MyI-Net: Fully Automatic Detection and Quantification of Myocardial Infarction from Cardiovascular MRI Images
Abstract
:1. Introduction
2. Materials
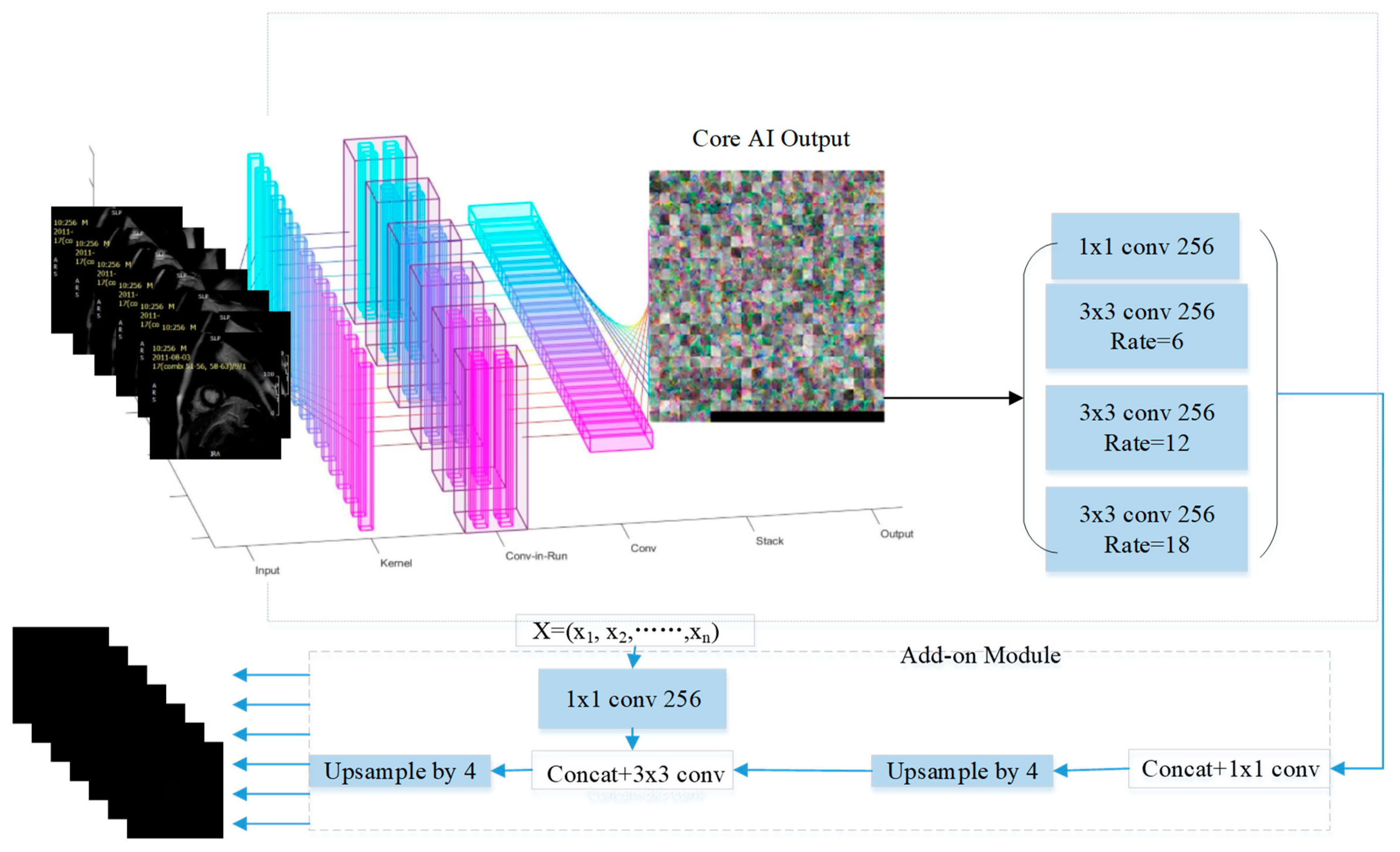
3. Automated Segmentation of Myocardial Infarction: Myocardial Infarction-Net (MyI-NET)
3.1. Feature Extraction via MI-ResNet
3.2. Feature Extraction via MI-MobileNet
3.3. Atrous Spatial Pyramid Pooling
3.4. Weight Matrix
3.5. Data Augmentation
3.6. Performance Metrics
4. Experiments and Results
4.1. Data Preparation
4.2. Experiment Environment
4.3. Segmentation Result Based of Proposed Method
4.4. Segmentation Result Based on State of Art Methods
5. Conclusions
5.1. A Short Summary of Results
5.2. Limitations and Directions for Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tambe, V.; Dogra, M.; Shah, B.S.; Hegazy, H. Traumatic rca dissection as a cause of inferior wall st elevation mi. Chest 2018, 154, 87A–88A. [Google Scholar] [CrossRef]
- Jia, S.; Weng, L.; Zheng, M. Nogo-C causes post-MI arrhythmia through increasing calcium leakage from sarcoplasmic reticulum. J. Mol. Cell. Cardiol. 2020, 140, 43. [Google Scholar] [CrossRef]
- Heart Statistics. Available online: https://www.bhf.org.uk/what-we-do/our-research/heart-statistics (accessed on 25 October 2022).
- Fischer, K.; Obrist, S.J.; Erne, S.A.; Stark, A.W.; Marggraf, M.; Kaneko, K.; Dominik, P.; Guensch, A.T.; Huber, S.G.; Ayaz, A.; et al. Feature tracking myocardial strain incrementally improves prognostication in myocarditis beyond traditional CMR imaging features. JACC Cardiovasc. Imaging 2020, 13, 1891–1901. [Google Scholar] [CrossRef] [PubMed]
- Kuetting, D.L.R.; Homsi, R.; Sprinkart, A.M.; Luetkens, J.; Thomas, D.K.; Schild, H.H.; Dabir, D. Quantitative assessment of systolic and diastolic function in patients with LGE negative systemic amyloidosis using CMR. Int. J. Cardiol. 2017, 232, 336–341. [Google Scholar] [CrossRef]
- Kurt, M.; Yurtseven, H.; Kurt, A. Calculation of the Raman and IR frequencies as order parameters and the damping constant (FWHM) close to phase transitions in methylhydrazinium structures. J. Mol. Struct. 2019, 1181, 488–492. [Google Scholar] [CrossRef]
- Hien, N.D. Comparison of magneto-resonance absorption FWHM for the intrasubband/intersubband transition in quantum wells. Superlattices Microstruct. 2019, 131, 86–94. [Google Scholar] [CrossRef]
- Eitel, I.; Desch, S.; de Waha, S.; Fuernau, G.; Gutberlet, M.; Schuler, G.; Thiele, H. Long-term prognostic value of myocardial salvage assessed by cardiovascular magnetic resonance in acute reperfused myocardial infarction. Heart 2011, 97, 2038–2045. [Google Scholar] [CrossRef]
- Amado, L.C.; Gerber, B.L.; Gupta, S.N.; Rettmann, D.W.; Szarf, G.; Schock, R.; Khurram, N.; Dara, L.K.; Lima, J.A.C. Accurate and objective infarct sizing by contrast-enhanced magnetic resonance imaging in a canine myocardial infarction model. J. Am. Coll. Cardiol. 2004, 44, 2383–2389. [Google Scholar] [CrossRef] [Green Version]
- Flett, A.S.; Hasleton, J.; Cook, C.; Hausenloy, D.; Quarta, G.; Ariti, C.; Vivek, M.; James, C.M. Evaluation of techniques for the quantification of myocardial scar of differing etiology using cardiac magnetic resonance. JACC Cardiovasc. Imaging 2011, 4, 150–156. [Google Scholar] [CrossRef] [Green Version]
- Hsu, L.Y.; Natanzon, A.; Kellman, P.; Hirsch, G.A.; Aletras, A.H.; Arai, A.E. Quantitative myocardial infarction on delayed enhancement MRI. Part I: Animal validation of an automated feature analysis and combined thresholding infarct sizing algorithm. J. Magn. Reson. Imaging 2006, 23, 298–308. [Google Scholar] [CrossRef]
- Tong, Q.; Li, C.; Si, W.; Liao, X.; Tong, Y.; Yuan, Z.; Heng, P.A. RIANet: Recurrent interleaved attention network for cardiac MRI segmentation. Comput. Biol. Med. 2019, 109, 290–302. [Google Scholar] [CrossRef] [PubMed]
- Shan, F.; Gao, Y.; Wang, J.; Shi, W.; Shi, N.; Han, M.; Zhang, Y.; Xue, Z.; Shen, D.; Shi, Y. Lung Infection Quantification of COVID-19 in CT Images with Deep Learning. arXiv 2020, arXiv:2003.04655. [Google Scholar]
- Xu, C.; Xu, L.; Gao, Z.; Zhao, S.; Zhang, H.; Zhang, Y.; Du, X.; Zhao, S.; Ghista, D.; Li, S. Direct Detection of Pixel-Level Myocardial Infarction Areas via a Deep-Learning Algorithm. In Proceedings of the Medical Image Computing and Computer Assisted Intervention (MICCAI), Quebec City, QC, Canada, 11–13 September 2017; pp. 240–249. [Google Scholar]
- Bleton, H.; Margeta, J.; Lombaert, H.; Delingette, H.; Ayache, N. Myocardial Infarct Localization Using Neighbourhood Approximation Forests. In Proceedings of the Statistical Atlases and Computational Models of the Heart (STACOM 2015), Munich, Germany, 9 October 2015; pp. 108–116. [Google Scholar]
- Fahmy, A.S.; Rausch, J.; Neisius, U.; Chan, R.H.; Maron, M.S.; Appelbaum, E.; Menze, B.; Nezafat, R. Automated cardiac MR Scar quantification in hypertrophic cardiomyopathy using deep convolutional neural networks. JACC Cardiovasc. Imaging 2018, 11, 1917–1918. [Google Scholar] [CrossRef] [PubMed]
- Bernard, O.; Lalande, A.; Zotti, C.; Cervenansky, F.; Yang, X.; Heng, P.; Cetin, I.; Lekadir, K.; Camara, O.; Ballester, M.A.G.; et al. Deep Learning Techniques for Automatic MRI Cardiac Multi-Structures Segmentation and Diagnosis: Is the Problem Solved? IEEE Trans. Med. Imaging 2018, 37, 2514–2525. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. In Computer Vision and Pattern Recognition; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Fahmy, A.S.; Neisius, U.; Chan, R.H.; Rowin, E.J.; Manning, W.J.; Maron, M.S.; Nezafat, R. Three-dimensional deep convolutional neural networks for automated myocardial scar quantification in hypertrophic cardiomyopathy: A multicenter multivendor study. Radiology 2020, 294, 52–60. [Google Scholar] [CrossRef]
- Kramer, C.M.; Barkhausen, J.; Bucciarelli-Ducci, C.; Flamm, S.D.; Kim, R.J.; Nagel, E. Standardized cardiovascular magnetic resonance imaging (CMR). J. Cardiovasc. Magn. Reson. 2020, 22, 17. [Google Scholar] [CrossRef]
- New Findings Confirm Predictions on Physician Shortage. Available online: https://www.aamc.org/news-insights/press-releases/new-findings-confirm-predictions-physician-shortage (accessed on 25 October 2022).
- Clinical Radiology UK Workforce Census Report 2018. Available online: https://www.rcr.ac.uk/publication/clinical-radiology-uk-workforce-census-report-2018 (accessed on 25 October 2022).
- Haque, I.R.I.; Neubert, J. Deep learning approaches to biomedical image segmentation. Inform. Med. Unlocked 2020, 18, 100297. [Google Scholar] [CrossRef]
- Luo, L.; Yang, Z.; Cao, M.; Wang, L.; Zhang, Y.; Lin, H. A neural network-based joint learning approach for biomedical entity and relation extraction from biomedical literature. J. Biomed. Inform. 2020, 103, 103384. [Google Scholar] [CrossRef]
- Moradi, M.; Dorffner, G.; Samwald, M. Deep contextualized embeddings for quantifying the informative content in biomedical text summarization. Comput. Methods Programs Biomed. 2020, 184, 105117. [Google Scholar] [CrossRef]
- Nam, J.G.; Park, S.; Hwang, E.J.; Lee, J.H.; Jin, K.N.; Lim, K.Y.; Vu, T.H.; Sohn, J.H.; Hwang, S.; Goo, J.M.; et al. Development and validation of deep learning–based automatic detection algorithm for malignant pulmonary nodules on chest radiographs. Radiology 2019, 290, 218–228. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.H.; McCann, G.; Tyukin, I. Myocardial Infarction Detection and Quantification Based on a Convolution Neural Network with Online Error Correction Capabilities. In Proceedings of the 2020 International Joint Conference on Neural Networks (IJCNN), Glasgow, UK, 19–24 July 2020; pp. 1–8. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef] [Green Version]
- Tyukin, I.Y.; Gorban, A.N.; Sofeykov, K.I.; Romanenko, I. Knowledge Transfer Between Artificial Intelligence Systems. Front. Neurorobotics 2018, 12, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L. MobileNetV2: Inverted Residuals and Linear Bottlenecks. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Li, H.; Qi, F.; Shi, G.; Lin, C. A multiscale dilated dense convolutional network for saliency prediction with instance-level attention competition. J. Vis. Commun. Image Represent. 2019, 64, 102611. [Google Scholar] [CrossRef]
- Friston, K.J.; Rosch, R.; Parr, T.; Price, C.; Bowman, H. Deep temporal models and active inference. Neurosci. Biobehav. Rev. 2018, 77, 388–402. [Google Scholar] [CrossRef] [PubMed]
- Tyukin, I.Y.; Gorban, A.N.; Green, S.; Prokhorov, D. Fast construction of correcting ensembles for legacy artificial intelligence systems: Algorithms and a case study. Inf. Sci. 2019, 485, 230–247. [Google Scholar] [CrossRef] [Green Version]
- Gorban, A.N.; Golubkov, A.; Grechuk, B.; Mirkes, E.M.; Tyukin, I.Y. Correction of AI systems by linear discriminants: Probabilistic foundations. Inf. Sci. 2018, 466, 303–322. [Google Scholar] [CrossRef] [Green Version]
- Poliak, M.; Tomicová, J.; Jaśkiewicz, M. Identification the risks associated with the neutralization of the CMR consignment note. Transp. Res. Procedia 2020, 44, 23–29. [Google Scholar] [CrossRef]
- Gorban, A.N.; Burton, R.; Romanenko, I.; Tyukin, I.Y. One-trial correction of legacy AI systems and stochastic separation theorems. Inf. Sci. 2019, 484, 237–254. [Google Scholar] [CrossRef] [Green Version]
















| Variables | Values | Number | Rate |
|---|---|---|---|
| Stemi | 52 | 17.28% | |
| Non-stemi | 249 | 82.72% | |
| Gender | Male | 172 | 57.14% |
| Female | 129 | 42.86% | |
| Age at MRI | 90–99 | 3 | 1% |
| 80–89 | 11 | 3.65% | |
| 70–79 | 44 | 14.62% | |
| 60–69 | 77 | 25.58% | |
| 50–59 | 74 | 24.58% | |
| 49–49 | 56 | 18.60% | |
| 30–39 | 35 | 11.63% | |
| 20–29 | 1 | 0.33% | |
| Average | 57 | Std | 13.67 |
| Name | Parameters |
|---|---|
| Training algorithm | Data |
| Learn rate drop period | 10 |
| Learn rate drop factors | 3 |
| Initial learn rate | |
| Max epochs | 50 |
| Mini batch size | 10 |
| Execute environment | GPU |
| Validation patience | 4 |
| Type | Weight |
|---|---|
| Training algorithm | SGDM |
| Learn rate drop period | 10 |
| Learn rate drop factors | 3 |
| Initial learn rate | |
| Max epochs | 50 |
| Mini batch size | 10 |
| Execute environment | GPU |
| Validation patience | 4 |
| Model | Global Accuracy | Mean Accuracy | wIoU | Bfscore |
|---|---|---|---|---|
| MI-MobileNet-AC | 0.9569 | 0.8202 | 0.9463 | 0.5351 |
| MI-ResNet50-AC | 0.9738 | 0.8601 | 0.9647 | 0.6446 |
| MI-ResNet18-AC | 0.9679 | 0.8483 | 0.9584 | 0.5839 |
| Category | LGE | Blood | Muscle | Background | |
|---|---|---|---|---|---|
| MI-ResNet50-AC | Accuracy | 0.6429 | 0.8402 | 0.8779 | 0.9686 |
| bfscore | 0.4634 | 0.6837 | 0.4022 | 0.8552 | |
| MI-ResNet18-AC | Accuracy | 0.7441 | 0.8255 | 0.8511 | 0.9724 |
| bfscore | 0.4221 | 0.6226 | 0.4187 | 0.8559 | |
| MI-MobileNet-AC | Accuracy | 0.4245 | 0.8809 | 0.8567 | 0.9664 |
| bfscore | 0.3669 | 0.5996 | 0.3729 | 0.8411 |
| Target Class | |||||
|---|---|---|---|---|---|
| LGE | Blood | Muscle | Background | ||
| MI-ResNet50-AC Output class | LGE | 0.6429 | 0.1557 | 0.1982 | 0.0031 |
| Blood | 0.0543 | 0.8402 | 0.0980 | 0.0074 | |
| Muscle | 0.0352 | 0.0640 | 0.8779 | 0.0229 | |
| Background | 0 | 0.0016 | 0.0291 | 0.9686 | |
| MI-ResNet18-AC Output class | LGE | 0.7441 | 0.1180 | 0.1371 | 0 |
| Blood | 0.0774 | 0.8255 | 0.0904 | 0.0068 | |
| Muscle | 0.0676 | 0.0621 | 0.8511 | 0.0193 | |
| Background | 0.0023 | 0.0023 | 0.0230 | 0.9724 | |
| MI-MobileNet-AC Output class | LGE | 0.4245 | 0.3429 | 0.2326 | 0 |
| Blood | 0.0242 | 0.8809 | 0.0870 | 0.0079 | |
| Muscle | 0.0266 | 0.0964 | 0.8567 | 0.0203 | |
| Background | 0.0011 | 0.0051 | 0.0273 | 0.9664 | |
| Model | Training Time Cost |
|---|---|
| MI-MobileNet-AC | 24′1″ |
| MI-ReNet50-AC | 57′35″ |
| MI-ResNet18-AC | 24′50″ |
| Model | Global Accuracy | Mean Accuracy | wIoU | Bfscore |
|---|---|---|---|---|
| CNN | 0.6021 | 0.5632 | 0.4367 | 0.1574 |
| MI-ResNet50-AC | 0.9738 | 0.8601 | 0.9647 | 0.6446 |
| Unet | 0.6332 | 0.6222 | 0.6117 | 0.1626 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Abdelaty, A.M.S.E.K.; Parke, K.; Arnold, J.R.; McCann, G.P.; Tyukin, I.Y. MyI-Net: Fully Automatic Detection and Quantification of Myocardial Infarction from Cardiovascular MRI Images. Entropy 2023, 25, 431. https://doi.org/10.3390/e25030431
Wang S, Abdelaty AMSEK, Parke K, Arnold JR, McCann GP, Tyukin IY. MyI-Net: Fully Automatic Detection and Quantification of Myocardial Infarction from Cardiovascular MRI Images. Entropy. 2023; 25(3):431. https://doi.org/10.3390/e25030431
Chicago/Turabian StyleWang, Shuihua, Ahmed M. S. E. K. Abdelaty, Kelly Parke, Jayanth Ranjit Arnold, Gerry P. McCann, and Ivan Y. Tyukin. 2023. "MyI-Net: Fully Automatic Detection and Quantification of Myocardial Infarction from Cardiovascular MRI Images" Entropy 25, no. 3: 431. https://doi.org/10.3390/e25030431
APA StyleWang, S., Abdelaty, A. M. S. E. K., Parke, K., Arnold, J. R., McCann, G. P., & Tyukin, I. Y. (2023). MyI-Net: Fully Automatic Detection and Quantification of Myocardial Infarction from Cardiovascular MRI Images. Entropy, 25(3), 431. https://doi.org/10.3390/e25030431








