8583 | 0.8514 | 0.8240 | 0.8641 | |
| 5 | 0.9932 | 0.9880 | 0.8684 | 0.8923 | 0.8401 | 0.9880 | 0.9876 | |
| #102 | 2 | 0.9898 | 0.9880 | 0.9383 | 0.9250 | 0.9250 | 0.9250 | 0.9250 |
| 3 | 0.9951 | 0.9892 | 0.8768 | 0.8977 | 0.9097 | 0.9179 | 0.9179 | |
| 4 | 0.9916 | 0.9863 | 0.9256 | 0.9410 | 0.9050 | 0.9871 | 0.9871 | |
| 5 | 0.9967 | 0.9896 | 0.8446 | 0.9180 | 0.9181 | 0.9907 | 0.9895 | |
| #112 | 2 | 0.9923 | 0.9884 | 0.9356 | 0.9403 | 0.9404 | 0.9404 | 0.9404 |
| 3 | 0.9940 | 0.9890 | 0.9147 | 0.9666 | 0.9769 | 0.9863 | 0.9890 | |
| 4 | 0.9946 | 0.9901 | 0.9751 | 0.9824 | 0.9825 | 0.9885 | 0.9896 | |
| 5 | 0.9961 | 0.9913 | 0.9735 | 0.9822 | 0.9830 | 0.9915 | 0.9914 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, Y.; Liu, W.; Zhang, X.; Liu, Z.; Liu, Y.; Wang, G. An Interval Iteration Based Multilevel Thresholding Algorithm for Brain MR Image Segmentation. Entropy 2021, 23, 1429. https://doi.org/10.3390/e23111429
Feng Y, Liu W, Zhang X, Liu Z, Liu Y, Wang G. An Interval Iteration Based Multilevel Thresholding Algorithm for Brain MR Image Segmentation. Entropy. 2021; 23(11):1429. https://doi.org/10.3390/e23111429
Chicago/Turabian StyleFeng, Yuncong, Wanru Liu, Xiaoli Zhang, Zhicheng Liu, Yunfei Liu, and Guishen Wang. 2021. "An Interval Iteration Based Multilevel Thresholding Algorithm for Brain MR Image Segmentation" Entropy 23, no. 11: 1429. https://doi.org/10.3390/e23111429
APA StyleFeng, Y., Liu, W., Zhang, X., Liu, Z., Liu, Y., & Wang, G. (2021). An Interval Iteration Based Multilevel Thresholding Algorithm for Brain MR Image Segmentation. Entropy, 23(11), 1429. https://doi.org/10.3390/e23111429






