Entropy Measurements for Leukocytes’ Surrounding Informativeness Evaluation for Acute Lymphoblastic Leukemia Classification
Abstract
1. Introduction
1.1. Summary of Surveyed Research Works
1.2. The Aim of This Work
1.3. Summary of Our Contributions
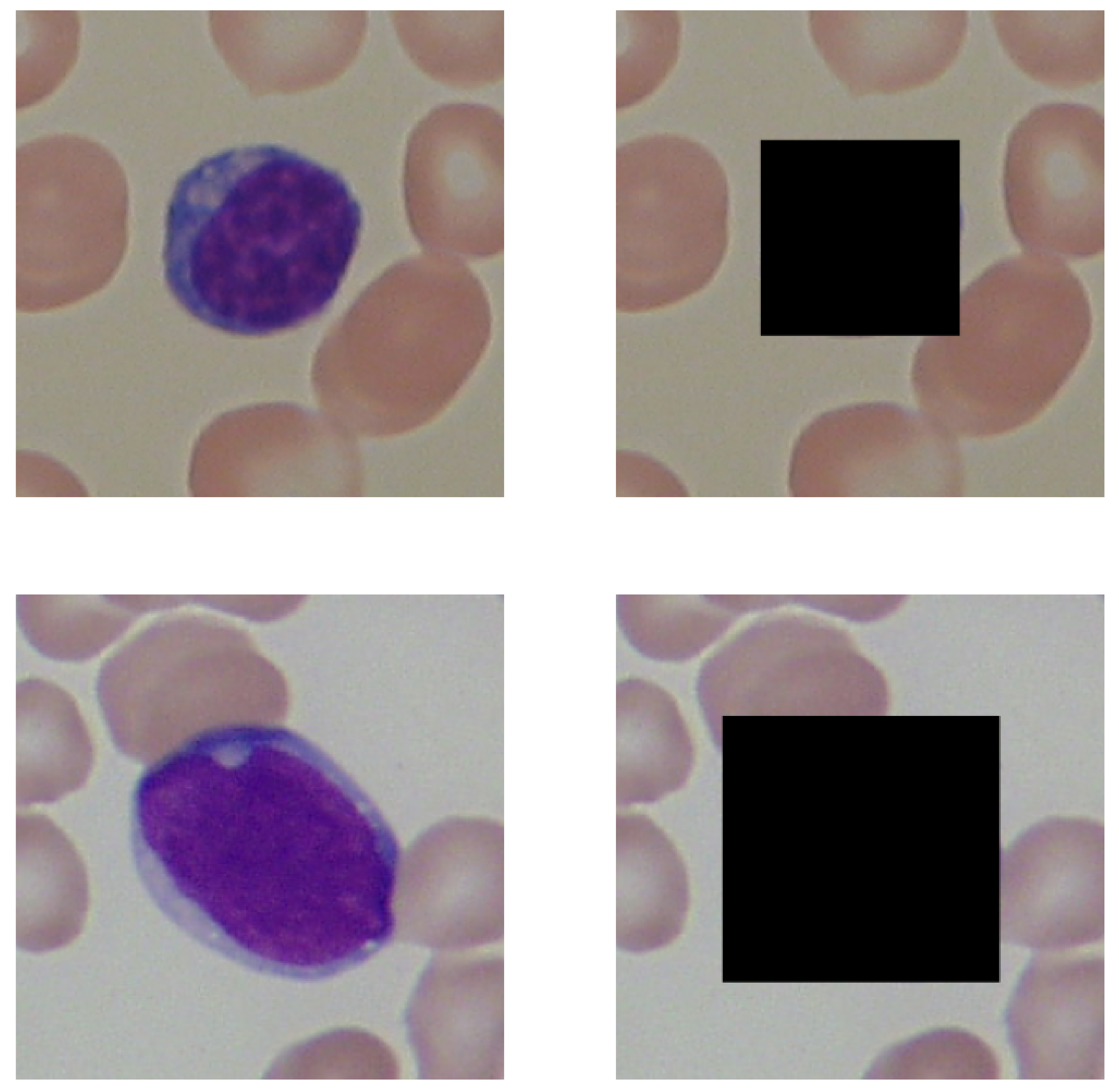
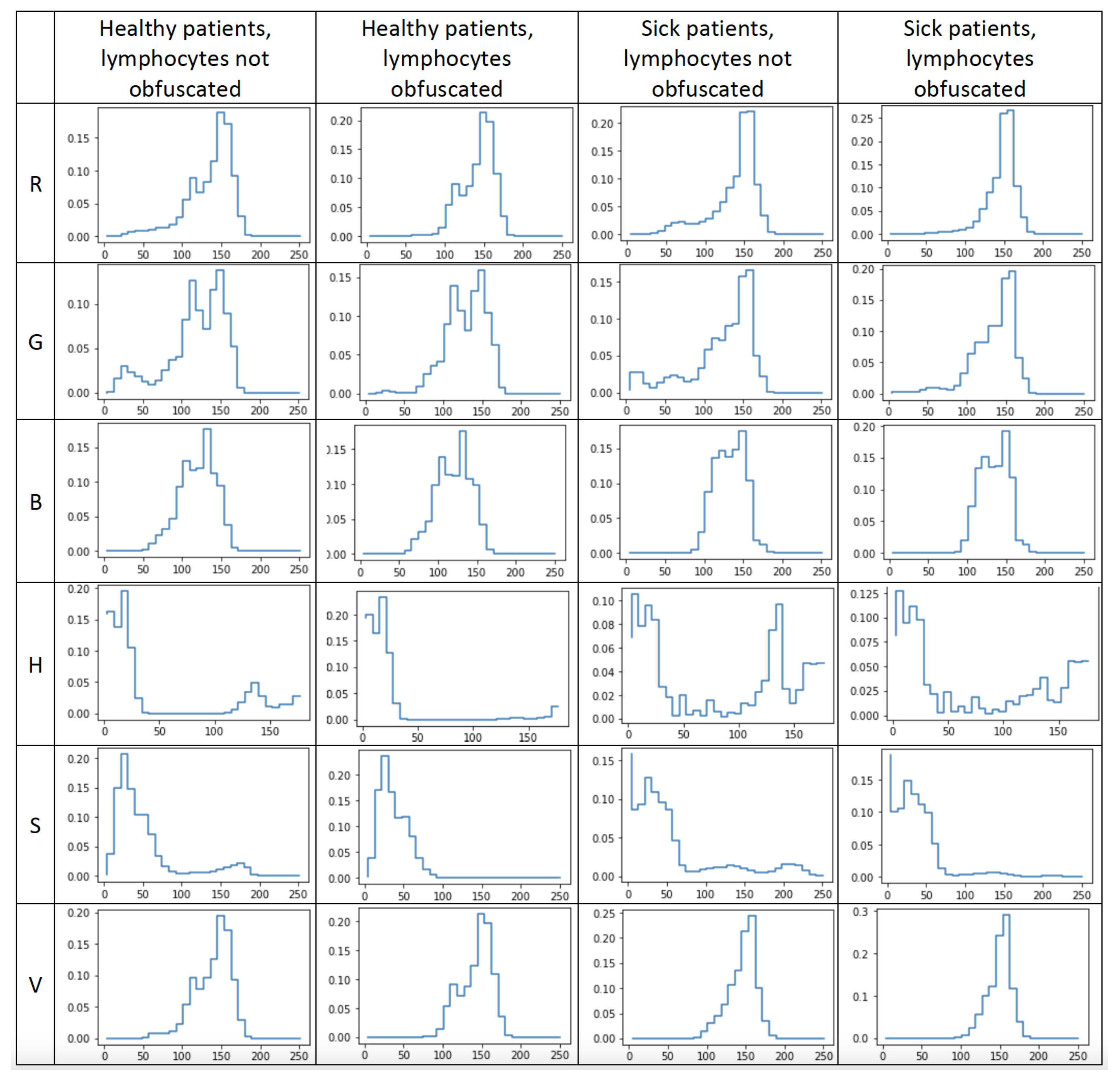
- We examined the influence of lymphocyte obfuscation on acute lymphoblastic leukemia classification to evaluate its surroundings’ informativeness. The hue distribution of lymphocytes’ surroundings processed by the XGBoost algorithm resulted in classification with 93% accuracy.
- We evaluated the informativeness of channels’ value distributions of both the RGB and HSV color encodings. We determined that the channel encoding color green contained the most information, with an XGBoost classification accuracy of 96%. The same evaluation of red and blue color channels resulted in classification accuracies of 87% and 83%, respectively. The hue, saturation, and value channels obtained classification results of 94%, 94%, and 84%, respectively.
- The classification results of the XGBoost algorithm interpreting the distributions of individual channel values resulted in a classification quality similar to the effects of deep learning application on raw images performed by other researchers. As a result, we reduced the amount of input information by three orders of magnitude while achieving comparable results.
- We evaluated the informativeness of the entropy measurements of each channel’s values distribution using the Shannon entropy. The Shannon entropy computed for the hue distribution of images with lymphocytes obfuscated resulted in a classification accuracy of 81% and 68% accuracy when using images without the lymphocytes being obfuscated. The results suggest that lymphocytes’ surroundings contain essential information for acute lymphoblastic leukemia classification.
1.4. Paper Organization
2. Materials and Methods
2.1. ALL-IDB Database
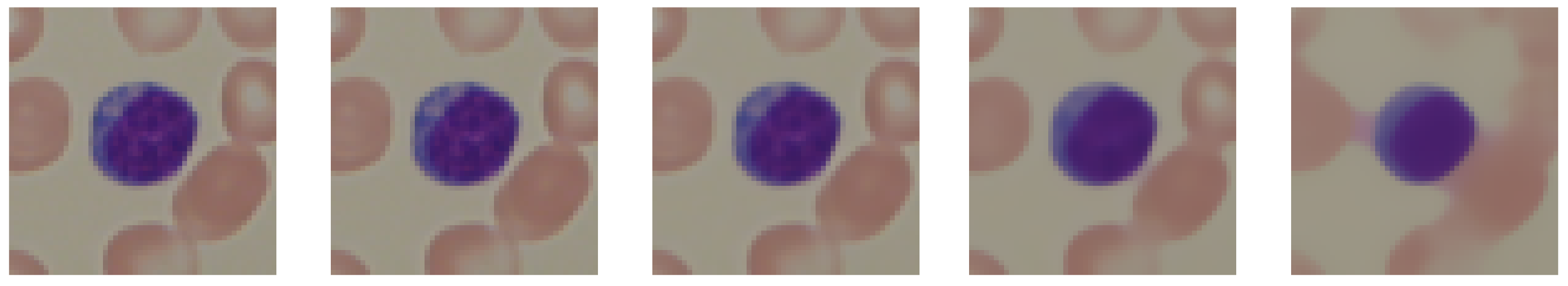
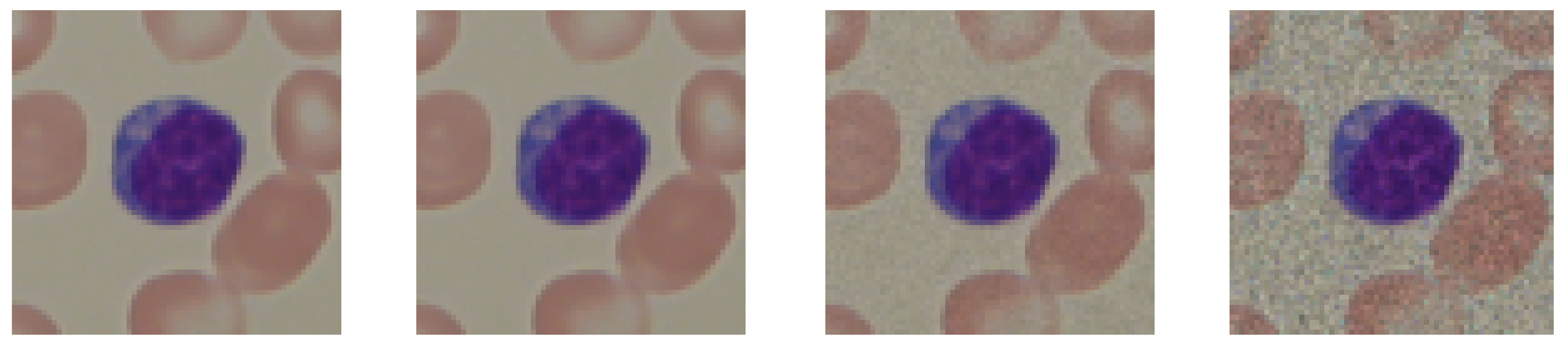
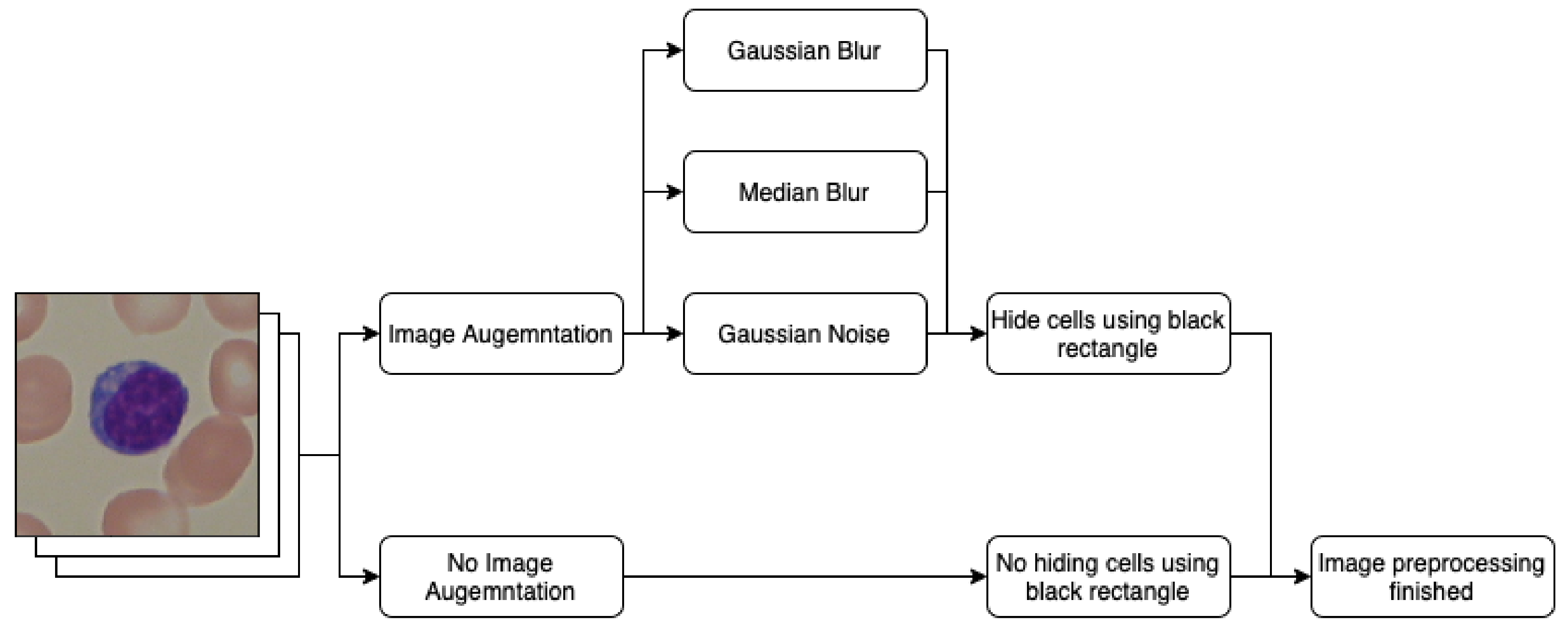
2.2. Image Preprocessing
- Gaussian blur;
- Median blur;
- Gaussian noise.
2.3. Image Vectorization
2.4. Distribution Difference Measurement
2.4.1. Cross-Entropy
2.4.2. Mean Sqaured Error
2.4.3. Algorithm
| Algorithm 1: The mathematical formulation of experimental procedure examining distribution difference measurements |
|
2.5. Shannon Entropy
2.6. Machine Learning Algorithms
2.7. Metrics
- = true positive;
- = true negative;
- = false positive;
- = false negative.
3. Results
3.1. Background Information Measurement
3.2. Comparison with the Literature
3.3. Influence of Data Augmentation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ALL | Acute lymphoblastic leukemia |
| AML | Acute myleoid leukemia |
| CLL | Chronic lymphocytic leukemia |
| CLM | Chronic myleoid leukemia |
| CNN | Convolutional neural network |
References
- Andrade, A.R.; Vogado, L.H.; Veras, R.D.M.S.; Silva, R.R.; Araujo, F.H.; Medeiros, F.N. Recent computational methods for white blood cell nuclei segmentation: A comparative study. Comput. Methods Programs Biomed. 2019, 173, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Labati, R.D.; Piuri, V.; Scotti, F. The Acute Lymphoblastic Leukemia Image Database for Image Processing. In Proceedings of the 2011 18th IEEE International Conference on Image Processing, Brussels, Belgium, 11–14 September 2011. [Google Scholar]
- Mohamed, M.; Far, B.; Guaily, A. An efficient technique for white blood cells nuclei automatic segmentation. In Proceedings of the 2012 IEEE International Conference on Systems, Man, and Cybernetics (SMC), Seoul, Korea, 14–17 October 2012; IEEE: Piscataway, NJ, USA, 2012; pp. 220–225. [Google Scholar]
- Sarrafzadeh, O.; Rabbani, H.; Talebi, A.; Banaem, H.U. Selection of the best features for leukocytes classification in blood smear microscopic images. In Proceedings of the Medical Imaging 2014: Digital Pathology, San Diego, CA, USA, 15–20 February 2014; SPIE: Bellingham, WA, USA, 2014; Volume 9041, pp. 159–166. [Google Scholar]
- Zheng, X.; Wang, Y.; Wang, G.; Liu, J. Fast and robust segmentation of white blood cell images by self-supervised learning. Micron 2018, 107, 55–71. [Google Scholar] [CrossRef] [PubMed]
- Madhloom, H.; Kareem, S.; Ariffin, H.; Zaidan, A.; Alanazi, H.; Zaidan, B. An automated white blood cell nucleus localization and segmentation using image arithmetic and automatic threshold. J. Appl. Sci. 2010, 10, 959–966. [Google Scholar] [CrossRef]
- Arslan, S.; Ozyurek, E.; Gunduz-Demir, C. A color and shape based algorithm for segmentation of white blood cells in peripheral blood and bone marrow images. Cytom. Part A 2014, 85, 480–490. [Google Scholar] [CrossRef]
- Nazlibilek, S.; Karacor, D.; Ercan, T.; Sazli, M.H.; Kalender, O.; Ege, Y. Automatic segmentation, counting, size determination and classification of white blood cells. Measurement 2014, 55, 58–65. [Google Scholar] [CrossRef]
- Prinyakupt, J.; Pluempitiwiriyawej, C. Segmentation of white blood cells and comparison of cell morphology by linear and naïve Bayes classifiers. Biomed. Eng. Online 2015, 14, 63. [Google Scholar] [CrossRef]
- Nasir, A.A.; Mashor, M.; Rosline, H. Unsupervised colour segmentation of white blood cell for acute leukaemia images. In Proceedings of the 2011 IEEE International Conference on Imaging Systems and Techniques, Penang, Malaysia, 17–18 May 2011; IEEE: Piscataway, NJ, USA, 2011; pp. 142–145. [Google Scholar]
- Mohapatra, S.; Samanta, S.S.; Patra, D.; Satpathi, S. Fuzzy based blood image segmentation for automated leukemia detection. In Proceedings of the 2011 International Conference on Devices and Communications (ICDeCom), Ranchi, India, 24–25 February 2011; IEEE: Piscataway, NJ, USA, 2011; pp. 1–5. [Google Scholar]
- Madhukar, M.; Agaian, S.; Chronopoulos, A.T. New decision support tool for acute lymphoblastic leukemia classification. In Proceedings of the Image Processing: Algorithms and Systems X; and Parallel Processing for Imaging Applications II, Burlingame, CA, USA, 23–25 January 2012; SPIE: Bellingham, WA, USA, 2012; Volume 8295, pp. 367–378. [Google Scholar]
- Amin, M.M.; Kermani, S.; Talebi, A.; Oghli, M.G. Recognition of acute lymphoblastic leukemia cells in microscopic images using k-means clustering and support vector machine classifier. J. Med. Signals Sens. 2015, 5, 49. [Google Scholar]
- Sarrafzadeh, O.; Dehnavi, A.M.; Rabbani, H.; Talebi, A. A simple and accurate method for white blood cells segmentation using K-means algorithm. In Proceedings of the 2015 IEEE Workshop on Signal Processing Systems (SiPS), Hangzhou, China, 14–16 October 2015; IEEE: Piscataway, NJ, USA, 2015; pp. 1–6. [Google Scholar]
- Vincent, I.; Kwon, K.R.; Lee, S.H.; Moon, K.S. Acute lymphoid leukemia classification using two-step neural network classifier. In Proceedings of the 2015 21st Korea-Japan Joint Workshop on Frontiers of Computer Vision (FCV), Mokpo, Korea, 28–30 January 2015; IEEE: Piscataway, NJ, USA, 2015; pp. 1–4. [Google Scholar]
- Vogado, L.H.; Veras, R.D.M.S.; Andrade, A.R.; e Silva, R.R.; De Araujo, F.H.; De Medeiros, F.N. Unsupervised leukemia cells segmentation based on multi-space color channels. In Proceedings of the 2016 IEEE International Symposium on Multimedia (ISM), San Jose, CA, USA, 11–13 December 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 451–456. [Google Scholar]
- Kumar, P.; Vasuki, S. Automated diagnosis of acute lymphocytic leukemia and acute myeloid leukemia using multi-SV. J. Biomed. Imaging Bioeng. 2017, 1, 20–24. [Google Scholar]
- Mohammed, E.A.; Mohamed, M.M.; Naugler, C.; Far, B.H. Chronic lymphocytic leukemia cell segmentation from microscopic blood images using watershed algorithm and optimal thresholding. In Proceedings of the 2013 26th IEEE Canadian Conference on Electrical and Computer Engineering (CCECE), Regina, SK, Canada, 5–8 May 2013; IEEE: Piscataway, NJ, USA, 2013; pp. 1–5. [Google Scholar]
- Abdeldaim, A.M.; Sahlol, A.T.; Elhoseny, M.; Hassanien, A.E. Computer-aided acute lymphoblastic leukemia diagnosis system based on image analysis. In Advances in Soft Computing and Machine Learning in Image Processing; Springer: Berlin/Heidelberg, Germany, 2018; pp. 131–147. [Google Scholar]
- Rehman, A.; Abbas, N.; Saba, T.; Rahman, S.I.U.; Mehmood, Z.; Kolivand, H. Classification of acute lymphoblastic leukemia using deep learning. Microsc. Res. Tech. 2018, 81, 1310–1317. [Google Scholar] [CrossRef]
- Prellberg, J.; Kramer, O. Acute lymphoblastic leukemia classification from microscopic images using convolutional neural networks. In ISBI 2019 C-NMC Challenge: Classification in Cancer Cell Imaging; Springer: Berlin/Heidelberg, Germany, 2019; pp. 53–61. [Google Scholar]
- Ahmed, N.; Yigit, A.; Isik, Z.; Alpkocak, A. Identification of leukemia subtypes from microscopic images using convolutional neural network. Diagnostics 2019, 9, 104. [Google Scholar] [CrossRef]
- Guo, Z.; Wang, Y.; Liu, L.; Sun, S.; Feng, B.; Zhao, X. Siamese Network-Based Few-Shot Learning for Classification of Human Peripheral Blood Leukocyte. In Proceedings of the 2021 IEEE 4th International Conference on Electronic Information and Communication Technology (ICEICT), Xi’an, China, 18–20 August 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 818–822. [Google Scholar]
- Abhishek, A.; Jha, R.K.; Sinha, R.; Jha, K. Automated classification of acute leukemia on a heterogeneous dataset using machine learning and deep learning techniques. Biomed. Signal Process. Control 2022, 72, 103341. [Google Scholar] [CrossRef]
- LeCun, Y.; Boser, B.; Denker, J.S.; Henderson, D.; Howard, R.E.; Hubbard, W.; Jackel, L.D. Backpropagation applied to handwritten zip code recognition. Neural Comput. 1989, 1, 541–551. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Xie, S.; Girshick, R.; Dollár, P.; Tu, Z.; He, K. Aggregated Residual Transformations for Deep Neural Networks. arXiv 2016, arXiv:1611.05431. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef]
- Koch, G.; Zemel, R.; Salakhutdinov, R. Siamese neural networks for one-shot image recognition. In Proceedings of the ICML deep learning workshop, Lille, France, 6–11 July 2015; Volume 2. [Google Scholar]
- Wang, Y.; Yao, Q.; Kwok, J.T.; Ni, L.M. Generalizing from a few examples: A survey on few-shot learning. ACM Comput. Surv. 2020, 53, 63. [Google Scholar] [CrossRef]
- Ojala, T.; Pietikainen, M.; Maenpaa, T. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans. Pattern Anal. Mach. Intell. 2002, 24, 971–987. [Google Scholar] [CrossRef]
- Dalal, N.; Triggs, B. Histograms of oriented gradients for human detection. In Proceedings of the 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’05), San Diego, CA, USA, 20–25 June 2005; IEEE: Piscataway, NJ, USA, 2005; Volume 1, pp. 886–893. [Google Scholar]
- Rodrigues, L.F.; Backes, A.R.; Travençolo, B.A.N.; de Oliveira, G.M.B. Optimizing a Deep Residual Neural Network with Genetic Algorithm for Acute Lymphoblastic Leukemia Classification. J. Digit. Imaging 2022, 35, 623–637. [Google Scholar] [CrossRef]
- Hinton, G.; Vinyals, O.; Dean, J. Distilling the Knowledge in a Neural Network. arXiv 2015, arXiv:1503.02531. [Google Scholar]
- Pałczyński, K.; Śmigiel, S.; Gackowska, M.; Ledziński, D.; Bujnowski, S.; Lutowski, Z. IoT Application of Transfer Learning in Hybrid Artificial Intelligence Systems for Acute Lymphoblastic Leukemia Classification. Sensors 2021, 21, 8025. [Google Scholar] [CrossRef]
- Golik, P.; Doetsch, P.; Ney, H. Cross-Entropy vs. Squared Error Training: A Theoretical and Experimental Comparison. In Proceedings of the International Sport and Culture Association, Lyon, France, 25–29 August 2013; pp. 1756–1760. [Google Scholar] [CrossRef]
- Peng, J.; Lee, K.; Ingersoll, G. An Introduction to Logistic Regression Analysis and Reporting. J. Educ. Res. 2002, 96, 3–14. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. arXiv 2016, arXiv:1603.02754. [Google Scholar]



| Ref. | Image Encoding | Methods |
|---|---|---|
| [3] | Grayscale | Arithmetical operations |
| [6] | Grayscale | Otsu threshold, arithmetical operations |
| [7] | RGB | Otsu threshold, region growing |
| [8] | Grayscale | Otsu threshold |
| [9] | RGB | Otsu threshold, arithmetical operations |
| [10] | HSI | K-means, region growing |
| [11] | L*a*b* | K-means |
| [12] | L*a*b* | K-means |
| [13] | HSV | K-means |
| [14] | L*a*b* | K-means, arithmetical operations |
| [15] | L*a*b* | K-means |
| [16] | CMYK + L*a*b* | K-means |
| [17] | HSV | K-means |
| [18] | Grayscale | Region growing, edge detectors |
| [19] | CMYK | Zack’s algorithm |
| [20] | RGB | AlexNet |
| [21] | RGB | ResNeXt50 |
| [22] | RGB | CNN |
| [23] | RGB | Siamese networks |
| [24] | RGB | CNN |
| [33] | RGB | ResNet50 v2 |
| Lymphocytes Obfuscated | Channel | Cross Entropy Acc. | Cross Entropy (Healthy) | Cross Entropy (Sick) | MSE Acc. | MSE (Healthy) | MSE (Sick) |
|---|---|---|---|---|---|---|---|
| False | B | 0.50 | 0.56 | 0.41 | 0.68 | 0.68 | 0.68 |
| True | B | 0.55 | 0.59 | 0.49 | 0.69 | 0.68 | 0.70 |
| False | G | 0.81 | 0.81 | 0.81 | 0.62 | 0.63 | 0.61 |
| True | G | 0.47 | 0.53 | 0.38 | 0.60 | 0.62 | 0.58 |
| False | R | 0.53 | 0.51 | 0.54 | 0.46 | 0.49 | 0.43 |
| True | R | 0.37 | 0.49 | 0.18 | 0.45 | 0.48 | 0.42 |
| False | Hue | 0.85 | 0.86 | 0.83 | 0.77 | 0.79 | 0.73 |
| True | Hue | 0.83 | 0.85 | 0.81 | 0.82 | 0.84 | 0.80 |
| False | Saturation | 0.79 | 0.80 | 0.77 | 0.79 | 0.83 | 0.72 |
| True | Saturation | 0.73 | 0.72 | 0.74 | 0.79 | 0.83 | 0.73 |
| False | Value | 0.45 | 0.51 | 0.36 | 0.50 | 0.51 | 0.48 |
| True | Value | 0.38 | 0.45 | 0.28 | 0.52 | 0.53 | 0.49 |
| Lymphocytes Obfuscated | Channel | Avg. Shannon Entropy (Healthy) | Std. Shannon Entropy (Healthy) | Avg. Shannon Entropy (Sick) | Std. Shannon Entropy (Sick) | Acc | (Healthy) | (Sick) |
|---|---|---|---|---|---|---|---|---|
| False | B | 0.43 | 0.01 | 0.45 | 0.02 | 0.51 | 0.31 | 0.51 |
| True | B | 0.43 | 0.02 | 0.44 | 0.02 | 0.51 | 0.30 | 0.51 |
| False | G | 0.48 | 0.02 | 0.50 | 0.02 | 0.55 | 0.37 | 0.55 |
| True | G | 0.43 | 0.03 | 0.46 | 0.03 | 0.54 | 0.39 | 0.54 |
| False | R | 0.44 | 0.02 | 0.47 | 0.02 | 0.54 | 0.38 | 0.54 |
| True | R | 0.39 | 0.03 | 0.42 | 0.04 | 0.54 | 0.39 | 0.54 |
| False | Hue | 0.63 | 0.03 | 0.71 | 0.07 | 0.68 | 0.70 | 0.65 |
| True | Hue | 0.57 | 0.03 | 0.69 | 0.10 | 0.81 | 0.84 | 0.78 |
| False | Saturation | 0.49 | 0.03 | 0.51 | 0.02 | 0.55 | 0.38 | 0.55 |
| True | Saturation | 0.44 | 0.03 | 0.47 | 0.04 | 0.51 | 0.36 | 0.51 |
| False | Value | 0.43 | 0.02 | 0.44 | 0.02 | 0.51 | 0.30 | 0.51 |
| True | Value | 0.39 | 0.03 | 0.41 | 0.03 | 0.52 | 0.34 | 0.53 |
| Lymphocytes Obfuscated | Channel | XGBoost Acc | XGBoost (Healthy) | XGBoost (Sick) | Logistic Regression Acc. | Logistic Regression (Healthy) | Logistic Regression (Sick) |
|---|---|---|---|---|---|---|---|
| False | B | 0.83 | 0.83 | 0.83 | 0.53 | 0.32 | 0.53 |
| True | B | 0.82 | 0.82 | 0.82 | 0.55 | 0.37 | 0.55 |
| False | G | 0.96 | 0.96 | 0.96 | 0.50 | 0.28 | 0.51 |
| True | G | 0.86 | 0.87 | 0.85 | 0.50 | 0.28 | 0.51 |
| False | R | 0.87 | 0.87 | 0.86 | 0.47 | 0.23 | 0.49 |
| True | R | 0.80 | 0.80 | 0.79 | 0.48 | 0.28 | 0.49 |
| False | Hue | 0.94 | 0.95 | 0.94 | 0.57 | 0.41 | 0.57 |
| True | Hue | 0.93 | 0.94 | 0.93 | 0.60 | 0.46 | 0.59 |
| False | Saturation | 0.94 | 0.94 | 0.94 | 0.54 | 0.32 | 0.54 |
| True | Saturation | 0.88 | 0.89 | 0.88 | 0.55 | 0.36 | 0.55 |
| False | Value | 0.84 | 0.84 | 0.84 | 0.48 | 0.25 | 0.49 |
| True | Value | 0.80 | 0.81 | 0.80 | 0.50 | 0.30 | 0.50 |
| Article | Input Data | Input Size | Obfuscation | Parameters | Accuracy | Precision | Recall | |
|---|---|---|---|---|---|---|---|---|
| This work | Green dist. | 51 | False | 6.4 K | 0.960 | 0.960 | 0.960 | 0.959 |
| This work | Hue dist. | 36 | True | 6.4 K | 0.935 | 0.935 | 0.935 | 0.934 |
| [35] | RGB image | 150 K | False | 3.4 M | 0.948 | 0.950 | 0.951 | 0.948 |
| [33] | RGB image | 150 K | False | 25 M | 0.985 | 0.986 | 0.985 | 0.984 |
| Lymphocytes Obfuscated | Kernel | XGBoost Acc. | Logistic Regression Acc. | Cross-Entropy Acc.k | MSE Acc. | Shannon Acc. |
|---|---|---|---|---|---|---|
| False | 51 | 0.93 | 0.60 | 0.79 | 0.74 | 0.64 |
| False | 21 | 0.93 | 0.58 | 0.79 | 0.77 | 0.69 |
| False | 0 | 0.94 | 0.57 | 0.85 | 0.77 | 0.68 |
| False | 9 | 0.95 | 0.58 | 0.83 | 0.78 | 0.69 |
| False | 3 | 0.95 | 0.57 | 0.83 | 0.77 | 0.69 |
| True | 51 | 0.91 | 0.65 | 0.82 | 0.83 | 0.79 |
| True | 21 | 0.93 | 0.63 | 0.78 | 0.82 | 0.82 |
| True | 9 | 0.93 | 0.62 | 0.83 | 0.81 | 0.82 |
| True | 3 | 0.93 | 0.61 | 0.82 | 0.82 | 0.81 |
| True | 0 | 0.93 | 0.60 | 0.83 | 0.82 | 0.81 |
| Lymphocytes Obfuscated | Kernel | XGBoost Acc. | Logistic Regression Acc. | Cross-Entropy Acc.k | MSE Acc. | Shannon Acc. |
|---|---|---|---|---|---|---|
| False | 51 | 0.94 | 0.59 | 0.73 | 0.77 | 0.67 |
| False | 0 | 0.94 | 0.57 | 0.85 | 0.77 | 0.68 |
| False | 9 | 0.95 | 0.58 | 0.81 | 0.79 | 0.69 |
| False | 21 | 0.95 | 0.59 | 0.81 | 0.79 | 0.69 |
| False | 3 | 0.95 | 0.57 | 0.83 | 0.77 | 0.69 |
| True | 51 | 0.91 | 0.63 | 0.72 | 0.80 | 0.80 |
| True | 3 | 0.93 | 0.61 | 0.82 | 0.82 | 0.81 |
| True | 9 | 0.93 | 0.63 | 0.81 | 0.81 | 0.82 |
| True | 0 | 0.93 | 0.60 | 0.83 | 0.82 | 0.81 |
| True | 21 | 0.94 | 0.65 | 0.78 | 0.81 | 0.81 |
| Lymphocytes Obfuscated | Kernel | XGBoost Acc. | Logistic Regression Acc. | Cross-Entropy Acc.k | MSE Acc. | Shannon Acc. |
|---|---|---|---|---|---|---|
| False | 0 | 0.94 | 0.57 | 0.84 | 0.76 | 0.68 |
| False | 0.0001 | 0.95 | 0.57 | 0.84 | 0.76 | 0.68 |
| False | 0.01 | 0.95 | 0.56 | 0.84 | 0.76 | 0.68 |
| False | 0.1 | 0.95 | 0.56 | 0.82 | 0.76 | 0.68 |
| True | 0.01 | 0.93 | 0.59 | 0.82 | 0.82 | 0.80 |
| True | 0.0001 | 0.93 | 0.60 | 0.83 | 0.82 | 0.81 |
| True | 0 | 0.93 | 0.60 | 0.82 | 0.82 | 0.81 |
| True | 0.1 | 0.95 | 0.59 | 0.82 | 0.82 | 0.81 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pałczyński, K.; Ledziński, D.; Andrysiak, T. Entropy Measurements for Leukocytes’ Surrounding Informativeness Evaluation for Acute Lymphoblastic Leukemia Classification. Entropy 2022, 24, 1560. https://doi.org/10.3390/e24111560
Pałczyński K, Ledziński D, Andrysiak T. Entropy Measurements for Leukocytes’ Surrounding Informativeness Evaluation for Acute Lymphoblastic Leukemia Classification. Entropy. 2022; 24(11):1560. https://doi.org/10.3390/e24111560
Chicago/Turabian StylePałczyński, Krzysztof, Damian Ledziński, and Tomasz Andrysiak. 2022. "Entropy Measurements for Leukocytes’ Surrounding Informativeness Evaluation for Acute Lymphoblastic Leukemia Classification" Entropy 24, no. 11: 1560. https://doi.org/10.3390/e24111560
APA StylePałczyński, K., Ledziński, D., & Andrysiak, T. (2022). Entropy Measurements for Leukocytes’ Surrounding Informativeness Evaluation for Acute Lymphoblastic Leukemia Classification. Entropy, 24(11), 1560. https://doi.org/10.3390/e24111560








