A Gene-Based Algorithm for Identifying Factors That May Affect a Speaker’s Voice
Abstract
1. Introduction
1.1. A genomic-Based Approach to Detect the Existence of Biomarkers
- FOXP2: This gene codes for a protein called “Forkhead Box P2”, which is involved in the development and function of the brain, including the areas responsible for language and speech.
- TAFT: This gene codes for a protein called TAFT1, which is involved in the development and function of the larynx, a structure in the throat that is involved in vocal production.
- OTOF: This gene codes for a protein called Otoferlin, which is involved in the development and function of the auditory system, including the inner ear. Feedback from this system greatly influences vocal production.
- MYO15A: This gene codes for a protein called Myosin XVa, which is involved in the development and function of the auditory system, including the hair cells in the inner ear.
- SEMA3A: This gene codes for a protein called Semaphorin 3A, which is involved in the development and function of the auditory system, including the auditory nerve.
- Formalize the methodology to find a link between an influencing factor and this gene;
- Validate the methodology;
- Demonstrate its predictive potential.
1.2. Anomalies in Speech Production
- Absence of speech: Phrases (in clinical/scientific literature) referring to (a) no development of speech capabilities, (b) no expressive speech, which is mostly limited to vocalizations, or (c) almost absent speech with a severely limited vocabulary (0–4 words).
- Apraxia: Phrases referring to difficulty using language correctly while speaking, leading to speaking and communication difficulties.
- Delayed speech: Phrases referring to developmental delay, the retarded development of the ability to speak, or the retarded acquisition of language skills and communication skills (ability to use a vocabulary correctly to communicate in a cogent manner).
- Dysarthric speech or dysarthria: Phrases referring to speaking problems resultant from damaged, paralyzed, or weakened muscles of the articulators caused by motor problems. Dysarthria results in slurred words, poor phonation, etc. The speaker uses vocabulary as in normal speech but finds it difficult to move the articulators (tongue, lips, jaw, etc.) correctly to form the proper sounds to utter the words.
- Idiosyncratic speech: Phrases referring to poor conformance to cogent language or incoherent language with articulation abnormalities.
- Impaired speech: Phrases referring to poor articulation and phonation, as well as difficulties that result in sparse and disfluent speech.
2. Methodology
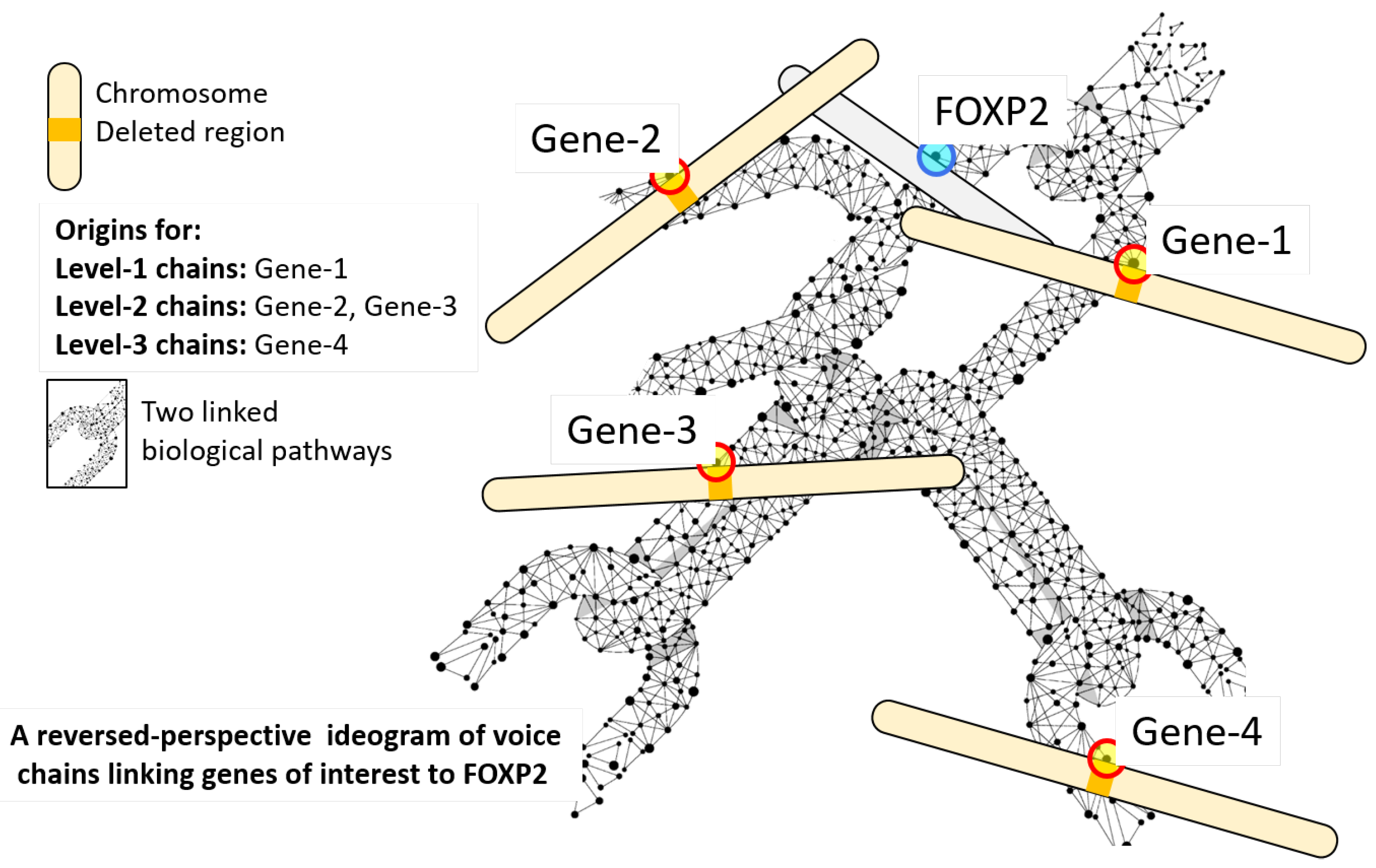
2.1. Voice Chains
| Algorithm 1: Pseudocode for a breadth-first algorithm for computing the set of chainlink genes that form level 1 and level 2 voice chains for FOXP2. |
 |
2.2. Ensemble Analysis
3. Analysis
4. Inferences
4.1. Voice Chains as Predictors of Speech Characteristics
4.2. Voice Chains as Information-Carrying Entities
- Level-1 voice chains co-occur with speech problems 100% of the time.
- For all instances where level-1 voice chains are present, severe symptoms occur 100% of the time (impaired, delayed or absent speech).
- For all instances of syndromes with no effect on speech (i.e., where normal is not just one of a range of other speech symptoms), level-1 chains are absent 100% of the the time.
4.3. Why Are There No Instances of Missing Voice Chains?
4.4. Ancillary Observations
- For each syndrome, some genes have been identified as largely important—i.e., these are implicated largely for the syndrome’s effect on the individual. Of the syndromes for which there is information about implicated genes, we see that in only 8 syndromes (2p16.1–p15, 2q23.1, 9p24.3, 11q23, 13q12.3, 17q23.1–q23.2, 19p13.13, and Yq11), none of the implicated genes appear in the two levels of voice chains shown. In all other cases, the implicated genes impact FOXP2 pathways and are likely to have a bearing on speech anomalies. We have noted earlier that FOXP2 is not the only gene known to be related to voice production. If we had chosen some other gene as an example in this paper (instead of FOXP2), it is likely that the implicated genes for the 8 exceptions mentioned above would appear as chainlink genes (while some others may not). This hypothesis can be easily tested in corresponding experiments.
- Identifying candidate genes for further investigation: Using only chainlink genes that appear on level-1 chains as illustrative examples (see Table A1 in the Appendix A for reference), we see that voice chains can be useful in identifying candidate genes for further investigation in the context of speech issues. Some examples are given below. The likely candidates are written in parentheses, while the already implicated genes are indicated in bold:
- 1p36 (ARID1A): Although not implicated for this syndrome in studies so far, ARID1A is located in 1p36.11, a region frequently deleted in human cancers [69]. Disruption in its function may lead to the co-occurrence of oncological and speech issues. This hypothesis is verifiable.
- 5q35.3 (NSD1): The gene NSD1 appears in a level-1 chain and is also an implicated gene. Ideally, this should not be a candidate for further investigation. However, paradoxically, while effects on speech are expected, the literature reports normal speech for some subjects for this case. This may be a result of biased sampling (the more severe cases may not be conducive to life due to other concurrent severe symptoms, which is a common occurrence in microdeletion syndromes; in some cases, only mosaic individuals survive). This warrants some investigation.
- 11p15.5 (HRAS): Although not implicated, and although two studies cited under OMIM: 130650 for this syndrome explicitly mention HRAS as not significant, HRAS has nevertheless been independently found to be extremely significant in RASopathy and cancer studies, e.g., [70]. Its role in this syndrome needs to be re-evaluated given its influence on 347 biological pathways and its strong influence on speech.
- 16p11.2 and 16p12.2–16p11.2 (SRCAP): Although not implicated, it connects to only one other pathway in the ensemble, and that is the ACC pathway of FOXP2. The effects on speech are expected to be strong if this gene is aberrant. This gene may be implicated in further investigations.
- 17p13.1 (KDM6B): Speech is absent in this syndrome. The gene TP53 is implicated, which also appears at level-1 and is associated with 206 pathways. KDM6B is the only other gene in the level-1 voice chains and connects to only 8 other pathways. It is likely that this gene also plays a strong role in influencing speech and merits investigation.
- 17q12 (ERBB2): The gene ERBB2 is associated with 124 pathways. It is a well-known oncogene [71], in that perturbations in its function have been observed to have deleterious effects. If it is also connected to FOXP2, then its appearance in the voice chain allows a surprising hypothesis—that biomarkers of some oncological conditions may also be present in voice.
- 19p13.3 (MAP2K2,UHRF1): MAP2K2 and URHF1 are not implicated. However their appearance as level-1 chainlink genes warrants investigation, especially for MAP2K2, which influences 257 pathways. Prompted by this, a literature search did reveal that MAP2K2 has been implicated in this syndrome recently [72], although this is not on the OMIM records, which were largely consulted for this study.
- 22q13.3 (BRD1): The gene BRD1 is not implicated and appears in 9 pathways only, but the effect on speech is severe in this syndrome. This warrants the investigation of BRD1 independently in relation to speech characteristics. A literature search reveals that BRD1 is indeed strongly associated with brain development and susceptibility to both schizophrenia and bipolar affective disorder [73], and consequent effects on speech are highly likely.
- Xp11.22 (SMC1A): Although SMC1A is not implicated, it appears in 33 pathways. The speech issues are severe and the gene warrants investigation for this effect. A recent report in the literature has implicated it in severe intellectual disability and therapy-resistant epilepsy in females [74]. The former is known to be associated with severe speech anomalies.
- Xp11.3 (KDM6A): Although not implicated, KDM6A warrants investigation. In the literature, it is independently known to be associated with delayed speech and psychomotor development [75].
- Expression of speech characteristics: The observation that deletions of genes on all chromosomes ultimately results in the expression of speech anomalies carries significance. From a much broader perspective, this suggests that the effect on speech may be supported by the action of multiple concurrent biological pathways. There may be no single gene or genes (on select chromosomes) that may code for speech capabilities per se, and FOXP2 may be one of a few genes that may consolidate and regulate the speech- and language-related emergent effects. It may be that genes directly code for structural elements in the range of phenotypes, while other properties, such as speech and language abilities, are emergent from the coordination of these (and epigenetic) factors.A more prosaic argument for this can also be presented. Within the ensemble of syndromes analyzed, there are three kinds of of cause-and-effect relationships: (a) syndromes with physical structures of the vocal tract (e.g., craniofacial anomalies that include cleft palate, changes in lip shape, etc.), which adversely affect the biomechanical aspects of voice and speech production, (b) syndromes in which auditory and motor functions are compromised, and (c) syndromes that affect the normal functions of the brain, causing cognitive, learning, memory, and other issues that are, in turn, likely to lead to speech problems. In no case do we see only speech aberrations in isolation of these factors. The associations between speech and other expressed factors have, in fact, been ubiquitously observed, e.g., [48]. This may support the hypothesis that speech abilities are likely to be emergent from an ensemble of factors (including other phenotypes), rather than expressed directly by any “speech” gene.
5. Conclusions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Appendix A
- In each row, the top left entry is the microdeletion syndrome.
- Below this, on the left, is the OMIM record for the cytogenetic region of the syndrome.
- Below the OMOM record, the common names by which the syndrome is referred to in the medical literature are listed.
- Below the list of common names, sets of chainlink genes that form level-1 and level-2 voice chains (denoted as and ) respectively, found by the proposed algorithm are listed. These sets are listed only if they are present.
- For each voice chain listed, the first entry denotes the level of the voice chain. Following this is the list of chainlink genes that belong to the corresponding microdeletion region, which are linked to FOXP2 through the corresponding voice chain level.
- For each gene, the number of pathways that a gene connects to (in general, inclusive of connections to the ACC pathway of FOXP2) is written as its subscript.
- All chainlink genes are not named. For brevity, the first 10 genes with the greatest number of links are listed, and the total number of the rest of the genes (total chainlink gene count) is indicated. At the end, the genes with a single pathway link are explicitly listed. They are included in the total chainlink gene count mentioned above. Thus, for example, the level-2 voicechain for the syndrome 2p16.1–p15 has a total chainlink gene count of 17, including all genes that are explicitly listed in the table.
- In each row, the genes in the voice chains that have been implicated for the syndrome’s phenotypic expression in prior studies are shown in parentheses on the top right. The genes that are also present as chainlink genes in the voicechains found for the syndrome are shown in bold font.
- The second column in this table lists the corresponding speech characteristics, with references. Where no reference is cited, the information is found in the OMIM record for the syndrome (where possible, OMIM references are used for brevity).
| Syndrome | (Implicated Genes from Microarray Studies) | Reported Effects on Speech | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Other Information | ||||||||||||||||||||||||||||||
| Voice Chains and Their Member Chainlink Genes | ||||||||||||||||||||||||||||||
| Chromosome 1 deletions | ||||||||||||||||||||||||||||||
| 1p36 | (Multiple incl. RERE, SPEN) | Delayed [9] or absent speech | ||||||||||||||||||||||||||||
| OMIM: 607872; Cytogenetic location: 1p36; Genomic coordinates (GRCh38): 1:23,600,000–27,600,000 Names: Chromosome 1p36 deletion sydrome; monosomy 1p36 syndrome ARID1A : PIK3CD MTOR GNB1 CDC42 CASP9 RPS6KA1 PRKCZ CNKSR1 SFN DVL1 SSU72 RBP7 PLEKHN1 PLEKHM2 PEX10 NBL1 MTCO3P12 MMP23B MMP23A HSPB7 HMGN2 GPR3 EXOSC10 DISP3 CROCCP2 CAMTA1 AHDC1 ACAP3 | ||||||||||||||||||||||||||||||
| 1q21.1–q21.2 | (Multiple incl. RBM8A , GJAS ) | Delayed [10] or Impaired [11] speech | ||||||||||||||||||||||||||||
| OMIM: 612474/274000, Cytogenetic location: 1q21.1 Genomic coordinates (GRCh38): 1:143,200,000–147,500,000
Names: Thrombocytopenia with absent radius (TAR) syndrome (OMIM 274000) : H4C14 H4C15 H2BC21 PRKAB2 H3C15 H3C14 H3C13 H2AC20 H2AC19 H2AC18 | ||||||||||||||||||||||||||||||
| 1q41–q42 | (Multiple incl. DISP1, HPE10, LEFTY1, LEFTY2, WDR26, TSEN2, BPNT1) | Apraxia [12] | ||||||||||||||||||||||||||||
| OMIM: 612530; Cytogenetic location: 1q41–q42 Genomic coordinates (GRCh38): 1:214,400,000–236,400,000 Names: Chromosome 1q41–q42 deletion syndrome; holoprosencephaly 10; HPE10 : NUP133 H2BU1 TGFB2 DUSP10 H3-3A WNT3A ARF1 PARP1 MIR3620 PSEN2 DISP1 MIR215 MIR194-1 DNAH14 CDC42BPA | ||||||||||||||||||||||||||||||
| 1q43–q44 | (AKT3, ZBTB18) | Delayed, impaired, or absent speech | ||||||||||||||||||||||||||||
| OMIM: 612337 Names: Mental retardation, autosomal dominant 22; MRD22; chromosome 1q43–q44 deletion syndrome (included); chromosome 1qter deletion syndrome (included) : AKT3 RYR2 ACTN2 CHRM3 MTR FH ADSS2 EXO1 RGS7 KMO FMN2 | ||||||||||||||||||||||||||||||
| Chromosome 2 deletions | ||||||||||||||||||||||||||||||
| 2p16.1–p15 | (Multiple incl. BCL11A) | Dysarthria, apraxia, or impaired speech [13] | ||||||||||||||||||||||||||||
| OMIM: 612513; Cytogenetic location: 2p16.1–p15 Genomic coordinates (GRCh38): 2:54,700,000–63,900,000 Names: Chromosome 2p16.1–p15 deletion syndrome : RPS27A XPO1 UGP2 MDH1 REL CCT4 RTN4 B3GNT2 VRK2 USP34 OTX1 CCDC88A | ||||||||||||||||||||||||||||||
| 2p21 | (Multiple incl. SLC3A1, PREPL) | Apraxia or idiosyncratic speech [14] | ||||||||||||||||||||||||||||
| OMIM: 606407; Cytogenetic location: 2p21 Genomic coordinates (GRCh38): 2:41,500,000–47,500,000 Names: Hypotonia–cystinuria syndrome; cystinuria with mitochondrial disease; homozygous 2p21 deletion syndrome : CALM2 PRKCE SLC3A1 HAAO ATP6V1E2 ABCG5 MSH2 ABCG8 EPAS1 COX7A2L SIX3 EML4 | ||||||||||||||||||||||||||||||
| 2q23.1 | (MBD5) | Delayed or impaired speech | ||||||||||||||||||||||||||||
| OMIM: 156200 Names: Mental retardation, autosomal dominant 1; MRD1; chromosome 2q23.1 deletion syndrome : ORC4 KIF5C | ||||||||||||||||||||||||||||||
| 2q32–q33 | (SATB2) (HOXD cluster and regulatory elements, COL3A1 COL5A2, GTF3C3, CASP8, CASP10) | Absent speech | ||||||||||||||||||||||||||||
| OMIM: 612313 Names: Glass syndrome; GLASS; chromosome 2q32–q33 deletion syndrome; SATB2-associated syndrome : CREB1 STAT1 CASP8 NUP35 ITGAV CD28 SUMO1 AOX1 FZD5 FZD7 TMEFF2 KLF7 HSPE1 DUSP19 DNAH7 | ||||||||||||||||||||||||||||||
| 2q37.3 | (HDAC4) | Impaired speech [15] | ||||||||||||||||||||||||||||
| OMIM: 600430 (2q37); Cytogenetic location: 2q37 Genomic coordinates (GRCh38): 2:236,400,000–242,193,529 Names: Chromosome 2q37 deletion syndrome, brachydactyly–intellectual disability syndrome; Albright hereditary osteodystrophy-like syndrome Type 3 : HDAC4 GPC1 AGXT COL6A3 ACKR3 NEU4 NDUFA10 PRLH PER2 DTYMK TWIST2 ILKAP | ||||||||||||||||||||||||||||||
| Chromosome 3 deletions | ||||||||||||||||||||||||||||||
| 3p13 | (FOXP1) | Delayed, idiosyncratic, and impaired speech and dysarthria (all severe), apraxia [16] | ||||||||||||||||||||||||||||
| OMIM: 613670 Names: Mental retardation with language impairment with or without autistic features : MITF PROK2 PPP4R2 GPR27 EIF4E3 FOXP1 GXYLT2 RYBP | ||||||||||||||||||||||||||||||
| 3q13.31 | (DRD3, ZBTB20 , GAP43, LSAMP) | Impaired [17] or absent speech | ||||||||||||||||||||||||||||
| OMIM: 615433; Cytogenetic location: 3q13.31 Genomic coordinates (GRCh38): 3:113,700,000–117,600,000 Names: Chromosome 3q13.31 deletion syndrome : DRD3 ATP6V1A GAP43 QTRT2 LSAMP | ||||||||||||||||||||||||||||||
| 3q29 | (Multiple incl. PAK2, DLG1) | Delayed speech | ||||||||||||||||||||||||||||
| OMIM: 609425; Cytogenetic location: 3q29 Genomic coordinates (GRCh38): 3:192,600,000–198,295,559 Names: Chromosome 3q29 deletion syndrome; microdeletion 3q29 syndrome; 3qter deletion syndrome : PAK2 DLG1 NCBP2 HES1 RPL35A TFRC RNF168 BDH1 PCYT1A MUC20 FBXO45 | ||||||||||||||||||||||||||||||
| Chromosome 4 deletions | ||||||||||||||||||||||||||||||
| 4p16.3 | (Multiple incl. FGFR3, MSX1) | Delayed [18] or absent speech | ||||||||||||||||||||||||||||
| OMIM: 194190; Cytogenetic location: 4p16.3 Genomic coordinates (GRCh38): 4:0–4,500,000 Names: Wolf–Hirschhorn syndrome; Pitt–Rogers–Danks syndrome; Pitt syndrome; Wittwer syndrome; Dillan 4p syndrome CTBP1 : FGFR3 TNIP2 NELFA MIR943 CTBP1 ADRA2C NSD2 GRK4 DGKQ PDE6B HAUS3 SLC26A1 SLBP ATP5ME RNF4 CPLX1 RGS12 ZFYVE28 PCGF3 NSG1 MXD4 ABCA11P | ||||||||||||||||||||||||||||||
| 4q21 | (Multiple) | Delayed or absent speech; impaired speech [19,20] | ||||||||||||||||||||||||||||
| OMIM: 613509; Cytogenetic location: 4q21 Genomic coordinates (GRCh38): 4:86,000,000–87,100,000 Names: Chromosome 4q21 deletion syndrome : MAPK10 FGF5 NUP54 PAQR3 PRKG2 CXCL10 CXCL9 ABRAXAS1 SEC31A CXCL11 SHROOM3 SEPTIN11 HNRNPDL G3BP2 BMP2K AFF1 | ||||||||||||||||||||||||||||||
| Chromosome 5 deletions | ||||||||||||||||||||||||||||||
| 5p (5p15.2 and/or (5p15.3 or 5p15.33)) | (Multiple incl. TERT, CTNND2) | Delayed or absent speech and apraxia [21] | ||||||||||||||||||||||||||||
| OMIM: 123450 Names: Chromosome 5p deletion syndrome; cri-du-chat syndrome; cat cry syndrome; Lejeune syndrome; 5p monosomy; partial monosomy 5 : ADCY2 SLC6A3 SDHA TERT TRIO MTRR LPCAT1 CEP72 SRD5A1 SLC9A3 OTULINL CTNND2 CLPTM1L | ||||||||||||||||||||||||||||||
| 5q14.3 | (MEF2C) | Absent speech | ||||||||||||||||||||||||||||
| OMIM: 612881 (distal version 5q14.3–q15); Cytogenetic location: 5q14.3–q15 Genomic coordinates (GRCh38): 5:83,500,000–98,900,000 Names: Distal chromosome 5q14.3 deletion syndrome; periventricular heterotopia associated with chromosome 5q deletion; periventricular nodular heterotopia 5; PVNH5 : RASA1 CCNH MEF2C POLR3G COX7C MIR9-2 EDIL3 | ||||||||||||||||||||||||||||||
| 5q33.1 | (RPS14) | Dysarthria [22] | ||||||||||||||||||||||||||||
| OMIM: 153550 Names: Chromosome 5q deletion syndrome; 5q syndrome; refractory macrocytic anemia due to 5q deletion; MAR : RPS14 GPX3 DCTN4 SPARC SLC36A1 NMUR2 CD74 NDST1 GM2A TNIP1 SYNPO IRGM | ||||||||||||||||||||||||||||||
| 5q35.3 | (NSD1) | Normal [23] or delayed speech | ||||||||||||||||||||||||||||
| OMIM: 117550 Names: Sotos syndrome 1; Sotos1; Nevo syndrome; cerebral gigantism, Nevo type; chromosome 5q35 deletion syndrome NSD1 : MAPK9 LTC4S F12 SQSTM1 MAML1 RACK1 FLT4 CANX GRK6 GRM6 CLK4 | ||||||||||||||||||||||||||||||
| Chromosome 6 deletions | ||||||||||||||||||||||||||||||
| 6pter–p24 | (Multiple incl. FOXC1, GMDS) | Delayed speech | ||||||||||||||||||||||||||||
| OMIM: 612582; Cytogenetic location: 6pter–p24 Genomic coordinates (GRCh38): 6:0–13,400,000 Names: Chromosome 6pter-p24 deletion syndrome : RIPK1 EDN1 F13A1 TFAP2A DSP BMP6 IRF4 TUBB2A GMDS ELOVL2 PAK1IP1 MAK FOXQ1 DUSP22 C6orf201 | ||||||||||||||||||||||||||||||
| 6q25.3 | (Multiple incl. ARID1B) | Delayed speech, apraxia, dysarthria [24] | ||||||||||||||||||||||||||||
| OMIM: N.A. : SLC22A2 GTF2H5 ACAT2 SLC22A1 EZR SOD2 IGF2R SYNJ2 SLC22A3 TCP1 | ||||||||||||||||||||||||||||||
| Chromosome 7 deletions | ||||||||||||||||||||||||||||||
| 7p21 | (TWIST1 (7p21.1)) | Delayed speech [25] | ||||||||||||||||||||||||||||
| OMIM: 101400 Names: Saethre–Chotzen syndrome; acrocephalosyndactyly III; ACS3; ACS III; Chotzen syndrome; acrocephaly, skull asymmetry, and mild syndactyly : RPA3 PRPS1L1 HDAC9 ITGB8 AHR POLR1F NDUFA4 DGKB TWIST1 THSD7A MIOS MEOX2 ARL4A | ||||||||||||||||||||||||||||||
| 7q11.23 | (Multiple incl. ELN, LIMK1, GTF2IRD1, GTF2I) | Normal or delayed speech | ||||||||||||||||||||||||||||
| OMIM: 194050; Cytogenetic location: 7q11.23 Genomic coordinates (GRCh38): 7:72,700,000–77,900,000 Names: Williams syndrome; WS; WMS; chromosome 7q11.23 deletion syndrome, 1.5- to 1.8-MB; Williams–Beuren syndrome; WBS : YWHAG POM121 RFC2 MDH2 LIMK1 HSPB1 STX1A FZD9 NCF1 POR ABHD11 | ||||||||||||||||||||||||||||||
| Chromosome 8 deletions | ||||||||||||||||||||||||||||||
| 8p23.1 | (GATA4) | No significant speech anomaly reports | ||||||||||||||||||||||||||||
| OMIM: 179613 (not exclusive to syndrome) Names: Recombinant chromosome 8 syndrome; REC8 syndrome; chromosome 8q22.1-qter duplication and 8pter-p23.1 deletion; San Luis Valley syndrome : FDFT1 CTSB BLK TNKS GATA4 AGPAT5 ANGPT2 MIR124-1 NEIL2 CLDN23 PINX1 MIR598 | ||||||||||||||||||||||||||||||
| 8q22.1 | (CCNE2, TMEM67, FAM92A1) | Delayed speech | ||||||||||||||||||||||||||||
| OMIM: 608156; Cytogenetic location: 8q22.1 Genomic coordinates (GRCh38): 8:92,300,000–97,900,000 Names: Nablus mask-like facial syndrome; NMLFS; chromosome 8q22.1 deletion syndrome : CCNE2 SDC2 TP53INP1 GDF6 UQCRB PTDSS1 ESRP1 PDP1 NDUFAF6 CDH17 | ||||||||||||||||||||||||||||||
| 8q24.11-q24.13 | (TRPS1, EXT1) | Delayed speech | ||||||||||||||||||||||||||||
| OMIM: 150230; Cytogenetic location: 8q24.11–q24.13 Genomic coordinates (GRCh38): 8:116,700,000–126,300,000 Names: Langer–Giedion syndrome; LGS; chromosome 8q24.1 deletion syndrome; trichorhinophalangeal syndrome type II; TRPS2 : SQLE TAF2 RAD21 MIR3610 TNFRSF11B DERL1 NDUFB9 EXT1 SLC30A8 FBXO32 WASHC5 FAM91A1 DSCC1 | ||||||||||||||||||||||||||||||
| Chromosome 9 deletions | ||||||||||||||||||||||||||||||
| 9p24.3 | (DMRT1, DMRT2) | Delayed speech, dysarthria, apraxia [26] | ||||||||||||||||||||||||||||
| OMIM: 154230; Cytogenetic location: 9p24.3 Genomic coordinates (GRCh38): 9:0–2,200,000 Names: 46,XY sex reversal 4; SRXY4; 46,XY gonadal dysgenesis, partial or complete, with 9p24.3 deletion; chromosome 9p24.3 deletion syndrome SMARCA2 : SMARCA2 DOCK8 WASHC1 | ||||||||||||||||||||||||||||||
| 9q34.3 | (EHMT1) | Delayed speech [27], Apraxia [28] and Absent speech | ||||||||||||||||||||||||||||
| OMIM: 610253 Names: Kleefstra syndrome; 9q syndrome; 9q subtelometric deletion syndrome; chromosome 9q34.3 deletion syndrome NOTCH1 : GRIN1 TRAF2 NOTCH1 PTGDS ANAPC2 TUBB4B NELFB ENTPD8 CACNA1B AGPAT2 UAP1L1 MIR602 FUT7 CYSRT1 | ||||||||||||||||||||||||||||||
| Chromosome 10 deletions | ||||||||||||||||||||||||||||||
| 10pter–p13 or 10p14–p15.1 | (Multiple incl. GATA3) | Sensorineural hearing loss | ||||||||||||||||||||||||||||
| OMIM: 146255 Names: Barakat syndrome; hypoparathyroidism, sensorineural deafness, and renal disease/dysplasia syndrome; HDRS; nephrosis, nerve deafness, and hypoparathyroidism : IL2RA PRKCQ AKR1C3 IDI1 CALML5 AKR1C4 CALML3 GATA3 NUDT5 TAF3 ZMYND11 CELF2 | ||||||||||||||||||||||||||||||
| 10q23 | (PTEN, BMPR1A) | Delayed or absent speech; impaired speech [29] | ||||||||||||||||||||||||||||
|
OMIM: 612242 (10q22.3–q23.2) Names: Chromosome 10q22.3–q23.2 deletion syndrome PTEN : PTEN CYP2C8 NRG3 CYP2C9 FAS CYP2C19 GLUD1 LIPA BMPR1A PLCE1 PDLIM1 PCGF5 MMRN2 IFIT2 | ||||||||||||||||||||||||||||||
| 10q26 | (HMX3, DOCK1, C10ORF90) | Impaired speech; delayed speech [30] | ||||||||||||||||||||||||||||
| OMIM: 609625; Cytogenetic location: 10q26 Genomic coordinates (GRCh38): 10:128,800,000–133,797,422 Names: Chromosome 10q26 deletion syndrome; terminal chromosome 10q26 deletion syndrome : FGFR2 CYP2E1 ECHS1 ACADSB DOCK1 BUB3 OAT RPL21P16 EIF3A GRK5 RAB11FIP2 MKI67 EBF3 CUZD1 CALY | ||||||||||||||||||||||||||||||
| Chromosome 11 deletions | ||||||||||||||||||||||||||||||
| 11p11.2–p12 | (EXT2, ALX4) | Idiosyncratic speech, dysarthria, delayed speech, apraxia [31] | ||||||||||||||||||||||||||||
| OMIM: 601224 (11p11.2); Cytogenetic location: 11p11.2 Genomic coordinates (GRCh38): 11:43,400,000–48,800,000 Names: Potocki–Shaffer syndrome; PSS; chromosome 11p11.2 deletion syndrome: proximal 11p deletion syndrome; DEFECT11 syndrome : TRAF6 NUP160 F2 PSMC3 NR1H3 CREB3L1 CKAP5 DDB2 SPI1 HSD17B12 | ||||||||||||||||||||||||||||||
| 11p13-p12 | (Multiple incl. WT1, PAX6, BDNF) (SLC1A2, PRRG4), | Impaired speech [32] | ||||||||||||||||||||||||||||
| OMIM: 612469; Cytogenetic location: 11p13–p12 Genomic coordinates (GRCh38): 11:31,000,000–43,400,000 Names: Wilms tumor, aniridia, genitourinary anomalies, mental retardation and obesity syndrome; WAGRO; WAGRO syndrome; WAGR syndrome with obesity; chromosome 11p13-p12 deletion syndrome : TRAF6 CAT CD44 CD59 CSTF3 SLC1A2 EIF3M PDHX WT1 APIP PRR5L ELF5 | ||||||||||||||||||||||||||||||
| 11p15.5 | (Multiple incl. CDKN1C, H19, IGF2) | Impaired speech [33] | ||||||||||||||||||||||||||||
| OMIM: 130650; Names: 130650: Beckwith–Wiedemann syndrome; BWS; exomphalos–macroglossia–gigantism syndrome; EMG syndrome; Wiedemann–Beckwith syndrome; WBS : HRAS : HRAS INS POLR2L KCNQ1 DUSP8 TNNT3 TNNI2 AP2A2 IRF7 TH RIC8A RASSF7 NLRP6 MIR483 KRTAP5-4 KRTAP5-1 DEAF1 CRACR2B BRSK2 | ||||||||||||||||||||||||||||||
| 11q13.3 | (FGF4, FGF3, FADD) | Delayed speech [34]; Impaired speech [35] | ||||||||||||||||||||||||||||
| OMIM: 166750 Names: Chromosome 11q13 deletion syndrome; ododental syndrome; otodental dysplasia and coloboma due to 11q13.3 microdeletion : FGF4 CCND1 FGF3 FGF19 FADD CPT1A CTTN PPFIA1 TPCN2 SHANK2 ANO1 | ||||||||||||||||||||||||||||||
| 11q23 | (Multiple incl. FLI1) | Dysarthric speech, absent speech [36]; impaired speech [37] | ||||||||||||||||||||||||||||
| OMIM: 188025; Cytogenetic location: 11q23 Genomic coordinates (GRCh38): 11:114,600,000–121,300,000 Names: Chromosome 11q23 deletion syndrome; thrombocytopenia, Paris–Trousseau type; TCPT; Paris–Trousseau syndrome; 11q terminal deletion syndrome : FXYD2 PPP2R1B CBL H2AX NCAM1 CD3G CD3D SC5D DLAT APOA1 ZPR1 TMPRSS4 TMPRSS13 TAGLN SIK3 POU2F3 MPZL2 C11orf52 | ||||||||||||||||||||||||||||||
| 11q23.3-q25 | (FLI1, BSX, NRGN, FRA11B, JAM3) | Impaired speech, delayed speech, apraxia [38] | ||||||||||||||||||||||||||||
| OMIM: 147791; Cytogenetic location: 11q23.3–q25 Genomic coordinates (GRCh38): 11:114,600,000–135,086,622 Names: Jacobsen syndrome; JBS; Del(11)(qter); distal deletion of 11q; distal monosomy 11q; monosomy 11qter : FXYD2 CBL KCNJ5 H2AX HSPA8 CHEK1 CD3G CD3D SC5D APOA1 ZPR1 VPS26B TMPRSS4 TMPRSS13 TAGLN SLC37A2 SIK3 POU2F3 MPZL2 IGSF9B GRAMD1B EI24 | ||||||||||||||||||||||||||||||
| Chromosome 12 deletions | ||||||||||||||||||||||||||||||
| 12q14.3 | (HMGA2) | Absent, impaired [38] or delayed speech [39] | ||||||||||||||||||||||||||||
| OMIM: 618908 Names: Silver–Russell syndrome 5; SRS5 : IRAK3 WIF1 MIR6502 GNS MSRB3 LEMD3 GRIP1 HMGA2 CAND1 | ||||||||||||||||||||||||||||||
| Chromosome 13 deletions | ||||||||||||||||||||||||||||||
| 13q12.3 | (POMP) | Delayed speech [40] | ||||||||||||||||||||||||||||
| OMIM: 601952 Names: Keratosis linearis with ichthyosis congenita and sclerosing keratoderma syndrome; KLICK syndrome : HMGB1 FLT1 ALOX5AP SLC7A1 HSPH1 B3GLCT | ||||||||||||||||||||||||||||||
| 13q14 | (RB1) | Normal speech [41] | ||||||||||||||||||||||||||||
| OMIM: 613884; Cytogenetic location: 13q14 Genomic coordinates (GRCh38): 13:50,300,000–54,700,000 Names: Chromosome 13q14 deletion syndrome : RB1 FOXO1 GTF2F2 KBTBD7 SLC25A15 HTR2A TNFSF11 LPAR6 CYSLTR2 DGKH WDFY2 ITM2B DLEU2 DLEU1 AKAP11 | ||||||||||||||||||||||||||||||
| 13q22.3 | (EDNRB) | Normal speech [41] | ||||||||||||||||||||||||||||
| OMIM: 277580; Names: Waardenburg syndrome, Type 4A; WS4A; Waardenburg syndrome with Hirschsprung disease, Type 4A; Waardenburg–Shah syndrome; Shah–Waardenburg syndrome; WS4 : EDNRB FBXL3 MYCBP2 | ||||||||||||||||||||||||||||||
| 13q33–q34 | (SOX1, ARHGEF7) | Apraxia | ||||||||||||||||||||||||||||
| OMIM: 619148; Cytogenetic location: 13q33–q34 Genomic coordinates (GRCh38): 13:106,400,000–114,364,328 Names: Chromosome 13q33–q34 deletion syndrome : IRS2 RASA3 TFDP1 F7 F10 CDC16 COL4A1 COL4A2 ARHGEF7 ATP4B ING1 | ||||||||||||||||||||||||||||||
| Chromosome 14 deletions | ||||||||||||||||||||||||||||||
| 14q11–q22 | (Multiple incl. PAX9, SUPT16H, CHD8, RALGAPA1] | Delayed [42] or Absent speech | ||||||||||||||||||||||||||||
| OMIM: 613457; Cytogenetic location: 14q11–q22 Genomic coordinates (GRCh38): 14:18,200,000–57,600,000 Names: Chromosome 14q11-q22 deletion syndrome : NFKBIA ADCY4 GNG2 SOS2 POLE2 PNP SLC7A7 SLC7A8 BMP4 GZMB ZNF219 WDHD1 TRD TEP1 RALGAPA1 PAX9 NDRG2 MIR208A DLGAP5 CEBPE BAZ1A AKAP6 | ||||||||||||||||||||||||||||||
| 14q22.1–q23.1 | (Multiple incl. PTGDR, BMP4) | Impaired speech [43] | ||||||||||||||||||||||||||||
| OMIM: 609640; Cytogenetic location: 14q22.1–q22.3 Genomic coordinates (GRCh38): 14:50,400,000–57,600,000 Names: Frias syndrome; Chromosome 14q22 deletion syndrome; growth deficiency, facial anomalies, and brachydactyly : GNG2 MNAT1 PRKCH BMP4 PSMA3 GNPNAT1 PELI2 PPM1A PYGL PTGER2 WDHD1 SIX6 SIX1 DLGAP5 | ||||||||||||||||||||||||||||||
| 14q32.2 | (DLK1, MEG3, RTL1) | Delayed speech; idiosyncratic speech [44] | ||||||||||||||||||||||||||||
| OMIM: 608149 (paternal)/ 616222 (maternal); Cytogenetic location: 14q32 Genomic coordinates (GRCh38): 14:103,500,000–107,043,718 Names: 14q32.2 Kagami–Ogata syndrome; uniparental disomy, paternal, chromosome 14/14q32.2; Temple syndrome; uniparental disomy, maternal, chromosome 14 : BDKRB2 CCNK BDKRB1 YY1 MIR6764 CYP46A1 PAPOLA DEGS2 EVL WARS1 MIR345 MIR342 MEG3 | ||||||||||||||||||||||||||||||
| Chromosome 15 deletions | ||||||||||||||||||||||||||||||
| 15q11.2 | (NIPA1, NIPA2, CYFIP1, TUBGCP5) | Delayed speech | ||||||||||||||||||||||||||||
| OMIM: 615656; Cytogenetic location: 15q11.2 Genomic coordinates (GRCh38): 15:20,500,000–25,500,000 Names: Burnside–Butler syndrome; 15q11.2 BP1–BP2 microdeletion : CYFIP1 UBE3A TUBGCP5 SNURF OR4N4 SNRPN OR4N4C OR4M2 NIPA2 NIPA1 NDN | ||||||||||||||||||||||||||||||
| 15q11–q13 | (NDN, SNRPN) | Delayed or Impaired speech | ||||||||||||||||||||||||||||
| OMIM: 176270 (for 5q11.2) Names: Prader-Willi syndrome; Prader-Lambhart-Willi syndrome; Labhart-Willi syndrome; Prader’s syndrome; Prader-Labhart-Willi-Fanconi syndrome : HERC2 TJP1 RYR3 GABRA5 CYFIP1 UBE3A GABRB3 CHRNA7 GABRG3 TUBGCP5 MTMR10 HMGN2P5 FMN1 | ||||||||||||||||||||||||||||||
| 15q11–q13 | (UBE3A) | Absent speech; impaired speech [45] | ||||||||||||||||||||||||||||
| OMIM: 105830 (for 15q11.2) Names: Angelman syndrome; happy puppet syndrome –Same as above– | ||||||||||||||||||||||||||||||
| 15q13.3 | (CHRNA7, CHRFAM7A, OTUD7A) | Impaired or idiosyncratic speech | ||||||||||||||||||||||||||||
| OMIM: 612001; Cytogenetic location: 15q13.3 Genomic coordinates (GRCh38): 15:30,900,000–33,400,000 Names: Chromosome 15q13.3 microdeletion syndrome : RYR3 CHRNA7 TRPM1 OTUD7A FAN1 ARHGAP11A MTMR10 FMN1 | ||||||||||||||||||||||||||||||
| 15q24 | (Multiple incl. SIN3A) | Delayed or impaired speech | ||||||||||||||||||||||||||||
| OMIM: 613406 Names: Witteveen–Kolk syndrome; WITKOS; chromosome 15q24 deletion syndrome (included) : CSK NRG4 CYP1A2 HCN4 CYP1A1 PML CYP11A1 SIN3A COX5A MPI SNX33 PTPN9 CLK3 | ||||||||||||||||||||||||||||||
| Chromosome 16 deletions | ||||||||||||||||||||||||||||||
| 16p11.2 | (SH2B1 , TBX6, CORO1A) | Apraxia [46]; dysarthria [47]; delayed or impaired speech | ||||||||||||||||||||||||||||
| OMIM: 611913; Cytogenetic location: 16p11.2 Genomic coordinates (GRCh38): 16:28,500,000–35,300,000 Names: Chromosome 16p11.2 deletion syndrome (593 kb) or 16p11.2 deletion syndrome (220 kb) SRCAP : MAPK3 ALDOA CD19 ITGAM VKORC1 SULT1A1 ITGAL STX4 PYCARD CDIPT SRCAP RNF40 PYDC1 ORAI3 MAZ HIRIP3 AHSP | ||||||||||||||||||||||||||||||
| 16p12.2–p11.2 | (Multiple incl. SH2B1) [76] | Delayed or impaired speech | ||||||||||||||||||||||||||||
| OMIM: 613604; Cytogenetic location: 16p12.2–p11.2 Genomic coordinates (GRCh38): 16:21,200,000–35,300,000 Names: Chromosome 16p12.2–p11.2 deletion syndrome, 7.1 to 8.7 MB SRCAP : MAPK3 PRKCB PLK1 ALDOA CD19 SCNN1G SCNN1B TNRC6A ITGAM VKORC1 XPO6 USP31 SRCAP RNF40 PYDC1 ORAI3 MAZ IGSF6 HIRIP3 CLN3 CHP2 AHSP | ||||||||||||||||||||||||||||||
| 16p12.1 | (Multiple) | Delayed speech | ||||||||||||||||||||||||||||
| OMIM: 136570; Cytogenetic location: 16p12 Genomic coordinates (GRCh38): 16:24,200,000–28,500,000 Names: Chromosome 16p12.1 deletion syndrome, 520 KB : TNRC6A IL4R CACNG3 EIF3CL HS3ST4 GTF3C1 SLC5A11 IL27 AQP8 NSMCE1 XPO6 CLN3 | ||||||||||||||||||||||||||||||
| 16p13.11 | (Multiple incl. MYH11) | Delayed speech [48] | ||||||||||||||||||||||||||||
| OMIM: 619351 Names: 16p13.1 microdeletion predisposing to autism and/or ID; megacystis–microcolon–intestinal hypoperistalsis syndrome 2; MMIHS2 : ABCC1 NDE1 MIR484 MYH11 RRN3 ABCC6 | ||||||||||||||||||||||||||||||
| 16p13.3 | (CREBBP, DNASE1, TRAP1) | Apraxia, dysarthria, impaired or delayed speech [49] | ||||||||||||||||||||||||||||
| OMIM: 610543; Cytogenetic location: 16p13.3 Genomic coordinates (GRCh38): 16:0–7,800,000 Names: Chromosome 16p13.3 deletion syndrome, proximal; severe Rubinstein–Taybi syndrome (RTS); broad thumb–hallux syndrome; Rubinstein syndrome; Rubinstein–Taybi deletion syndrome; RSTS deletion syndrome CREBBP : PDPK1 CREBBP ADCY9 TSC2 MLST8 GNG13 CACNA1H AXIN1 UBE2I ELOB RPS2 WDR24 TFAP4 SOX8 SEPTIN12 RHBDL1 RBFOX1 PGP NPRL3 NAGPA MIR3176 HAGHL GNPTG GLIS2 FAHD1 E4F1 CHTF18 BAIAP3 | ||||||||||||||||||||||||||||||
| 16q22 | (CBFB) | –Not available– | ||||||||||||||||||||||||||||
| OMIM: 614541; Cytogenetic location: 16q22 Genomic coordinates (GRCh38): 16:72,800,000–74,100,000 Names: Chromosome 16q22 deletion syndrome : CMTR2 : CDH1 TRADD SNTB2 SLC7A6 NFATC3 TAT PARD6A E2F4 NQO1 ST3GAL2 WWP2 PSKH1 PKD1L3 NUTF2 NOB1 CHTF8 CDH16 | ||||||||||||||||||||||||||||||
| 16q24.3–q24.2 | (CDH15, ZNF778, ANKRD11, ZFPM1) | Delayed or impaired speech [50] | ||||||||||||||||||||||||||||
| : SLC7A5 MVD CYBA APRT RPL13 CDT1 TUBB3 MC1R DPEP1 TRAPPC2L SPIRE2 CHMP1A | ||||||||||||||||||||||||||||||
| Chromosome 17 deletions | ||||||||||||||||||||||||||||||
| 17p11.2 | (LLGL1, RAI1, UBB) | Delayed speech; dysarthria [51] | ||||||||||||||||||||||||||||
| OMIM: 182290
Names: Smith–Magenis syndrome; chromosome 17p11.2 deletion syndrome : UBB MAP2K3 MAPK7 ALDH3A1 SHMT1 ALDH3A2 SREBF1 TOP3A MIR6778 KCNJ12 | ||||||||||||||||||||||||||||||
| 17p13.1 | (Multiple incl. KCNAB3, GUCY2D, TP53, TRAPPC1, MPDU1, CDG1F, FXR2, FMRP, EFNB3) | Absent speech | ||||||||||||||||||||||||||||
| OMIM: 613776; Cytogenetic location: 17p13.1 Genomic coordinates (GRCh38): 17:6,500,000–10,800,000 Names: Chromosome 17p13.1 deletion syndrome TP53 KDM6B : TP53 ATP1B2 PIK3R5 DLG4 POLR2A ALOX12 ALOX15B ALOX12B DVL2 SLC2A4 VAMP2 TNK1 SHBG MYH4 MYH13 MYH1 MIR497 MIR324 DNAH2 | ||||||||||||||||||||||||||||||
| 17p13.3 | (LIS1, PAFAH1B1, YWHAE) †‡ | Delayed speech [52] | ||||||||||||||||||||||||||||
| OMIM: 247200; Cytogenetic location: 17p13.3 Genomic coordinates (GRCh38): 17:0–3,400,000 Names: Miller–Dieker lissencephaly syndrome; MDLS; chromosome 17p13.3 deletion syndrome (included) : CRK RPA1 YWHAE PAFAH1B1 INPP5K ABR SERPINF2 SLC43A2 OR3A1 OR1G1 SMYD4 SERPINF1 | ||||||||||||||||||||||||||||||
| 17q11.2 | (NF1)‡ | No significant speech issues | ||||||||||||||||||||||||||||
| OMIM: 162200 Names: Neurofibromatosis type I; NF1; Von Recklinghausen disease; neurofibromatosis, peripheral type; Morbus–Recklinghausen : SLC6A4 KSR1 NF1 NOS2 PSMD11 VTN NLK CDK5R1 RPL23A ALDOC RAB11FIP4 MIR451A MIR423 | ||||||||||||||||||||||||||||||
| 17q12 | (Multiple incl. HNF1B, LHX1, CCL3L3, SNIP) | Delayed or impaired speech | ||||||||||||||||||||||||||||
| OMIM: 614527; Cytogenetic location: 17q12 Genomic coordinates (GRCh38): 17:33,500,000–39,800,000 Names: Chromosome 17q12 deletion syndrome ERBB2 : CACNB1 ERBB2 AP2B1 PIP4K2B ACACA CCL5 CCL2 PSMB3 CCL4 RPL23 PEX12 MMP28 DUSP14 | ||||||||||||||||||||||||||||||
| 17q21.31 | (KANSL1, MAPT, CRHR1) | Delayed or absent speech | ||||||||||||||||||||||||||||
| OMIM: 610443 Names: Koolen–De Vries syndrome; KDVS; chromosome 17q21.31 deletion syndrome; Microdeletion 17q21.31 syndrome : KANSL1 BRCA1 : ITGA2B BRCA1 MAP3K14 G6PC1 FZD2 WNT3 DUSP3 HDAC5 NSF AOC3 TMEM106A RND2 HEXIM1 | ||||||||||||||||||||||||||||||
| 17q23.1-q23.2 | (TBX4) | Delayed speech [53] | ||||||||||||||||||||||||||||
| OMIM: 613355; Cytogenetic location: 17q23.1–q23.2 Genomic coordinates (GRCh38): 17:59,500,000–63,100,000 Names: Chromosome 17q23.1–q23.2 deletion syndrome : RPS6KB1 CLTC BRIP1 MIR21 SKA2 MRC2 PTRH2 CA4 MED13 DHX40 BCAS3 APPBP2 | ||||||||||||||||||||||||||||||
| 17q24.3–q24.2 | (Multiple incl. ABCA5, MAP2K6, SOX9) | Delayed speech [54] | ||||||||||||||||||||||||||||
| OMIM: 135400; Cytogenetic location: 17q24.2–q24.3 Genomic coordinates (GRCh38): 17:66,200,000–72,900,000 Names: Hypertrichosis, congenital generalized, with or without gingival hyperplasia; HTC3; fibromatosis, gingival, with hypertrichosis; chromosome 17q24.2–q24.3 deletion syndrome : PRKCA MAP2K6 PRKAR1A KCNJ2 PSMD12 CACNG4 KPNA2 KCNJ16 BPTF | ||||||||||||||||||||||||||||||
| Chromosome 18 deletions | ||||||||||||||||||||||||||||||
| 18q | (Multiple incl. MBP, TSHZ1) | Delayed [55] or impaired [56] speech | ||||||||||||||||||||||||||||
| OMIM: 601808; Cytogenetic location: 18q Genomic coordinates (GRCh38): 18:18,500,000–80,373,285 Names: Chromosome 18q deletion syndrome; 18q syndrome : BCL2 SMAD4 ROCK1 SMAD2 ACAA2 NFATC1 PIK3C3 SMAD7 LAMA3 SLC14A2 SS18 SETBP1 SERPINB4 SERPINB13 RIOK3 NETO1 MIR122 MBD1 LINC-ROR GAREM1 CELF4 | ||||||||||||||||||||||||||||||
| Chromosome 19 deletions | ||||||||||||||||||||||||||||||
| 19p13.13 | (Multiple incl. NFIX, MAST1, CALR) | Delayed speech; impaired speech [57] | ||||||||||||||||||||||||||||
| OMIM: 613638; Cytogenetic location: 19p13.13 Genomic coordinates (GRCh38): 19:12,600,000–13,800,000 Names: Chromosome 19p13.13 deletion syndrome MAP2K2 UHRF1 : MAP2K2 FGF22 VAV1 POLR2E SHC2 GNG7 PIP5K1C GTF2F1 GNA11 PSPN ZBTB7A TJP3 SEMA6B REXO1 PLIN4 MYDGF MIR7-3 DPP9 DAZAP1 | ||||||||||||||||||||||||||||||
| 19q13.11 | (Multiple incl. LSM14A, UBA2, WTIP, TSHZ3) | Delayed or absent speech; impaired speech [58] | ||||||||||||||||||||||||||||
| OMIM: 613026 (distal)/ 617219 (proximal); Cytogenetic location: 19q13.11 Genomic coordinates (GRCh38): 19:31,900,000–35,100,000
Names: Chromosome 19q13.11 deletion syndrome, distal; chromosome 19q13.11 deletion syndrome, proximal CEBPA : SCN1B GPI SLC7A9 PSMC4 CEBPA SLC7A10 UBA2 CHST8 WTIP RGS9BP | ||||||||||||||||||||||||||||||
| Chromosome 20 deletions | ||||||||||||||||||||||||||||||
| 20p12.3 | (Multiple incl. BMP2) | Delayed or Impaired speech [59] | ||||||||||||||||||||||||||||
| : PLCB1 PCNA PLCB4 BMP2 MCM8 PROKR2 CRLS1 CDS2 GPCPD1 RPS18P1 HAO1 TRMT6 LRRN4 | ||||||||||||||||||||||||||||||
| Chromosome 22 deletions | ||||||||||||||||||||||||||||||
| 22q11.2 | (Multiple incl. TBX1 , COMT , INI1 , TOP3B ) | Apraxia, dysarthria, delayed, or impaired speech [60] | ||||||||||||||||||||||||||||
| OMIM: 611867; Cytogenetic location: 22q11.2 Genomic coordinates (GRCh38): 22:23,100,000–25,500,000 Names: Chromosome 22q11.2 deletion syndrome; distal chromosome 22q11.2 deletion syndrome : MAPK1 CRKL GGT1 BID ADORA2A COMT GNAZ UPB1 ATP6V1E1 IGL PIWIL3 MIR185 | ||||||||||||||||||||||||||||||
| 22q12.2 | (NF2) | Delayed or impaired speech [61] | ||||||||||||||||||||||||||||
| OMIM: 101000 Names: Neurofibromatosis type II; neurofibromatosis, central type; acoustic schwannomas, bilateral; bilateral acoustic neurofibromatosis; BANF; acoustic neurinoma, bilateral; ACN : LIF PLA2G3 AP1B1 LIMK2 INPP5J OSM PISD SFI1 RNF185 MTMR3 SEC14L2 PIK3IP1 MIR3200 DEPDC5 | ||||||||||||||||||||||||||||||
| 22q13.3 | (ARSA, SHANK3) | Delayed or absent speech | ||||||||||||||||||||||||||||
| OMIM: 606232 Names: Phelan–McDermid syndrome; PHMDS; chromosome 22q13.3 deletion syndrome; telomeric 22q13 monosomy syndrome BRD1 : MAPK11 MAPK12 NUP50 PRR5 PPARA WNT7B TYMP HDAC10 CHKB ARSA UPK3A PLXNB2 GRAMD4 CELSR1 | ||||||||||||||||||||||||||||||
| Chromosome X deletions | ||||||||||||||||||||||||||||||
| Xp11.3 | (Multiple incl. RP2, ZNF674) | Impaired speech [62] | ||||||||||||||||||||||||||||
| OMIM: 300578; Cytogenetic location: Xp11.3 Genomic coordinates (GRCh38): X:42,500,000–47,600,000 Names: Chromosome Xp11.3 deletion syndrome; mental retardation, X-linked, with retinitis pigmentosa KDM6A : ARAF MAOA MAOB UBA1 TIMP1 NDUFB11 MIR221 CHST7 MIR222 USP11 | ||||||||||||||||||||||||||||||
| Xp21 | (GK, DMD, NR0B1) †‡ | Delayed speech [63] | ||||||||||||||||||||||||||||
| OMIM: 300679; Cytogenetic location: Xp21 Genomic coordinates (GRCh38): X:31,500,000–37,800,000 Names: Chromosome Xp21 deletion syndrome; Complex glycerol kinase deficiency : TAB3 CYBB GK DMD NR0B1 XK ARX | ||||||||||||||||||||||||||||||
| Xq28 (a) | (ABCD1, BCAP31, SLC6A8) | Delayed speech [64] | ||||||||||||||||||||||||||||
| OMIM: 300475 Names: Deafness, dystonia, and cerebral hypomyelination; DDCH; contiguous ABCD1/DXS1375E deletion syndrome, included; CADDS, included : IKBKG IRAK1 MIR718 DUSP9 F8 H2AB1 NSDHL G6PD FLNA IDH3G MPP1 MIR224 MIR105-1 MAGEA11 | ||||||||||||||||||||||||||||||
| Xq28 (b) | (MECP2) | Normal to absent speech | ||||||||||||||||||||||||||||
| OMIM: 312750 Names: Rett syndrome; RTT; RTS autism, dementia, ataxia, and loss of purposeful hand use –same as above– | ||||||||||||||||||||||||||||||
| Chromosome Y deletions | ||||||||||||||||||||||||||||||
| Yq11 | (USP9Y, BPY2, CDY1) | Normal speech | ||||||||||||||||||||||||||||
| OMIM: 415000 Names: Spermatogenic failure, Y-linked, 2; SPGFY2; Sertoli-cell-only syndrome; Del Castillo syndrome; germ cell aplasia, spermatogenic failure : RPS4Y2 UTY NLGN4Y KDM5D CD24P4 TMSB4Y | ||||||||||||||||||||||||||||||
References
- Singh, R. Profiling Humans from Their Voice; Springer-Nature: Singapore, 2019. [Google Scholar]
- Sataloff, R.T. Genetics of the voice. J. Voice 1995, 9, 16–19. [Google Scholar] [CrossRef] [PubMed]
- Ganapathiraju, M.K.; Thahir, M.; Handen, A.; Sarkar, S.N.; Sweet, R.A.; Nimgaonkar, V.L.; Loscher, C.E.; Bauer, E.M.; Chaparala, S. Schizophrenia interactome with 504 novel protein–protein interactions. NPJ Schizophr. 2016, 2, 16012. [Google Scholar] [CrossRef] [PubMed]
- Morgan, A.; Fisher, S.E.; Scheffer, I.; Hildebrand, M. FOXP2-Related Speech and Language Disorders. In GeneReviews®; University of Washington: Seattle, WA, USA, 2017. [Google Scholar]
- Fisher, S.E.; Scharff, C. FOXP2 as a molecular window into speech and language. Trends Genet. 2009, 25, 166–177. [Google Scholar] [CrossRef] [PubMed]
- FOXP2. Online Mendelian Inheritance in Man (OMIM) Entry 605317. Available online: https://omim.org/entry/605317 (accessed on 21 September 2022).
- Den Hoed, J.; Fisher, S.E. Genetic pathways involved in human speech disorders. Curr. Opin. Genet. Dev. 2020, 65, 103–111. [Google Scholar] [CrossRef]
- FOXP2. The Human Protein Atlas. Available online: https://www.proteinatlas.org/ENSG00000128573-FOXP2 (accessed on 21 September 2022).
- Bac, C. Investigation of Speech Delay in Individuals with 1p36 Deletion Syndrome. Ph.D. Thesis, University of Cincinnati, Cincinnati, OH, USA, 2015. [Google Scholar]
- Pang, H.; Yu, X.; Kim, Y.M.; Wang, X.; Jinkins, J.K.; Yin, J.; Li, S.; Gu, H. Disorders Associated With Diverse, Recurrent Deletions and Duplications at 1q21.1. Front. Genet. 2020, 11, 577. [Google Scholar] [CrossRef]
- Brazil, A.; Stanford, K.; Smolarek, T.; Hopkin, R. Delineating the phenotype of 1p36 deletion in adolescents and adults. Am. J. Med. Genet. Part A 2014, 164, 2496–2503. [Google Scholar] [CrossRef]
- He, J.; Xie, Y.; Kong, S.; Qiu, W.; Wang, X.; Wang, D.; Sun, X.; Sun, D. Psychomotor retardation with a 1q42.11–q42.12 deletion. Hereditas 2017, 154, 6. [Google Scholar] [CrossRef][Green Version]
- Peter, B.; Matsushita, M.; Oda, K.; Raskind, W. De novo microdeletion of BCL11A is associated with severe speech sound disorder. Am. J. Med. Genet. Part A 2014, 164, 2091–2096. [Google Scholar] [CrossRef]
- Eggermann, T.; Spengler, S.; Venghaus, A.; Denecke, B.; Zerres, K.; Baudis, M.; Ensenauer, R. 2p21 Deletions in hypotonia–cystinuria syndrome. Eur. J. Med. Genet. 2012, 55, 561–563. [Google Scholar] [CrossRef]
- Chen, C.P.; Lin, S.P.; Chern, S.R.; Tsai, F.J.; Wu, P.C.; Lee, C.C.; Chen, L.F.; Lee, M.S.; Wang, W. Deletion 2q37.3 ⟶ qter and duplication 15q24.3 ⟶ qter characterized by array CGH in a girl with epilepsy and dysmorphic features. Genet. Couns. 2010, 21, 263. [Google Scholar]
- Palumbo, O.; D’Agruma, L.; Minenna, A.F.; Palumbo, P.; Stallone, R.; Palladino, T.; Zelante, L.; Carella, M. 3p14.1 de novo microdeletion involving the FOXP1 gene in an adult patient with autism, severe speech delay and deficit of motor coordination. Gene 2013, 516, 107–113. [Google Scholar] [CrossRef]
- Lowther, C.; Costain, G.; Melvin, R.; Stavropoulos, D.J.; Lionel, A.C.; Marshall, C.R.; Scherer, S.W.; Bassett, A.S. Adult expression of a 3q13.31 microdeletion. Mol. Cytogenet. 2014, 7, 23. [Google Scholar] [CrossRef][Green Version]
- Van Borsel, J.; De Grande, S.; Van Buggenhout, G.; Fryns, J.P. Speech and language in Wolf-Hirschhorn syndrome: A case-study. J. Commun. Disord. 2004, 37, 21–33. [Google Scholar] [CrossRef]
- Bonnet, C.; Andrieux, J.; Beri-Dexheimer, M.; Leheup, B.; Boute, O.; Manouvrier, S.; Delobel, B.; Copin, H.; Receveur, A.; Mathieu, M.; et al. Microdeletion at chromosome 4q21 defines a new emerging syndrome with marked growth restriction, mental retardation and absent or severely delayed speech. J. Med. Genet. 2010, 47, 377–384. [Google Scholar] [CrossRef]
- Tran, T.M.; Sherwood, J.K.; Doolittle, M.J.; Sathler, M.F.; Hofmann, F.; Stone-Roy, L.M.; Kim, S. Loss of cGMP-dependent protein kinase II alters ultrasonic vocalizations in mice, a model for speech impairment in human microdeletion 4q21 syndrome. Neurosci. Lett. 2021, 759, 136048. [Google Scholar] [CrossRef]
- Kristoffersen, K.E. Speech and language development in cri du chat syndrome: A critical review. Clin. Linguist. Phon. 2008, 22, 443–457. [Google Scholar] [CrossRef] [PubMed]
- Flax, J.F.; Hare, A.; Azaro, M.A.; Vieland, V.J.; Brzustowicz, L.M. Combined linkage and linkage disequilibrium analysis of a motor speech phenotype within families ascertained for autism risk loci. J. Neurodev. Disord. 2010, 2, 210–223. [Google Scholar] [CrossRef]
- Rauch, A.; Beese, M.; Mayatepek, E.; Dörr, H.G.; Wenzel, D.; Reis, A.; Trautmann, U. A novel 5q35.3 subtelomeric deletion syndrome. Am. J. Med. Genet. Part A 2003, 121, 1–8. [Google Scholar] [CrossRef]
- Peter, B.; Lancaster, H.; Vose, C.; Fares, A.; Schrauwen, I.; Huentelman, M. Two unrelated children with overlapping 6q25.3 deletions, motor speech disorders, and language delays. Am. J. Med. Genet. Part A 2017, 173, 2659–2669. [Google Scholar] [CrossRef]
- Bianchi, E.; Aricŏ, M.; Podestă, A.F.; Grana, M.; Fiori, P.; Beluffi, G.; Opitz, J.M.; Reynolds, J.F. A family with the Saethre-Chotzen syndrome. Am. J. Med. Genet. 1985, 22, 649–658. [Google Scholar] [CrossRef]
- Vanzo, R.J.; Martin, M.M.; Sdano, M.R.; South, S.T. Familial KANK1 deletion that does not follow expected imprinting pattern. Eur. J. Med. Genet. 2013, 56, 256–259. [Google Scholar] [CrossRef] [PubMed]
- Yatsenko, S.; Cheung, S.; Scott, D.; Nowaczyk, M.; Tarnopolsky, M.; Naidu, S.; Bibat, G.; Patel, A.; Leroy, J.; Scaglia, F.; et al. Deletion 9q34.3 syndrome: Genotype-phenotype correlations and an extended deletion in a patient with features of Opitz C trigonocephaly. J. Med. Genet. 2005, 42, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Samango-Sprouse, C.; Lawson, P.; Sprouse, C.; Stapleton, E.; Sadeghin, T.; Gropman, A. Expanding the phenotypic profile of Kleefstra syndrome: A female with low-average intelligence and childhood apraxia of speech. Am. J. Med. Genet. Part A 2016, 170, 1312–1316. [Google Scholar] [CrossRef] [PubMed]
- Septer, S.; Zhang, L.; Lawson, C.E.; Cocjin, J.; Attard, T.; Ardinger, H.H. Aggressive juvenile polyposis in children with chromosome 10q23 deletion. World J. Gastroenterol. WJG 2013, 19, 2286. [Google Scholar] [CrossRef]
- Nishi, E.; Uehara, T.; Yanagi, K.; Hasegawa, Y.; Ueda, K.; Kaname, T.; Yamamoto, T.; Kosaki, K.; Okamoto, N. Clinical spectrum of individuals with de novo EBF3 variants or deletions. Am. J. Med. Genet. Part A 2021, 185, 2913–2921. [Google Scholar] [CrossRef]
- Kim, H.G.; Rosenfeld, J.A.; Scott, D.A.; Bénédicte, G.; Labonne, J.D.; Brown, J.; McGuire, M.; Mahida, S.; Naidu, S.; Gutierrez, J.; et al. Disruption of PHF21A causes syndromic intellectual disability with craniofacial anomalies, epilepsy, hypotonia, and neurobehavioral problems including autism. Mol. Autism 2019, 10, 35. [Google Scholar] [CrossRef]
- Xu, S.; Han, J.; Morales, A.; Menzie, C.; Williams, K.; Fan, Y.S. Characterization of 11p14-p12 deletion in WAGR syndrome by array CGH for identifying genes contributing to mental retardation and autism. Cytogenet. Genome Res. 2008, 122, 181–187. [Google Scholar] [CrossRef]
- Borsel, J.V.; Morlion, B.; Snick, K.V.; Leroy, J.S. Articulation in Beckwith-Wiedemann syndrome: Two case studies. Am. J. Speech-Lang. Pathol. 2000, 9, 202–213. [Google Scholar] [CrossRef]
- Kim, Y.S.; Kim, G.H.; Byeon, J.H.; Eun, S.H.; Eun, B.L. Chromosome 11q13 deletion syndrome. Korean J. Pediatr. 2016, 59, S10. [Google Scholar] [CrossRef]
- Chilian, B.; Abdollahpour, H.; Bierhals, T.; Haltrich, I.; Fekete, G.; Nagel, I.; Rosenberger, G.; Kutsche, K. Dysfunction of SHANK2 and CHRNA7 in a patient with intellectual disability and language impairment supports genetic epistasis of the two loci. Clin. Genet. 2013, 84, 560–565. [Google Scholar] [CrossRef]
- Takahashi, I.; Takahashi, T.; Sawada, K.; Shimojima, K.; Yamamoto, T. Jacobsen syndrome due to an unbalanced translocation between 11q23 and 22q11.2 identified at age 40 years. Am. J. Med. Genet. Part A 2012, 158, 220–223. [Google Scholar] [CrossRef]
- Penny, L.A.; Dell’Aquila, M.; Jones, M.C.; Bergoffen, J.; Cunniff, C.; Fryns, J.P.; Grace, E.; Graham, J.M.; Kousseff, B.; Mattina, T.; et al. Clinical and molecular characterization of patients with distal 11q deletions. Am. J. Hum. Genet. 1995, 56, 676. [Google Scholar]
- Manolakos, E.; Orru, S.; Neroutsou, R.; Kefalas, K.; Louizou, E.; Papoulidis, I.; Thomaidis, L.; Peitsidis, P.; Sotiriou, S.; Kitsos, G.; et al. Detailed molecular and clinical investigation of a child with a partial deletion of chromosome 11 (Jacobsen syndrome). Mol. Cytogenet. 2009, 2, 26. [Google Scholar] [CrossRef]
- Lynch, S.A.; Foulds, N.; Thuresson, A.C.; Collins, A.L.; Annerén, G.; Hedberg, B.O.; Delaney, C.A.; Iremonger, J.; Murray, C.M.; Crolla, J.A.; et al. The 12q14 microdeletion syndrome: Six new cases confirming the role of HMGA2 in growth. Eur. J. Hum. Genet. 2011, 19, 534–539. [Google Scholar] [CrossRef][Green Version]
- Bartholdi, D.; Stray-Pedersen, A.; Azzarello-Burri, S.; Kibaek, M.; Kirchhoff, M.; Oneda, B.; Rødningen, O.; Schmitt-Mechelke, T.; Rauch, A.; Kjaergaard, S. A newly recognized 13q12.3 microdeletion syndrome characterized by intellectual disability, microcephaly, and eczema/atopic dermatitis encompassing the HMGB1 and KATNAL1 genes. Am. J. Med. Genet. Part A 2014, 164, 1277–1283. [Google Scholar] [CrossRef]
- Tüysüz, B.; Collin, A.; Arapoğlu, M.; Suyugül, N. Clinical variability of Waardenburg–Shah syndrome in patients with proximal 13q deletion syndrome including the endothelin-B receptor locus. Am. J. Med. Genet. Part A 2009, 149, 2290–2295. [Google Scholar] [CrossRef]
- Fonseca, D.J.; Prada, C.F.; Siza, L.M.; Angel, D.; Gomez, Y.M.; Restrepo, C.M.; Douben, H.; Rivadeneira, F.; de Klein, A.; Laissue, P. A de novo 14q12q13.3 interstitial deletion in a patient affected by a severe neurodevelopmental disorder of unknown origin. Am. J. Med. Genet. Part A 2012, 158, 689–693. [Google Scholar] [CrossRef]
- Martínez-Fernández, M.L.; Bermejo-Sánchez, E.; Fernández, B.; MacDonald, A.; Fernández-Toral, J.; Martínez-Frías, M.L. Haploinsufficiency of BMP4 gene may be the underlying cause of Frias syndrome. Am. J. Med. Genet. Part A 2014, 164, 338–345. [Google Scholar] [CrossRef]
- Huang, H.; Mikami, Y.; Shigematsu, K.; Uemura, N.; Shinsaka, M.; Iwatani, A.; Miyake, F.; Kabe, K.; Takai, Y.; Saitoh, M.; et al. Kagami–Ogata syndrome in a fetus presenting with polyhydramnios, malformations, and preterm delivery: A case report. J. Med. Case Rep. 2019, 13, 340. [Google Scholar] [CrossRef]
- Murthy, S.; Nygren, A.; El Shakankiry, H.; Schouten, J.; Al Khayat, A.; Ridha, A.; Al Ali, M. Detection of a novel familial deletion of four genes between BP1 and BP2 of the Prader-Willi/Angelman syndrome critical region by oligo-array CGH in a child with neurological disorder and speech impairment. Cytogenet. Genome Res. 2007, 116, 135–140. [Google Scholar] [CrossRef]
- Mei, C.; Fedorenko, E.; Amor, D.J.; Boys, A.; Hoeflin, C.; Carew, P.; Burgess, T.; Fisher, S.E.; Morgan, A.T. Deep phenotyping of speech and language skills in individuals with 16p11.2 deletion. Eur. J. Hum. Genet. 2018, 26, 676–686. [Google Scholar] [CrossRef] [PubMed]
- Demopoulos, C.; Kothare, H.; Mizuiri, D.; Henderson-Sabes, J.; Fregeau, B.; Tjernagel, J.; Houde, J.F.; Sherr, E.H.; Nagarajan, S.S. Abnormal speech motor control in individuals with 16p11.2 deletions. Sci. Rep. 2018, 8, 1274. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, T.; Theisen, A.; Rosenfeld, J.A.; Lamb, A.N.; Ravnan, J.B.; Schultz, R.A.; Torchia, B.S.; Neill, N.; Casci, I.; Bejjani, B.A.; et al. Copy number variants of schizophrenia susceptibility loci are associated with a spectrum of speech and developmental delays and behavior problems. Genet. Med. 2011, 13, 868–880. [Google Scholar] [CrossRef] [PubMed]
- Hennekam, R.C.; Baselier, A.C.; Beyaert, E.; Bos, A.; Blok, J.; Jansma, H.; Thorbecke-Nilsen, V.; Veerman, H. Psychological and speech studies in Rubinstein-Taybi syndrome. Am. J. Ment. Retard. 1992, 96, 645–660. [Google Scholar] [PubMed]
- Novara, F.; Rinaldi, B.; Sisodiya, S.M.; Coppola, A.; Giglio, S.; Stanzial, F.; Benedicenti, F.; Donaldson, A.; Andrieux, J.; Stapleton, R.; et al. Haploinsufficiency for ANKRD11-flanking genes makes the difference between KBG and 16q24.3 microdeletion syndromes: 12 new cases. Eur. J. Hum. Genet. 2017, 25, 694–701. [Google Scholar] [CrossRef]
- Gropman, A.L.; Duncan, W.C.; Smith, A.C. Neurologic and developmental features of the Smith-Magenis syndrome (del 17p11.2). Pediatr. Neurol. 2006, 34, 337–350. [Google Scholar] [CrossRef]
- Schiff, M.; Delahaye, A.; Andrieux, J.; Sanlaville, D.; Vincent-Delorme, C.; Aboura, A.; Benzacken, B.; Bouquillon, S.; Elmaleh-Berges, M.; Labalme, A.; et al. Further delineation of the 17p13.3 microdeletion involving YWHAE but distal to PAFAH1B1: Four additional patients. Eur. J. Med. Genet. 2010, 53, 303–308. [Google Scholar] [CrossRef]
- Schönewolf-Greulich, B.; Ronan, A.; Ravn, K.; Baekgaard, P.; Lodahl, M.; Nielsen, K.; Rendtorff, N.D.; Tranebjaerg, L.; Brøndum-Nielsen, K.; Tümer, Z. Two new cases with microdeletion of 17q23.2 suggest presence of a candidate gene for sensorineural hearing loss within this region. Am. J. Med. Genet. Part A 2011, 155, 2964–2969. [Google Scholar] [CrossRef]
- Vergult, S.; Dauber, A.; Delle Chiaie, B.; Van Oudenhove, E.; Simon, M.; Rihani, A.; Loeys, B.; Hirschhorn, J.; Pfotenhauer, J.; Phillips, J.A.; et al. 17q24.2 microdeletions: A new syndromal entity with intellectual disability, truncal obesity, mood swings and hallucinations. Eur. J. Hum. Genet. 2012, 20, 534–539. [Google Scholar] [CrossRef]
- Cody, J.D.; Sebold, C.; Malik, A.; Heard, P.; Carter, E.; Crandall, A.; Soileau, B.; Semrud-Clikeman, M.; Cody, C.M.; Hardies, L.J.; et al. Recurrent interstitial deletions of proximal 18q: A new syndrome involving expressive speech delay. Am. J. Med. Genet. Part A 2007, 143, 1181–1190. [Google Scholar] [CrossRef]
- Marseglia, G.; Scordo, M.R.; Pescucci, C.; Nannetti, G.; Biagini, E.; Scandurra, V.; Gerundino, F.; Magi, A.; Benelli, M.; Torricelli, F. 372 kb Microdeletion in 18q12.3 causing SETBP1 haploinsufficiency associated with mild mental retardation and expressive speech impairment. Eur. J. Med. Genet. 2012, 55, 216–221. [Google Scholar] [CrossRef]
- Bonaglia, M.C.; Marelli, S.; Novara, F.; Commodaro, S.; Borgatti, R.; Minardo, G.; Memo, L.; Mangold, E.; Beri, S.; Zucca, C.; et al. Genotype–phenotype relationship in three cases with overlapping 19p13.12 microdeletions. Eur. J. Hum. Genet. 2010, 18, 1302–1309. [Google Scholar] [CrossRef]
- Melo, J.B.; Estevinho, A.; Saraiva, J.; Ramos, L.; Carreira, I.M. Cutis Aplasia as a clinical hallmark for the syndrome associated with 19q13.11 deletion: The possible role for UBA2 gene. Mol. Cytogenet. 2015, 8, 21. [Google Scholar] [CrossRef]
- Amasdl, S.; Natiq, A.; Sbiti, A.; Zerkaoui, M.; Lyahyai, J.; Amzazi, S.; Liehr, T.; Sefiani, A. 20p12.3 deletion is rare cause of syndromic cleft palate: Case report and review of literature. BMC Res. Notes 2016, 9, 5. [Google Scholar] [CrossRef][Green Version]
- Solot, C.B.; Sell, D.; Mayne, A.; Baylis, A.L.; Persson, C.; Jackson, O.; McDonald-McGinn, D.M. Speech-language disorders in 22q11.2 deletion syndrome: Best practices for diagnosis and management. Am. J. Speech-Lang. Pathol. 2019, 28, 984–999. [Google Scholar] [CrossRef]
- Davidson, T.B.; Sanchez-Lara, P.A.; Randolph, L.M.; Krieger, M.D.; Wu, S.Q.; Panigrahy, A.; Shimada, H.; Erdreich-Epstein, A. Microdeletion del (22)(q12.2) encompassing the facial development-associated gene, MN1 (meningioma 1) in a child with Pierre-Robin sequence (including cleft palate) and neurofibromatosis 2 (NF2): A case report and review of the literature. BMC Med. Genet. 2012, 13, 19. [Google Scholar] [CrossRef]
- Hayashi, S.; Mizuno, S.; Migita, O.; Okuyama, T.; Makita, Y.; Hata, A.; Imoto, I.; Inazawa, J. The CASK gene harbored in a deletion detected by array-CGH as a potential candidate for a gene causative of X-linked dominant mental retardation. Am. J. Med. Genet. Part A 2008, 146, 2145–2151. [Google Scholar] [CrossRef]
- Fries, M.H.; Lebo, R.V.; Schonberg, S.A.; Golabi, M.; Seltzer, W.K.; Gitelman, S.E.; Golbus, M.S. Mental retardation locus in Xp21 chromosome microdeletion. Am. J. Med. Genet. 1993, 46, 363–368. [Google Scholar] [CrossRef]
- Gedeon, A.; Meinänen, M.; Ades, L.; Kääriäinen, H.; Gecz, J.; Baker, E.; Sutherland, G.; Mulley, J. Overlapping submicroscopic deletions in Xq28 in two unrelated boys with developmental disorders: Identification of a gene near FRAXE. Am. J. Hum. Genet. 1995, 56, 907. [Google Scholar]
- National Institute on Deafness and Other Communication Disorders. Statistics on voice, speech, and language. IEEE/ACM Trans. Audio Speech Lang. Process. 2008, 25, 2098–2111. [Google Scholar]
- American Speech-Language-Hearing Association. Speech Sound Disorders: Articulation and Phonology. Practice Portal. 2017. Available online: www.asha.org/Practice-Portal/Clinical-Topics/Articulation-and-Phonology (accessed on 21 September 2022).
- Shmueli, G.; Minka, T.P.; Kadane, J.B.; Borle, S.; Boatwright, P. A useful distribution for fitting discrete data: Revival of the Conway–Maxwell–Poisson distribution. J. R. Stat. Soc. Ser. C (Appl. Stat.) 2005, 54, 127–142. [Google Scholar] [CrossRef]
- Sellers, K.F.; Borle, S.; Shmueli, G. The COM-Poisson model for count data: A survey of methods and applications. Appl. Stoch. Model. Bus. Ind. 2012, 28, 104–116. [Google Scholar] [CrossRef]
- Huang, J.; Zhao, Y.L.; Li, Y.; Fletcher, J.A.; Xiao, S. Genomic and functional evidence for an ARID1A tumor suppressor role. Genes Chromosom. Cancer 2007, 46, 745–750. [Google Scholar] [CrossRef] [PubMed]
- Sheffels, E.; Sealover, N.E.; Theard, P.L.; Kortum, R.L. Anchorage-independent growth conditions reveal a differential SOS2 dependence for transformation and survival in RAS-mutant cancer cells. Small GTPases 2021, 12, 67–78. [Google Scholar] [CrossRef]
- Bertucci, F.; Borie, N.; Ginestier, C.; Groulet, A.; Charafe-Jauffret, E.; Adélaïde, J.; Geneix, J.; Bachelart, L.; Finetti, P.; Koki, A.; et al. Identification and validation of an ERBB2 gene expression signature in breast cancers. Oncogene 2004, 23, 2564–2575. [Google Scholar] [CrossRef]
- Siggberg, L.; Olsén, P.; Näntö-Salonen, K.; Knuutila, S. 19p13.3 aberrations are associated with dysmorphic features and deviant psychomotor development. Cytogenet. Genome Res. 2011, 132, 8–15. [Google Scholar] [CrossRef]
- Severinsen, J.; Bjarkam, C.R.; Kiar-Larsen, S.; Olsen, I.M.; Nielsen, M.M.; Blechingberg, J.; Nielsen, A.L.; Holm, I.E.; Foldager, L.; Young, B.D.; et al. Evidence implicating BRD1 with brain development and susceptibility to both schizophrenia and bipolar affective disorder. Mol. Psychiatry 2006, 11, 1126–1138. [Google Scholar] [CrossRef]
- Jansen, S.; Kleefstra, T.; Willemsen, M.; De Vries, P.; Pfundt, R.; Hehir-Kwa, J.; Gilissen, C.; Veltman, J.; de Vries, B.; Vissers, L. De novo loss-of-function mutations in X-linked SMC1A cause severe ID and therapy-resistant epilepsy in females: Expanding the phenotypic spectrum. Clin. Genet. 2016, 90, 413–419. [Google Scholar] [CrossRef]
- Porntaveetus, T.; Abid, M.F.; Theerapanon, T.; Srichomthong, C.; Ohazama, A.; Kawasaki, K.; Kawasaki, M.; Suphapeetiporn, K.; Sharpe, P.T.; Shotelersuk, V. Expanding the oro-dental and mutational spectra of Kabuki syndrome and expression of KMT2D and KDM6A in human tooth germs. Int. J. Biol. Sci. 2018, 14, 381. [Google Scholar] [CrossRef]
- Sagi-Dain, L.; Maya, I.; Peleg, A.; Reches, A.; Banne, E.; Baris, H.N.; Tenne, T.; Singer, A.; Ben-Shachar, S. Microarray analysis in pregnancies with isolated unilateral kidney agenesis. Pediatr. Res. 2018, 83, 825–828. [Google Scholar] [CrossRef]
- Žilina, O.; Teek, R.; Tammur, P.; Kuuse, K.; Yakoreva, M.; Vaidla, E.; Mölter-Väär, T.; Reimand, T.; Kurg, A.; Õunap, K. Chromosomal microarray analysis as a first-tier clinical diagnostic test: Estonian experience. Mol. Genet. Genom. Med. 2014, 2, 166–175. [Google Scholar] [CrossRef]
- Hsu, F.; Kent, W.J.; Clawson, H.; Kuhn, R.M.; Diekhans, M.; Haussler, D. The UCSC known genes. Bioinformatics 2006, 22, 1036–1046. [Google Scholar] [CrossRef]



| Syndrome | Implicated Genes from Microarray Studies That Also form Voice Chains | No. of Genes in | No. of Genes in | Reported Effects on Speech |
|---|---|---|---|---|
| 1p36 | SPEN | 1 (10) | 226 (3152) | Del [9], or Abs |
| 1q21.1–q21.2 | RBM8A, GJAS | – | 41 (906) | Del [10] or Imp [11] |
| 1q41–q42 | DISP1, LEFTY1, LEFTY2, BPNT1 | – | 60 (1020) | Apr [12] |
| 1q43–q44 | AKT3 | – | 78 (903) | Del, Imp, or Abs |
| 2p16.1–p15 | – | 17 (508) | Dys, Apr, or Imp [13] | |
| 2p21 | SLC3A1 | – | 24 (557) | Apr or Idio [14] |
| 2q23.1 | – | 2 (31) | Del or Imp | |
| 2q32–q33 | COL3A1, COL5A2, GTF3C3, CASP8, CASP10 | – | 62 (1429) | Abs |
| 2q37.3 | HDAC4 | – | 38 (330) | Imp [15] |
| 3p13 | FOXP1 | – | 8 (48) | Del, Idio, Imp and Dys (all severe), Apr [16] |
| 3q13.31 | DRD3, GAP43, LSAMP | – | 5 (56) | Imp [17] or Abs |
| 3q29 | PAK2, DLG1 | – | 24 (413) | Del |
| 4p16.3 | FGFR3 | 1 (25) | 44 (495) | Del [18] or Abs |
| 4q21 | – | 41 (696) | Del or Abs; Imp [19,20] | |
| 5p (5p15.2 and/or (5p15.3 or 5p15.33)) | TERT, CTNND2 | – | 37 (500) | Del, Abs, and Apr [21] |
| 5q14.3 | MEF2C | – | 7 (275) | Abs |
| 5q33.1 | RPS14 | – | 17 (153) | Dys [22] |
| 5q35.3 | NSD1 | 1 (8) | 47 (653) | Norm [23] or Del |
| 6pter–p24 | FOXC1, GMDS | – | 41 (416) | Del |
| 6q25.3 | ARID1B | – | 21 (363) | Del, Apr, Dys [24] |
| 7p21 | TWIST1 | – | 18 (243) | Del [25] |
| 7q11.23 | ELN, LIMK1, GTF2IRD1, GTF2I | – | 35 (536) | Norm or Del |
| 8p23.1 | GATA4 | – | 49 (337) | No significant anomaly reports |
| 8q22.1 | CCNE2 | – | 13 (152) | Del |
| 8q24.11–q24.13 | EXT1 | – | 21 (211) | Del |
| 9p24.3 | 1 (9) | 3 (13) | Del, Dys, Apr [26] | |
| 9q34.3 | EHMT1 | 1 (62) | 46 (718) | Del [27], Apr [28] and Abs |
| 10pter–p13 or 10p14–p15.1 | GATA3 | – | 49 (796) | Sensorineural hearing loss |
| 10q23 | PTEN, BMPR1A | 1 (107) | 59 (1003) | Del or Abs; Imp [29] |
| 10q26 | DOCK1 | – | 48 (708) | Imp; Del [30] |
| 11p11.2–p12 | EXT2 | – | 46 (737) | Idio, Dys, Del, Apr [31] |
| 11p13–p12 | PAX6, SLC1A2, PRRG4 | – | 23 (342) | Imp [32] |
| 11p15.5 | IGF2 | 1 (347) | 50 (1243) | Imp [33] |
| 11q13.3 | FGF4, FGF3, FADD | – | 11 (608) | Del [34] Imp [35] |
| 11q23 | – | 77 (1421) | Dysarthric, Abs [36]; Imp [37] | |
| 11q23.3-q25 | FLI1, JAM3 | – | 119 (1522) | Imp, Del, Apr [38] |
| 12q14.3 | HMGA2 | – | 9 (108) | Abs, Imp [38] or Del [39] |
| 13q12.3 | – | 6 (119) | Del [40] | |
| 13q14 | RB1 | – | 46 (595) | Norm [41] |
| 13q22.3 | EDNRB | – | 3 (37) | Norm [41] |
| 13q33–q34 | SOX1, ARHGEF7 | – | 30 (635) | Apr |
| 14q11–q22 | PAX9, SUPT16H, CHD8, RALGAPA1 | – | 173 (2021) | Del [42] or Abs |
| 14q22.1–q23.1 | PTGDR, BMP4 | – | 41 (530) | Imp [43] |
| 14q32.2 | DLK1, MEG3 | – | 23 (242) | Del; Idio [44] |
| 15q11.2 | NIPA1, NIPA2, CYFIP1, TUBGCP5 | – | 11 (72) | Del |
| 15q11–q13 | NDN, SNRPN | – | 30 (230) | Del or Imp |
| 15q11–q13 | UBE3A | –Same as above– | Abs; Imp [45] | |
| 15q13.3 | CHRNA7, OTUD7A | – | 8 (47) | Imp or Idio |
| 15q24 | SIN3A | – | 38 (623) | Del or Imp |
| 16p11.2 | SH2B1, TBX6, CORO1A | 1 (1) | 70 (1067) | Apr [46]; Dys [47], Del or Imp |
| 16p12.2–p11.2 | SH2B1 | 1 (1) | 106 (1682) | Del or Imp |
| 16p12.1 | – | 17 (157) | Del | |
| 16p13.11 | MYH11 | – | 6 (116) | Del [48] |
| 16p13.3 | CREBBP, TRAP1 | 1 (140) | 122 (1532) | Apr, Dys, Imp or Del [49] |
| 16q22 | CBFB | 1 (2) | 79 (830) | –Not available– |
| 16q24.3–q24.2 | CDH15, ZNF778, ZFPM1 | – | 28 (307) | Del or Imp [50] |
| 17p11.2 | LLGL1, UBB | – | 36 (845) | Del; Dys [51] |
| 17p13.1 | KCNAB3, GUCY2D, TP53, TRAPPC1, MPDU1, FXR2, EFNB3 | 2 (214) | 74 (1387) | Abs |
| 17p13.3 | PAFAH1B1, YWHAE | – | 37 (487) | Del [52] |
| 17q11.2 | NF1 | – | 40 (620) | No significant issues |
| 17q12 | HNF1B, LHX1, CCL3L3 | 1 (124) | 59 (890) | Del or Imp |
| 17q21.31 | KANSL1, MAPT, CRHR1 | 2 (68) | 46 (740) | Del or Abs |
| 17q23.1–q23.2 | – | 28 (307) | Del [53] | |
| 17q24.3–q24.2 | ABCA5, MAP2K6, SOX9 | – | 22 (720) | Del [54] |
| 18q | MBP | – | 122 (1666) | Del [55] or Imp [56] |
| 19p13.13 | 2 (263) | 131 (2080) | Del; Imp [57] | |
| 19q13.11 | UBA2, WTIP | 1 (23) | 22 (283) | Del or Abs; Imp [58] |
| 20p12.3 | BMP2 | – | 13 (415) | Del or Imp [59] |
| 22q11.2 | TBX1, COMT, TOP3B | – | 66 (1294) | Apr, Dys, Del or Imp [60] |
| 22q12.2 | NF2 | – | 27 (253) | Del or Imp [61] |
| 22q13.3 | ARSA, SHANK3 | 1 (9) | 45 (670) | Del or Abs |
| Xp11.3 | RP2 | 1 (6) | 20 (335) | Imp [62] |
| Xp21 | GK, DMD, NR0B1 | – | 7 (121) | Del [63] |
| Xq28 (a) | ABCD1, BCAP31, SLC6A8 | – | 64 (1053) | Del [64] |
| Xq28 (b) | MECP2 | – | –same as above– | Norm to Abs |
| Yq11 | – | 6 (40) | Norm |
| Speech Type | Count | Chainlink Genes | Chainlink Connectivity | ||
|---|---|---|---|---|---|
| Mean | Median | Mean | Median | ||
| Normal | 6 | 32 | 40 | 404 | 595 |
| Apraxic | 19 | 43 | 30 | 675 | 557 |
| Dysarthric | 11 | 43 | 36 | 725 | 737 |
| Impaired | 31 | 46 | 38 | 734 | 608 |
| Delayed | 51 | 47 | 37 | 682 | 500 |
| Absent | 19 | 60 | 46 | 902 | 718 |
| (a) | ||||||
|---|---|---|---|---|---|---|
| Normal | Apraxic | Dysarthric | Impaired | Delayed | Absent | |
| Normal | 0 | 2.1 | 2.3 | 7.6 | 14.7 | 15.3 |
| Apraxic | 0 | 0.2 | 1.1 | 2.9 | 6.8 | |
| Dysarthric | 0 | 0.2 | 1.2 | 3.7 | ||
| Impaired | 0 | 0.5 | 3.7 | |||
| Delayed | 0 | 4.6 | ||||
| Absent | 0 | |||||
| (b) | ||||||
| Normal | Apraxic | Dysarthric | Impaired | Delayed | Absent | |
| Normal | 0 | 6.3 | 7.0 | 17.4 | 22.5 | 22.2 |
| Apraxic | 0 | 0.7 | 1.5 | 1.9 | 5.9 | |
| Dysarthric | 0 | 0.0 | 0.4 | 1.7 | ||
| Impaired | 0 | 0.9 | 2.4 | |||
| Delayed | 0 | 5.5 | ||||
| Absent | 0 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, R. A Gene-Based Algorithm for Identifying Factors That May Affect a Speaker’s Voice. Entropy 2023, 25, 897. https://doi.org/10.3390/e25060897
Singh R. A Gene-Based Algorithm for Identifying Factors That May Affect a Speaker’s Voice. Entropy. 2023; 25(6):897. https://doi.org/10.3390/e25060897
Chicago/Turabian StyleSingh, Rita. 2023. "A Gene-Based Algorithm for Identifying Factors That May Affect a Speaker’s Voice" Entropy 25, no. 6: 897. https://doi.org/10.3390/e25060897
APA StyleSingh, R. (2023). A Gene-Based Algorithm for Identifying Factors That May Affect a Speaker’s Voice. Entropy, 25(6), 897. https://doi.org/10.3390/e25060897





