Classification of Multiple H&E Images via an Ensemble Computational Scheme
Abstract
:1. Introduction
- A computational scheme capable of indicating the main ensembles of descriptors for the study of histological images, exploring the ReliefF algorithm and multiple classifiers;
- An optimized ensemble of deep-learned features with the best results for classifying colorectal cancer, liver tissue and oral dysplasia, using a reduced number of features (up to 25 descriptors);
- Indications of the discriminative power of ensembles based on fractal features from the LIME and CAM representations;
- Solutions without overfitting and a more robust baseline scheme, with the necessary details for comparisons and improvements of CAD systems focused on H&E images.
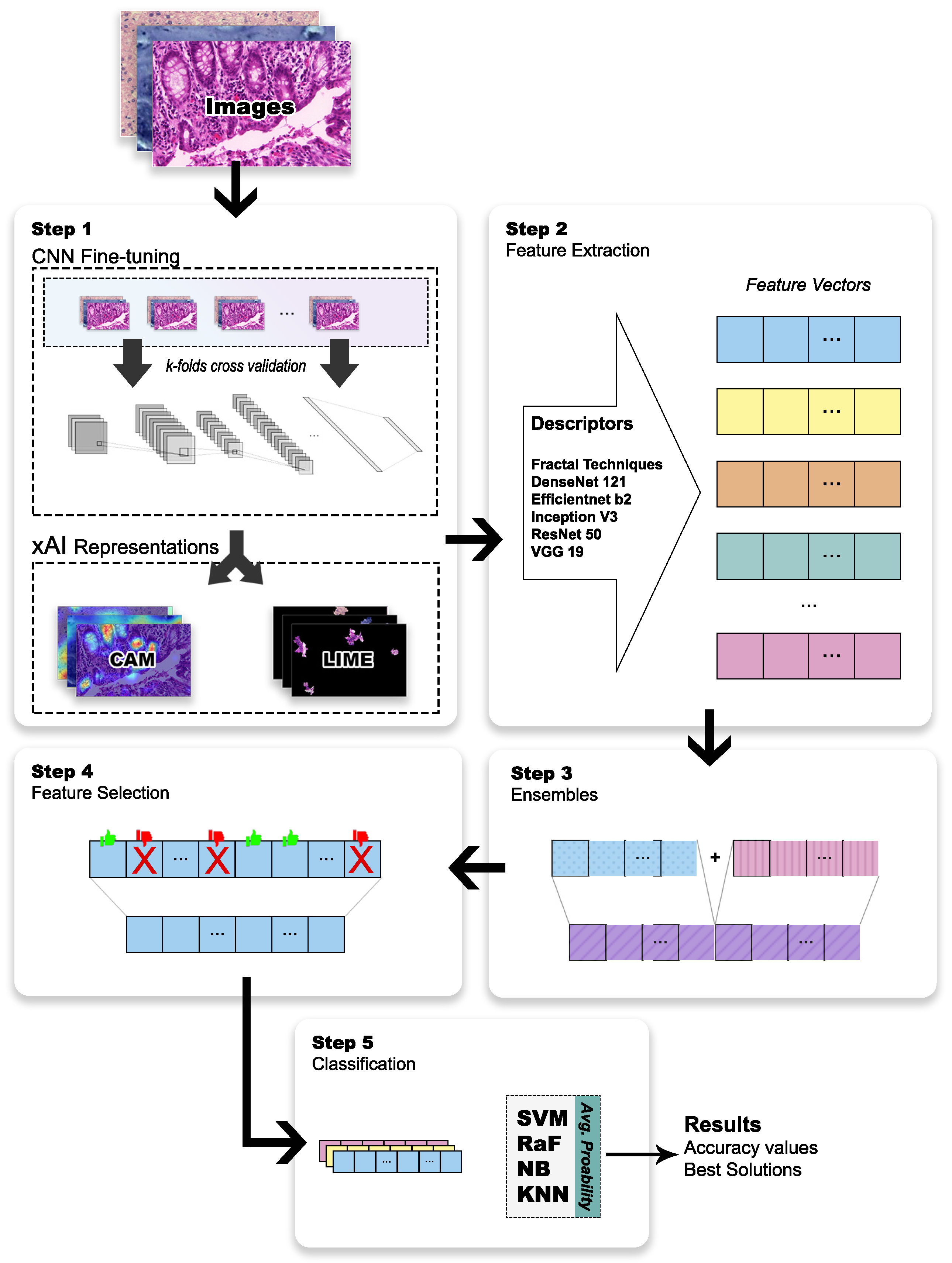
2. Materials and Methods
2.1. Datasets
2.2. Step 1—Fine-Tuning the CNN and xAI Representations

xAI Representations: LIME and Grad-CAM
2.3. Step 2—Feature Extraction
2.3.1. Handcrafted Features: Multiscale and Multidimensional Fractal Techniques
- Area under the curve (A): It indicates the complexity of the texture. For a discrete function consisting of N points defined in , this descriptor can be obtained via Equation (11), with a and b as the point indices that delimit the analysis range;
- Skewness (S): it is defined via Equation (12), where N is the number of points in the function, is the i-th point in the function, is the average of the function values, and a and b are the indices of the points that delimit the interval;
- Area ratio (R): From the asymmetry, the ratio between the halves of the area under the curve must also present similar values for similar classes. This descriptor was obtained through Equation (11), with a and b indicating the points that delimit the interval;
- Maximum point: It indicates the value in the largest heterogeneous area of the curve. Thus, images from the same class can present similar values, for both and x. Totally different values are expected for different classes.
2.3.2. Deep-Learned Features
2.4. Step 3—Feature Ensemble
2.5. Step 4—Feature Selection
2.6. Step 5—Classifier Ensemble and Evaluation Metrics
2.7. Software Packages and Execution Environment
3. Results and Discussion
3.1. Feature Ensemble Performance
3.1.1. Details of the Top 10 Solutions
3.1.2. Feature Summary
3.2. Proposed Scheme versus Fine-Tuned CNN Classifications
3.3. Performance Overview in Relation to the Literature
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| H&E | Hematoxylin and eosin |
| CAD | Computer-aided diagnosis |
| CNN | Convolutional neural network |
| xAI | Explainable artificial intelligence |
| CAM | Class activation mapping |
| Grad-CAM | Gradient-weighted class activation mapping |
| LIME | Local interpretable model-agnostic explanations |
| UCSB | Breast cancer dataset |
| CR | Colorectal cancer dataset |
| LG | Liver tissue dataset |
| OED | Oral epithelial dysplasia dataset |
| SGDM | Stochastic gradient descent |
| F | Fractal techniques |
| D | DenseNet-121 |
| E | EfficientNet-b2 |
| I | Inception-V3 |
| R | ResNet-50 |
| V | VGG-19 |
| Grad-CAM representation via DenseNet-121 | |
| Grad-CAM representation via EfficientNet-b2 | |
| Grad-CAM representation via ResNet-50 | |
| Grad-CAM representation via Inception-V3 | |
| Grad-CAM representation via VGG-19 | |
| LIME representation via DenseNet-121 | |
| LIME representation via EfficientNet-b2 | |
| LIME representation via Inception-V3 | |
| LIME representation via ResNet-50 | |
| LIME representation via VGG-19 | |
| VP | True positives |
| VN | True negatives |
| FP | False positives |
| FN | False negatives |
References
- Gurcan, M.N.; Boucheron, L.E.; Can, A.; Madabhushi, A.; Rajpoot, N.M.; Yener, B. Histopathological image analysis: A review. IEEE Rev. Biomed. Eng. 2009, 2, 147–171. [Google Scholar] [CrossRef] [PubMed]
- Titford, M. A short history of histopathology technique. J. Histotechnol. 2006, 29, 99–110. [Google Scholar] [CrossRef]
- Angel Arul Jothi, J.; Mary Anita Rajam, V. A survey on automated cancer diagnosis from histopathology images. Artif. Intell. Rev. 2017, 48, 31–81. [Google Scholar] [CrossRef]
- Shmatko, A.; Ghaffari Laleh, N.; Gerstung, M.; Kather, J.N. Artificial intelligence in histopathology: Enhancing cancer research and clinical oncology. Nat. Cancer 2022, 3, 1026–1038. [Google Scholar] [CrossRef] [PubMed]
- Carleton, H.M.; Drury, R.A.B.; Wallington, E.A. Carleton’s Histological Technique; Oxford University Press: New York, NY, USA, 1980. [Google Scholar]
- Titford, M. The long history of hematoxylin. Biotech. Histochem. 2005, 80, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.K.C. The Wonderful Colors of the Hematoxylin–Eosin Stain in Diagnostic Surgical Pathology. Int. J. Surg. Pathol. 2014, 22, 12–32. [Google Scholar] [CrossRef] [PubMed]
- Frick, C.; Rumgay, H.; Vignat, J.; Ginsburg, O.; Nolte, E.; Bray, F.; Soerjomataram, I. Quantitative estimates of preventable and treatable deaths from 36 cancers worldwide: A population-based study. Lancet Glob. Health 2023, 11, e1700–e1712. [Google Scholar] [CrossRef]
- Higgins, C. Applications and challenges of digital pathology and whole slide imaging. Biotech. Histochem. 2015, 90, 341–347. [Google Scholar] [CrossRef]
- Hu, W.; Li, X.; Li, C.; Li, R.; Jiang, T.; Sun, H.; Huang, X.; Grzegorzek, M.; Li, X. A state-of-the-art survey of artificial neural networks for whole-slide image analysis: From popular convolutional neural networks to potential visual transformers. Comput. Biol. Med. 2023, 161, 107034. [Google Scholar] [CrossRef]
- He, L.; Long, L.R.; Antani, S.; Thoma, G.R. Histology image analysis for carcinoma detection and grading. Comput. Methods Programs Biomed. 2012, 107, 538–556. [Google Scholar] [CrossRef]
- Roberto, G.F.; Lumini, A.; Neves, L.A.; do Nascimento, M.Z. Fractal Neural Network: A new ensemble of fractal geometry and convolutional neural networks for the classification of histology images. Expert Syst. Appl. 2021, 166, 114103. [Google Scholar] [CrossRef]
- Nanni, L.; Brahnam, S.; Ghidoni, S.; Maguolo, G. General purpose (GenP) bioimage ensemble of handcrafted and learned features with data augmentation. arXiv 2019, arXiv:1904.08084. [Google Scholar]
- Wang, W.; Yang, Y.; Wang, X.; Wang, W.; Li, J. Development of convolutional neural network and its application in image classification: A survey. Opt. Eng. 2019, 58, 040901. [Google Scholar] [CrossRef]
- Wang, Z.; Li, M.; Wang, H.; Jiang, H.; Yao, Y.; Zhang, H.; Xin, J. Breast Cancer Detection Using Extreme Learning Machine Based on Feature Fusion With CNN Deep Features. IEEE Access 2019, 7, 105146–105158. [Google Scholar] [CrossRef]
- Mahbod, A.; Schaefer, G.; Ellinger, I.; Ecker, R.; Pitiot, A.; Wang, C. Fusing fine-tuned deep features for skin lesion classification. Comput. Med. Imaging Graph. 2019, 71, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Alinsaif, S.; Lang, J. Histological Image Classification using Deep Features and Transfer Learning. In Proceedings of the 2020 17th Conference on Computer and Robot Vision (CRV), Ottawa, ON, Canada, 13–15 May 2020; pp. 101–108. [Google Scholar] [CrossRef]
- Zerouaoui, H.; Idri, A.; El Alaoui, O. A new approach for histological classification of breast cancer using deep hybrid heterogenous ensemble. Data Technol. Appl. 2023, 57, 245–278. [Google Scholar] [CrossRef]
- de Oliveira, C.I.; do Nascimento, M.Z.; Roberto, G.F.; Tosta, T.A.; Martins, A.S.; Neves, L.A. Hybrid models for classifying histological images: An association of deep features by transfer learning with ensemble classifier. Multimed. Tools Appl. 2023, 1–24. [Google Scholar] [CrossRef]
- Taino, D.F.; Ribeiro, M.G.; Roberto, G.F.; Zafalon, G.F.; do Nascimento, M.Z.; Tosta, T.A.; Martins, A.S.; Neves, L.A. Analysis of cancer in histological images: Employing an approach based on genetic algorithm. Pattern Anal. Appl. 2021, 24, 483–496. [Google Scholar] [CrossRef]
- Torrey, L.; Shavlik, J. Transfer learning. In Handbook of Research on Machine Learning Applications and Trends: Algorithms, Methods, and Techniques; IGI Global: Hershey, PA, USA, 2010; pp. 242–264. [Google Scholar]
- Nanni, L.; Ghidoni, S.; Brahnam, S. Handcrafted vs. non-handcrafted features for computer vision classification. Pattern Recognit. 2017, 71, 158–172. [Google Scholar] [CrossRef]
- Hagerty, J.R.; Stanley, R.J.; Almubarak, H.A.; Lama, N.; Kasmi, R.; Guo, P.; Drugge, R.J.; Rabinovitz, H.S.; Oliviero, M.; Stoecker, W.V. Deep learning and handcrafted method fusion: Higher diagnostic accuracy for melanoma dermoscopy images. IEEE J. Biomed. Health Inform. 2019, 23, 1385–1391. [Google Scholar] [CrossRef]
- Wei, L.; Su, R.; Wang, B.; Li, X.; Zou, Q.; Gao, X. Integration of deep feature representations and handcrafted features to improve the prediction of N6-methyladenosine sites. Neurocomputing 2019, 324, 3–9. [Google Scholar] [CrossRef]
- Hasan, A.M.; Jalab, H.A.; Meziane, F.; Kahtan, H.; Al-Ahmad, A.S. Combining Deep and Handcrafted Image Features for MRI Brain Scan Classification. IEEE Access 2019, 7, 79959–79967. [Google Scholar] [CrossRef]
- Li, S.; Xu, P.; Li, B.; Chen, L.; Zhou, Z.; Hao, H.; Duan, Y.; Folkert, M.; Ma, J.; Huang, S.; et al. Predicting lung nodule malignancies by combining deep convolutional neural network and handcrafted features. Phys. Med. Biol. 2019, 64, 175012. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, S.; Singh, S.K. Ensembling handcrafted features with deep features: An analytical study for classification of routine colon cancer histopathological nuclei images. Multimed. Tools Appl. 2020, 79, 34931–34954. [Google Scholar] [CrossRef]
- Fortin, C.S.; Kumaresan, R.; Ohley, W.J.; Hoefer, S. Fractal dimension in the analysis of medical images. IEEE Eng. Med. Biol. Mag. 1992, 11, 65–71. [Google Scholar] [CrossRef]
- Aralica, G.; Ivelj, M.Š.; Pačić, A.; Baković, J.; Periša, M.M.; Krištić, A.; Konjevoda, P. Prognostic Significance of Lacunarity in Preoperative Biopsy of Colorectal Cancer. Pathol. Oncol. Res. 2020, 26, 2567–2576. [Google Scholar] [CrossRef]
- Jain, A.K.; Duin, R.P.W.; Mao, J. Statistical pattern recognition: A review. IEEE Trans. Pattern Anal. Mach. Intell. 2000, 22, 4–37. [Google Scholar] [CrossRef]
- Qin, J.; Puckett, L.; Qian, X. Image Based Fractal Analysis for Detection of Cancer Cells. In Proceedings of the 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Seoul, Republic of Korea, 16–19 December 2020; pp. 1482–1486. [Google Scholar] [CrossRef]
- Roberto, G.F.; Neves, L.A.; Nascimento, M.Z.; Tosta, T.A.; Longo, L.C.; Martins, A.S.; Faria, P.R. Features based on the percolation theory for quantification of non-Hodgkin lymphomas. Comput. Biol. Med. 2017, 91, 135–147. [Google Scholar] [CrossRef]
- Roberto, G.F.; Nascimento, M.Z.; Martins, A.S.; Tosta, T.A.; Faria, P.R.; Neves, L.A. Classification of breast and colorectal tumors based on percolation of color normalized images. Comput. Graph. 2019, 84, 134–143. [Google Scholar] [CrossRef]
- Candelero, D.; Roberto, G.F.; Do Nascimento, M.Z.; Rozendo, G.B.; Neves, L.A. Selection of cnn, haralick and fractal features based on evolutionary algorithms for classification of histological images. In Proceedings of the 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Seoul, Republic of Korea, 16–19 December 2020; pp. 2709–2716. [Google Scholar]
- Khan, A.; Sohail, A.; Zahoora, U.; Qureshi, A.S. A survey of the recent architectures of deep convolutional neural networks. Artif. Intell. Rev. 2020, 53, 5455–5516. [Google Scholar] [CrossRef]
- Khedkar, S.; Subramanian, V.; Shinde, G.; Gandhi, P. Explainable AI in healthcare. In Proceedings of the 2nd International Conference on Advances in Science & Technology (ICAST), Mumbai, India, 8–9 April 2019. [Google Scholar]
- Wells, L.; Bednarz, T. Explainable ai and reinforcement learning—A systematic review of current approaches and trends. Front. Artif. Intell. 2021, 4, 550030. [Google Scholar] [CrossRef] [PubMed]
- Samek, W.; Müller, K.R. Towards explainable artificial intelligence. In Explainable AI: Interpreting, Explaining and Visualizing Deep Learning; Springer: Berlin/Heidelberg, Germany, 2019; pp. 5–22. [Google Scholar]
- Gunning, D.; Aha, D. DARPA’s explainable artificial intelligence (XAI) program. AI Mag. 2019, 40, 44–58. [Google Scholar]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning deep features for discriminative localization. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2921–2929. [Google Scholar]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why should i trust you?” Explaining the predictions of any classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 1135–1144. [Google Scholar]
- Bishop, C.M. Pattern Recognition and Machine Learning; Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Watanabe, S. Pattern Recognition: Human and Mechanical; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 1985. [Google Scholar]
- Bolón-Canedo, V.; Sánchez-Marono, N.; Alonso-Betanzos, A.; Benítez, J.M.; Herrera, F. A review of microarray datasets and applied feature selection methods. Inf. Sci. 2014, 282, 111–135. [Google Scholar] [CrossRef]
- Urbanowicz, R.J.; Meeker, M.; La Cava, W.; Olson, R.S.; Moore, J.H. Relief-based feature selection: Introduction and review. J. Biomed. Inform. 2018, 85, 189–203. [Google Scholar] [CrossRef]
- Manhrawy, I.I.; Qaraad, M.; El-Kafrawy, P. Hybrid feature selection model based on relief-based algorithms and regulizer algorithms for cancer classification. Concurr. Comput. Pract. Exp. 2021, 33, e6200. [Google Scholar] [CrossRef]
- Ghosh, P.; Azam, S.; Jonkman, M.; Karim, A.; Shamrat, F.J.M.; Ignatious, E.; Shultana, S.; Beeravolu, A.R.; De Boer, F. Efficient prediction of cardiovascular disease using machine learning algorithms with relief and LASSO feature selection techniques. IEEE Access 2021, 9, 19304–19326. [Google Scholar] [CrossRef]
- Zebari, R.; Abdulazeez, A.; Zeebaree, D.; Zebari, D.; Saeed, J. A comprehensive review of dimensionality reduction techniques for feature selection and feature extraction. J. Appl. Sci. Technol. Trends 2020, 1, 56–70. [Google Scholar] [CrossRef]
- Li, M.; Ma, X.; Chen, C.; Yuan, Y.; Zhang, S.; Yan, Z.; Chen, C.; Chen, F.; Bai, Y.; Zhou, P.; et al. Research on the auxiliary classification and diagnosis of lung cancer subtypes based on histopathological images. IEEE Access 2021, 9, 53687–53707. [Google Scholar] [CrossRef]
- Burçak, K.C.; Uğuz, H. A New Hybrid Breast Cancer Diagnosis Model Using Deep Learning Model and ReliefF. Trait. Signal 2022, 39, 521–529. [Google Scholar] [CrossRef]
- Silva, A.B.; De Oliveira, C.I.; Pereira, D.C.; Tosta, T.A.; Martins, A.S.; Loyola, A.M.; Cardoso, S.V.; De Faria, P.R.; Neves, L.A.; Do Nascimento, M.Z. Assessment of the association of deep features with a polynomial algorithm for automated oral epithelial dysplasia grading. In Proceedings of the 2022 35th SIBGRAPI Conference on Graphics, Patterns and Images (SIBGRAPI), Natal, Brazil, 24–27 October 2022; Volume 1, pp. 264–269. [Google Scholar]
- Dietterich, T.G. Ensemble methods in machine learning. In Proceedings of the International Workshop on Multiple Classifier Systems, Cagliari, Italy, 21–23 June 2000; Springer: Berlin/Heidelberg, Germany, 2000; pp. 1–15. [Google Scholar]
- Sagi, O.; Rokach, L. Ensemble learning: A survey. WIREs Data Min. Knowl. Discov. 2018, 8, e1249. [Google Scholar] [CrossRef]
- Lumini, A.; Nanni, L. Convolutional neural networks for ATC classification. Curr. Pharm. Des. 2018, 24, 4007–4012. [Google Scholar] [CrossRef] [PubMed]
- Kausar, T.; Wang, M.; Idrees, M.; Lu, Y. HWDCNN: Multi-class recognition in breast histopathology with Haar wavelet decomposed image based convolution neural network. Biocybern. Biomed. Eng. 2019, 39, 967–982. [Google Scholar] [CrossRef]
- Toğaçar, M.; Cömert, Z.; Ergen, B. Enhancing of dataset using DeepDream, fuzzy color image enhancement and hypercolumn techniques to detection of the Alzheimer’s disease stages by deep learning model. Neural Comput. Appl. 2021, 33, 9877–9889. [Google Scholar] [CrossRef]
- Maia, B.M.S.; de Assis, M.C.F.R.; de Lima, L.M.; Rocha, M.B.; Calente, H.G.; Correa, M.L.A.; Camisasca, D.R.; Krohling, R.A. Transformers, convolutional neural networks, and few-shot learning for classification of histopathological images of oral cancer. Expert Syst. Appl. 2023, 241, 122418. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Reddy, A.S.B.; Juliet, D.S. Transfer Learning with ResNet-50 for Malaria Cell-Image Classification. In Proceedings of the 2019 International Conference on Communication and Signal Processing (ICCSP), Tamil Nadu, India, 4–6 April 2019; pp. 0945–0949. [Google Scholar] [CrossRef]
- Ganguly, A.; Das, R.; Setua, S.K. Histopathological Image and Lymphoma Image Classification using customized Deep Learning models and different optimization algorithms. In Proceedings of the 2020 11th International Conference on Computing, Communication and Networking Technologies (ICCCNT), Kharagpur, India, 1–3 July 2020; pp. 1–7. [Google Scholar] [CrossRef]
- Al-Haija, Q.A.; Adebanjo, A. Breast Cancer Diagnosis in Histopathological Images Using ResNet-50 Convolutional Neural Network. In Proceedings of the 2020 IEEE International IOT, Electronics and Mechatronics Conference (IEMTRONICS), Vancouver, BC, Canada, 9–12 September 2020; pp. 1–7. [Google Scholar] [CrossRef]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, PMLR, Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]
- Gelasca, E.D.; Byun, J.; Obara, B.; Manjunath, B. Evaluation and Benchmark for Biological Image Segmentation. In Proceedings of the IEEE International Conference on Image Processing, San Diego, CA, USA, 12–15 October 2008. [Google Scholar]
- Sirinukunwattana, K.; Pluim, J.P.; Chen, H.; Qi, X.; Heng, P.A.; Guo, Y.B.; Wang, L.Y.; Matuszewski, B.J.; Bruni, E.; Sanchez, U.; et al. Gland segmentation in colon histology images: The glas challenge contest. Med. Image Anal. 2017, 35, 489–502. [Google Scholar] [CrossRef] [PubMed]
- AGEMAP. The Atlas of Gene Expression in Mouse Aging Project (AGEMAP). 2020.
- Silva, A.B.; Martins, A.S.; Tosta, T.A.A.; Neves, L.A.; Servato, J.P.S.; de Araújo, M.S.; de Faria, P.R.; do Nascimento, M.Z. Computational analysis of histological images from hematoxylin and eosin-stained oral epithelial dysplasia tissue sections. Expert Syst. Appl. 2022, 193, 116456. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. In Advances in Neural Information Processing Systems 32; Curran Associates, Inc.: New York, NY, USA, 2019; pp. 8024–8035. [Google Scholar]
- torchvision.models. 2021. Available online: https://pytorch.org/vision/stable/models.html (accessed on 24 December 2023).
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- dos Santos, F.P.; Ponti, M.A. Alignment of Local and Global Features from Multiple Layers of Convolutional Neural Network for Image Classification. In Proceedings of the 2019 32nd SIBGRAPI Conference on Graphics, Patterns and Images (SIBGRAPI), Rio de Janeiro, Brazil, 28–31 October 2019; pp. 241–248. [Google Scholar]
- Van der Walt, S.; Schönberger, J.L.; Nunez-Iglesias, J.; Boulogne, F.; Warner, J.D.; Yager, N.; Gouillart, E.; Yu, T. scikit-image: Image processing in Python. PeerJ 2014, 2, e453. [Google Scholar] [CrossRef]
- Cantrell, C.D. Modern Mathematical Methods for Physicists and Engineers; Cambridge University Press: Cambridge, MA, USA, 2000. [Google Scholar]
- Allain, C.; Cloitre, M. Characterizing the lacunarity of random and deterministic fractal sets. Phys. Rev. A 1991, 44, 3552. [Google Scholar] [CrossRef]
- Ivanovici, M.; Richard, N. Fractal dimension of color fractal images. IEEE Trans. Image Process. 2010, 20, 227–235. [Google Scholar] [CrossRef]
- Hoshen, J.; Kopelman, R. Percolation and cluster distribution. I. Cluster multiple labeling technique and critical concentration algorithm. Phys. Rev. B 1976, 14, 3438. [Google Scholar] [CrossRef]
- Căliman, A.; Ivanovici, M. Psoriasis image analysis using color lacunarity. In Proceedings of the 2012 13th International Conference on Optimization of Electrical and Electronic Equipment (OPTIM), Brasov, Romania, 24–26 May 2012; pp. 1401–1406. [Google Scholar]
- Segato dos Santos, L.F.; Neves, L.A.; Rozendo, G.B.; Ribeiro, M.G.; Zanchetta do Nascimento, M.; Azevedo Tosta, T.A. Multidimensional and fuzzy sample entropy (SampEnMF) for quantifying H&E histological images of colorectal cancer. Comput. Biol. Med. 2018, 103, 148–160. [Google Scholar] [CrossRef] [PubMed]
- Tosta, T.A.; Bruno, D.O.; Longo, L.C.; do Nascimento, M.Z. Colour Feature Extraction and Polynomial Algorithm for Classification of Lymphoma Images. In Proceedings of the Progress in Pattern Recognition, Image Analysis, Computer Vision, and Applications: 24th Iberoamerican Congress, CIARP 2019, Havana, Cuba, 28–31 October 2019; Springer Nature: Berlin/Heidelberg, Germany, 2019; Volume 11896, p. 262. [Google Scholar]
- Ribeiro, M.G.; Neves, L.A.; do Nascimento, M.Z.; Roberto, G.F.; Martins, A.S.; Azevedo Tosta, T.A. Classification of colorectal cancer based on the association of multidimensional and multiresolution features. Expert Syst. Appl. 2019, 120, 262–278. [Google Scholar] [CrossRef]
- Dasigi, V.; Mann, R.C.; Protopopescu, V.A. Information fusion for text classification—An experimental comparison. Pattern Recognit. 2001, 34, 2413–2425. [Google Scholar] [CrossRef]
- Kononenko, I.; Šimec, E.; Robnik-Šikonja, M. Overcoming the myopia of inductive learning algorithms with RELIEFF. Appl. Intell. 1997, 7, 39–55. [Google Scholar] [CrossRef]
- Demir, S.; Key, S.; Baygin, M.; Tuncer, T.; Dogan, S.; Belhaouari, S.B.; Poyraz, A.K.; Gurger, M. Automated knee ligament injuries classification method based on exemplar pyramid local binary pattern feature extraction and hybrid iterative feature selection. Biomed. Signal Process. Control 2022, 71, 103191. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Pedrini, H.; Schwartz, W.R. Análise de Imagens Digitais: Princípios, Algoritmos e Aplicações; Thomson Learning: Chicago, IL, USA, 2008. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Cunningham, P.; Delany, S.J. k-Nearest Neighbour Classifiers. arXiv 2020, arXiv:2004.04523. [Google Scholar]
- Martinez, E.; Louzada, F.; Pereira, B. A curva ROC para testes diagnósticos. Cad Saúde Coletiva 2003, 11, 7–31. [Google Scholar]
- Kohavi, R. A study of cross-validation and bootstrap for accuracy estimation and model selection. In Proceedings of the International Joint Conferences on Artificial Intelligence, Montreal, QC, Canada, 20–25 August 1995; Volume 14, pp. 1137–1145. [Google Scholar]
- Gildenblat, J. PyTorch Library for CAM Methods. 2021. Available online: https://github.com/jacobgil/pytorch-grad-cam (accessed on 24 December 2023).
- Ribeiro, M.T. Lime. 2016. Available online: https://github.com/marcotcr/lime (accessed on 20 December 2023).
- MATLAB, version 9.7.0 (R2019b); The MathWorks Inc.: Natick, MA, USA, 2019.
- Frank, E.; Hall, M.A.; Holmes, G.; Kirkby, R.; Pfahringer, B.; Witten, I.H. Weka: A machine learning workbench for data mining. In Data Mining and Knowledge Discovery Handbook: A Complete Guide for Practitioners and Researchers; Maimon, O., Rokach, L., Eds.; Springer: Berlin/Heidelberg, Germany, 2005; pp. 1305–1314. [Google Scholar]
- Tavolara, T.E.; Niazi, M.K.K.; Arole, V.; Chen, W.; Frankel, W.; Gurcan, M.N. A modular cGAN classification framework: Application to colorectal tumor detection. Sci. Rep. 2019, 9, 18969. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Chen, H.; Li, Y.; Peng, Y.; Li, J.; Yang, F. Breast cancer classification in pathological images based on hybrid features. Multimed. Tools Appl. 2019, 78, 21325–21345. [Google Scholar] [CrossRef]
- Zhang, R.; Zhu, J.; Yang, S.; Hosseini, M.S.; Genovese, A.; Chen, L.; Rowsell, C.; Damaskinos, S.; Varma, S.; Plataniotis, K.N. HistoKT: Cross Knowledge Transfer in Computational Pathology. In Proceedings of the ICASSP 2022—2022 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Singapore, 22–27 May 2022; pp. 1276–1280. [Google Scholar]
- Azarmehr, N.; Shephard, A.; Mahmood, H.; Rajpoot, N.; Khurram, S.A. A neural architecture search based framework for segmentation of epithelium, nuclei and oral epithelial dysplasia grading. In Proceedings of the Medical Imaging with Deep Learning, Zürich, Switzerland, 6–8 July 2022. [Google Scholar]
- Dabass, M.; Vig, R.; Vashisth, S. Five-grade cancer classification of colon histology images via deep learning. In Communication and Computing Systems; CRC Press: Boca Raton, FL, USA, 2019; pp. 18–24. [Google Scholar]
- Sena, P.; Fioresi, R.; Faglioni, F.; Losi, L.; Faglioni, G.; Roncucci, L. Deep learning techniques for detecting preneoplastic and neoplastic lesions in human colorectal histological images. Oncol. Lett. 2019, 18, 6101–6107. [Google Scholar] [CrossRef] [PubMed]
- Bentaieb, A.; Hamarneh, G. Adversarial Stain Transfer for Histopathology Image Analysis. IEEE Trans. Med. Imaging 2018, 37, 792–802. [Google Scholar] [CrossRef] [PubMed]
- Awan, R.; Al-Maadeed, S.; Al-Saady, R.; Bouridane, A. Glandular structure-guided classification of microscopic colorectal images using deep learning. Comput. Electr. Eng. 2020, 85, 106450. [Google Scholar] [CrossRef]
- Di Ruberto, C.; Putzu, L.; Arabnia, H.; Quoc-Nam, T. A feature learning framework for histology images classification. In Emerging Trends in Applications and Infrastructures for Computational Biology, Bioinformatics, and Systems Biology: Systems and Applications; Elsevier B.V.: Amsterdam, The Netherlands, 2016; pp. 37–48. [Google Scholar]
- Andrearczyk, V.; Whelan, P.F. Deep learning for biomedical texture image analysis. In Proceedings of the Irish Machine Vision & Image Processing Conference, Kildare, Ireland, 30 August–1 September 2017; Irish Pattern Recognition & Classification Society (IPRCS): Dublin, Ireland, 2017. [Google Scholar]
- Watanabe, K.; Kobayashi, T.; Wada, T. Semi-supervised feature transformation for tissue image classification. PLoS ONE 2016, 11, e0166413. [Google Scholar] [CrossRef] [PubMed]
- Adel, D.; Mounir, J.; El-Shafey, M.; Eldin, Y.A.; El Masry, N.; AbdelRaouf, A.; Abd Elhamid, I.S. Oral epithelial dysplasia computer aided diagnostic approach. In Proceedings of the 2018 13th International Conference on Computer Engineering and Systems (ICCES), Cairo, Egypt, 18–19 December 2018; pp. 313–318. [Google Scholar]
- Krishnan, M.M.R.; Shah, P.; Choudhary, A.; Chakraborty, C.; Paul, R.R.; Ray, A.K. Textural characterization of histopathological images for oral sub-mucous fibrosis detection. Tissue Cell 2011, 43, 318–330. [Google Scholar] [CrossRef]
- Li, Y.; Xie, X.; Shen, L.; Liu, S. Reverse active learning based atrous DenseNet for pathological image classification. BMC Bioinform. 2019, 20, 445. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, L.; Yi, Z. Breast cancer cell nuclei classification in histopathology images using deep neural networks. Int. J. Comput. Assist. Radiol. Surg. 2018, 13, 179–191. [Google Scholar] [CrossRef]
- Papastergiou, T.; Zacharaki, E.I.; Megalooikonomou, V. Tensor decomposition for multiple-instance classification of high-order medical data. Complexity 2018, 2018, 8651930. [Google Scholar] [CrossRef]
- Araújo, T.; Aresta, G.; Castro, E.; Rouco, J.; Aguiar, P.; Eloy, C.; Polónia, A.; Campilho, A. Classification of breast cancer histology images using convolutional neural networks. PLoS ONE 2017, 12, e0177544. [Google Scholar] [CrossRef] [PubMed]









| Dataset | Image Type | Number of Classes | Classes | Number of Samples | Resolution |
|---|---|---|---|---|---|
| UCSB [66] | Breast cancer | 2 | Malignant and benign | 58 (32/26) | |
| CR [67] | Colorectal cancer | 2 | Malignant and benign | 165 (74/91) | from to |
| LG [68] | Liver tissue | 2 | Male and female | 265 (150/115) | |
| OED [69] | Oral epithelial dysplasia | 2 | Healthy and severe | 148 (74/74) |
| Architecture | Parameter | Layers | Accuracy (ImageNet) |
|---|---|---|---|
| DenseNet-121 [58] | 121 | 91.97% | |
| EfficientNet-b2 [65] | 324 | 95.31% | |
| Inception-V3 [59] | 48 | 93.45% | |
| ResNet-50 [60] | 50 | 92.86% | |
| VGG-19 [61] | 19 | 90.87% |
| Origin | Composition | Number of Features |
|---|---|---|
| Handcrafted | Fractals (F) | 116 |
| Deep learned | DenseNet-121 (D) | 1024 |
| EfficientNet-b2 (E) | 1408 | |
| Inception-V3 (I) | 2048 | |
| ResNet-50 (R) | 2048 | |
| VGG-19 (V) | 4096 | |
| xAI | Grad-CAM DenseNet-121 () | 116 |
| Grad-CAM EfficientNet-b2 () | 116 | |
| Grad-CAM Inception-V3 () | 116 | |
| Grad-CAM ResNet-50 () | 116 | |
| Grad-CAM VGG-19 () | 116 | |
| LIME DenseNet-121 () | 116 | |
| LIME EfficientNet-b2 () | 116 | |
| LIME Inception-V3 () | 116 | |
| LIME ResNet-50 () | 116 | |
| LIME VGG-19 () | 116 |
| Composition | Number of Features |
|---|---|
| F + D | 1140 |
| F + E | 1524 |
| F + I | 2164 |
| F + R | 2164 |
| F + V | 4212 |
| F + D + E + I + R + V | 10,740 |
| Composition | Number of Features |
|---|---|
| D + E | 2432 |
| D + I | 3072 |
| D + R | 3072 |
| D + V | 5120 |
| E + I | 3456 |
| E + R | 3456 |
| E + V | 5504 |
| I + R | 4096 |
| I + V | 6144 |
| R + V | 6144 |
| D + E + I + R + V | 10,624 |
| Composition | Number of Features |
|---|---|
| + | 232 |
| + | 232 |
| + | 232 |
| + | 232 |
| + | 232 |
| + | 232 |
| + | 232 |
| + | 232 |
| + | 232 |
| + | 232 |
| + + + + | 580 |
| + | 232 |
| + | 232 |
| + | 232 |
| + | 232 |
| + | 232 |
| + | 232 |
| + | 232 |
| + | 232 |
| + | 232 |
| + | 232 |
| + + + + | 580 |
| CR | LG | OED | UCSB | |
|---|---|---|---|---|
| Handcrafted | 84.48% ± 2.05 | 90.42% ± 5.23 | 87.03% ± 1.78 | 72.41% ± 1.89 |
| Deep learned | 99.27% ± 0.76 | 98.49% ± 0.65 | 96.28% ± 0.55 | 91.38% ± 1.54 |
| xAI | 83.82% ± 0.90 | 86.98% ± 2.54 | 80.88% ± 0.43 | 78.28% ± 1.14 |
| Ensemble of handcrafted and deep learned | 99.76% ± 0.30 | 99.02% ± 0.51 | 96.49% ± 0.66 | 90.52% ± 0.86 |
| Ensemble of deep learned | 100% | 99.66% ± 0.11 | 97.23% ± 0.47 | 92.93% ± 0.93 |
| Ensemble of xAI | 86.18% ± 1.45 | 89.62% ± 0.42 | 78.85% ± 0.61 | 78.45% ± 1.16 |
| p-Value | Ensemble of Deep Learned | Ensemble of Handcrafted and Deep Learned | Deep Learned | Handcrafted | Ensemble of xAI | xAI | Average Ranking |
|---|---|---|---|---|---|---|---|
| Ensemble of deep learned | - | 0.3955 | 0.2396 | 0.0191 | 0.0191 | 0.0066 | 1 |
| Ensemble of handcrafted and deep learned | 0.3955 | - | 0.7313 | 0.1006 | 0.1006 | 0.0380 | 2.25 |
| Deep learned | 0.2396 | 0.7313 | - | 0.1819 | 0.1819 | 0.0735 | 2.75 |
| Handcrafted | 0.0191 | 0.1006 | 0.1819 | - | 1.0000 | 0.6073 | 4.75 |
| Ensemble of xAI | 0.0191 | 0.1006 | 0.1819 | 1.0000 | - | 0.6073 | 4.75 |
| xAI | 0.0066 | 0.0380 | 0.0735 | 0.6073 | 0.6073 | - | 5.5 |
| Feature Vector | Size | Accuracy (%) | F-1 Score |
|---|---|---|---|
| D + E | 10 | 100 | 1.000 |
| E + V | 10 | 100 | 1.000 |
| D + E + I + R + V | 10 | 100 | 1.000 |
| D + E | 15 | 100 | 1.000 |
| D + I | 15 | 100 | 1.000 |
| E + V | 15 | 100 | 1.000 |
| D + E + I + R + V | 15 | 100 | 1.000 |
| D + E | 20 | 100 | 1.000 |
| D + I | 20 | 100 | 1.000 |
| D + V | 20 | 100 | 1.000 |
| Feature Vector | Size | Accuracy (%) | F-1 Score |
|---|---|---|---|
| D + R | 25 | 100 | 1.000 |
| D + E | 10 | 99.62 | 0.996 |
| D + E | 15 | 99.62 | 0.996 |
| D + I | 15 | 99.62 | 0.996 |
| D + R | 15 | 99.62 | 0.996 |
| D + E | 20 | 99.62 | 0.996 |
| D + I | 20 | 99.62 | 0.996 |
| D + R | 20 | 99.62 | 0.996 |
| D + E | 25 | 99.62 | 0.996 |
| D + I | 25 | 99.62 | 0.996 |
| Feature Vector | Size | Accuracy (%) | F1-Score |
|---|---|---|---|
| I + V | 20 | 97.97 | 0.980 |
| E + I | 25 | 97.97 | 0.980 |
| D + E | 20 | 97.30 | 0.973 |
| D + I | 20 | 97.30 | 0.973 |
| I + R | 20 | 97.30 | 0.973 |
| D + R | 25 | 97.30 | 0.973 |
| I + V | 25 | 97.30 | 0.973 |
| D + I | 10 | 96.62 | 0.966 |
| D + E | 15 | 96.62 | 0.966 |
| I + V | 15 | 96.62 | 0.966 |
| Feature Vector | Size | Accuracy (%) | F1-Score |
|---|---|---|---|
| D + E | 25 | 94.83 | 0.948 |
| D + E | 15 | 93.10 | 0.931 |
| E + R | 15 | 93.10 | 0.931 |
| I + R | 15 | 93.10 | 0.931 |
| D + E | 20 | 93.10 | 0.931 |
| D + I | 25 | 93.10 | 0.931 |
| E + I | 25 | 93.10 | 0.931 |
| I + R | 25 | 93.10 | 0.931 |
| E + R | 10 | 91.38 | 0.914 |
| D + R | 15 | 91.38 | 0.914 |
| Method | Approach | Accuracy (%) |
|---|---|---|
| Proposed | DenseNet-121 and EfficientNet-b2 (ensemble of deep-learned features) | 100% |
| Roberto et al. [12] | ResNet-50, fractal dimension, lacunarity and percolation (ensemble of handcrafted and deep-learned features) | 99.39% |
| Dabass et al. [101] | 31-layer CNN (deep learning) | 96.97% |
| de Oliveira et al. [19] | ResNet50 (activation_48_relu layer), ReliefF and 35 deep-learned features | 98.00% |
| Tavolara et al. [97] | GAN and U-Net (deep learning) | 94.02% |
| Sena et al. [102] | 12-layer CNN (deep learning) | 93.28% |
| Segato dos Santos et al. [81] | Sample entropy and fuzzy logic (handcrafted) | 91.39% |
| Roberto et al. [33] | Percolation (handcrafted) | 90.90% |
| Bentaieb and Hamarneh [103] | U-Net and AlexNet (deep learning) | 87.50% |
| Zhang et al. [99] | ResNet deep-tuning (DL) | 86.67% |
| Awan et al. [104] | Color normalization, U-Net and GoogLeNet (deep learning) | 85.00% |
| Method | Approach | Accuracy (%) |
|---|---|---|
| Proposed | DenseNet-121 and ResNet-50 (ensemble of deep-learned features) | 100% |
| Di Ruberto et al. [105] | Statistical analysis and texture features (handcrafted) | 100% |
| Nanni et al. [13] | 6 CNNs and handcrafted features (ensemble of handcrafted and deep-learned features) | 100% |
| Roberto et al. [12] | ResNet-50, fractal dimension, lacunarity and percolation (ensemble of handcrafted and deep-learned features) | 99.62% |
| de Oliveira et al. [19] | ResNet50 (activation_48_relu layer), ReliefF and 5 deep-learned features | 99.32% |
| Andrearczyk and Whelan [106] | Texture CNN (deep learning) | 99.10% |
| Watanabe et al. [107] | GIST descriptor, PCA and LDA (handcrafted) | 93.70% |
| Method | Approach | Accuracy (%) |
|---|---|---|
| Proposed | Inception-V3 and VGG-19 (ensemble of deep-learned features) | 97.97% |
| Adel et al. [108] | SIFT, SURF, ORB (handcrafted) | 92.80% |
| Azarmehr et al. [100] | Neural architecture search and handcrafted descriptors (morphological and nonmorphological) | 95.20% |
| Silva et al. [69] | Morphological and nonmorphological features (handcrafted) | 92.40% |
| Maia et al. [57] | Densenet121 | 91.91% |
| Krishnan et al. [109] | Fractal dimension, wavelet, Brownian movement and Gabor filters (handcrafted) | 88.38% |
| Method | Approach | Accuracy (%) |
|---|---|---|
| Li et al. [110] | RefineNet and Atrous DenseNet (deep learning) | 97.63% |
| Yu et al. [98] | CNN, LBP, SURF, GLCM and other handcrafted features (ensemble handcrafted and deep-learned features) | 96.67% |
| Proposed | DenseNet-121 and EfficientNet-b2 (ensemble of deep-learned features) | 94.83% |
| Feng et al. [111] | Stacked denoising autoencoder (deep learning) | 94.41% |
| Kausar et al. [55] | Color normalization, Haar wavelet decomposition and 16-layer CNN (deep learning) | 91.00% |
| Roberto et al. [12] | ResNet-50, fractal dimension, lacunarity and percolation (ensemble of handcrafted and deep-learned features) | 89.66% |
| Roberto et al. [33] | Percolation (handcrafted) | 86.20% |
| Papastergiou et al. [112] | Spacial decomposition and tensors (deep learning) | 84.67% |
| Araújo et al. [113] | Color normalization, 13-layer CNN and SVM (deep learning) | 83.30% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Longo, L.H.d.C.; Roberto, G.F.; Tosta, T.A.A.; de Faria, P.R.; Loyola, A.M.; Cardoso, S.V.; Silva, A.B.; do Nascimento, M.Z.; Neves, L.A. Classification of Multiple H&E Images via an Ensemble Computational Scheme. Entropy 2024, 26, 34. https://doi.org/10.3390/e26010034
Longo LHdC, Roberto GF, Tosta TAA, de Faria PR, Loyola AM, Cardoso SV, Silva AB, do Nascimento MZ, Neves LA. Classification of Multiple H&E Images via an Ensemble Computational Scheme. Entropy. 2024; 26(1):34. https://doi.org/10.3390/e26010034
Chicago/Turabian StyleLongo, Leonardo H. da Costa, Guilherme F. Roberto, Thaína A. A. Tosta, Paulo R. de Faria, Adriano M. Loyola, Sérgio V. Cardoso, Adriano B. Silva, Marcelo Z. do Nascimento, and Leandro A. Neves. 2024. "Classification of Multiple H&E Images via an Ensemble Computational Scheme" Entropy 26, no. 1: 34. https://doi.org/10.3390/e26010034
APA StyleLongo, L. H. d. C., Roberto, G. F., Tosta, T. A. A., de Faria, P. R., Loyola, A. M., Cardoso, S. V., Silva, A. B., do Nascimento, M. Z., & Neves, L. A. (2024). Classification of Multiple H&E Images via an Ensemble Computational Scheme. Entropy, 26(1), 34. https://doi.org/10.3390/e26010034









