Brain Tumor Segmentation of MRI Images Using Processed Image Driven U-Net Architecture
Abstract
:1. Introduction
- Explored advanced deep learning models in depth and proved the conjectured performance evidence of U-Net deep learning models for brain tumor image segmentation for detection.
- Experimented segmentation with standard U-Net, further explored and incorporated subset division, category brain slicing, feature scaling, and narrow object region filtering prior to feeding to U-Net model and examined the performance enhancement of U-Net architecture by incorporating all these pre-learning processes.
- Specifically examined some challenging unsuccessful tumor segmentation test cases and observe a substantial parameter tuning for result refinement.
- The manuscript presented a state-of-the-art brain tumor detection technique which, in combination with a few image processing processes, followed by a deep learning model, can provide accurate brain tumor segmentation. Undoubtedly, this approach can assist in the medical workflow as well as give clinical direction in identification, therapy planning, and later evaluations.
2. Related Work
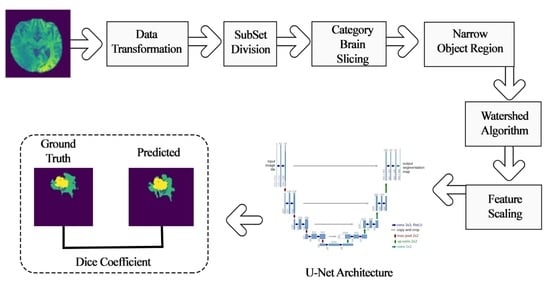
3. Material and Research Flow
3.1. Material
3.2. Research Flow
4. Methodology
4.1. Pre-Learning Process Techniques
4.1.1. Data Transformation
4.1.2. Subset Division
4.1.3. Category Brain Slicing
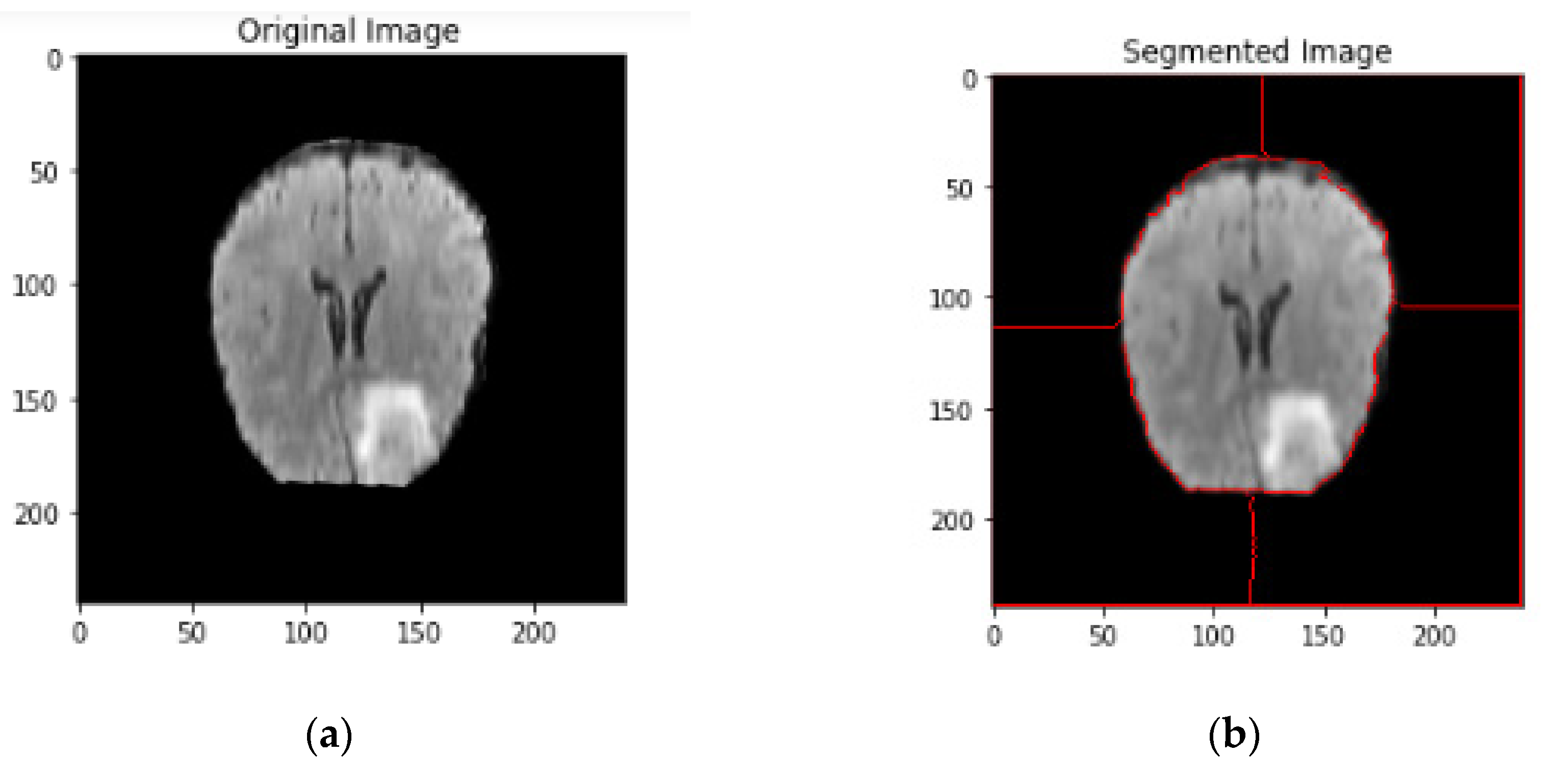
4.1.4. Narrow Object Region
4.1.5. Watershed Algorithm
4.1.6. Feature Scaling
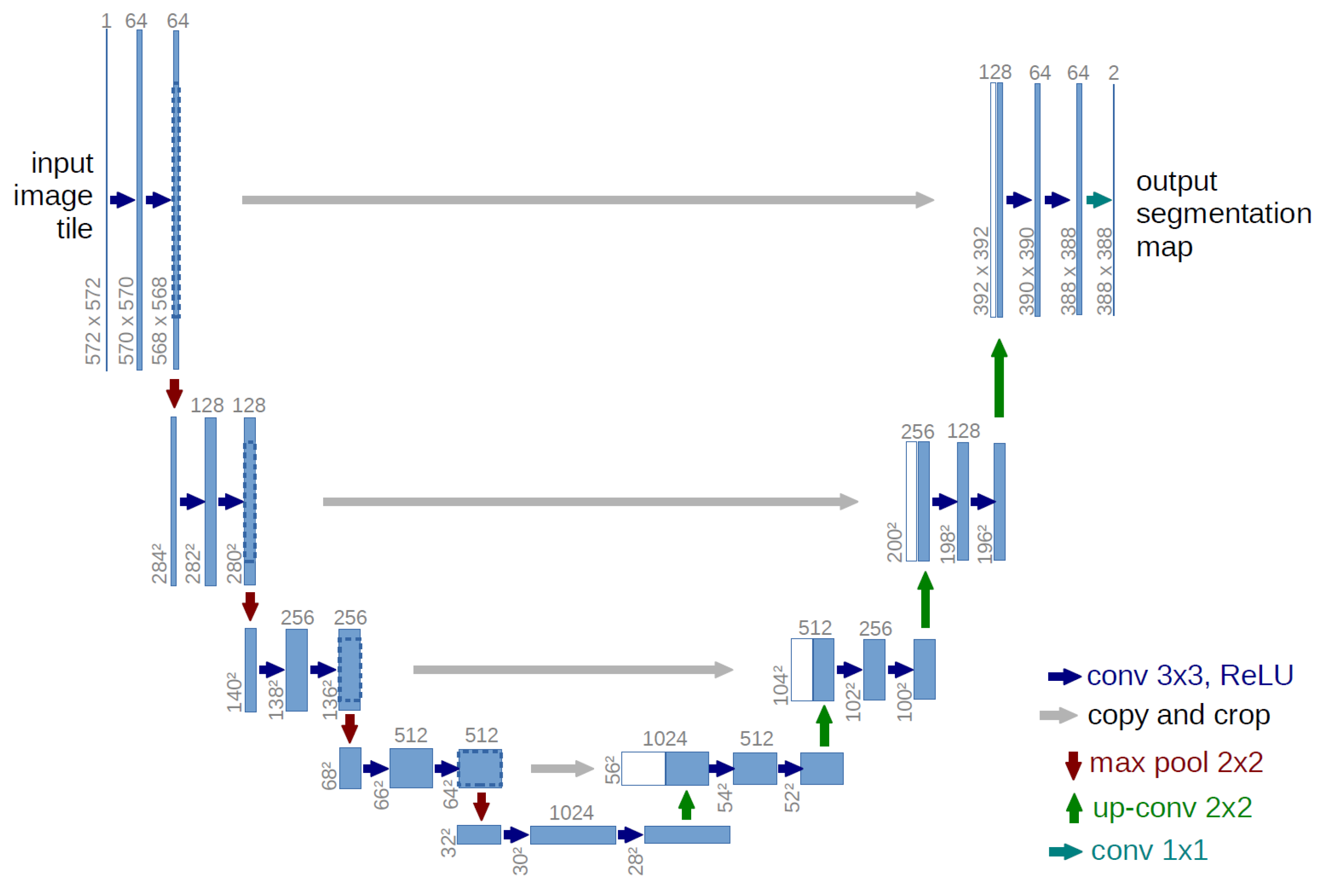
4.2. Deep Learning Model for Tumor Region Segmentation
- A contracting path similar to an encoder to capture the context from a compact feature representation.
- A symmetric expanding path that is similar to a decoder, which allows for accurate localization. This step is done to retain boundary information (spatial information) despite downsampling and max-pooling performed in the encoder stage.
5. Experimental Setup and Results
5.1. Performance Evaluation Measures
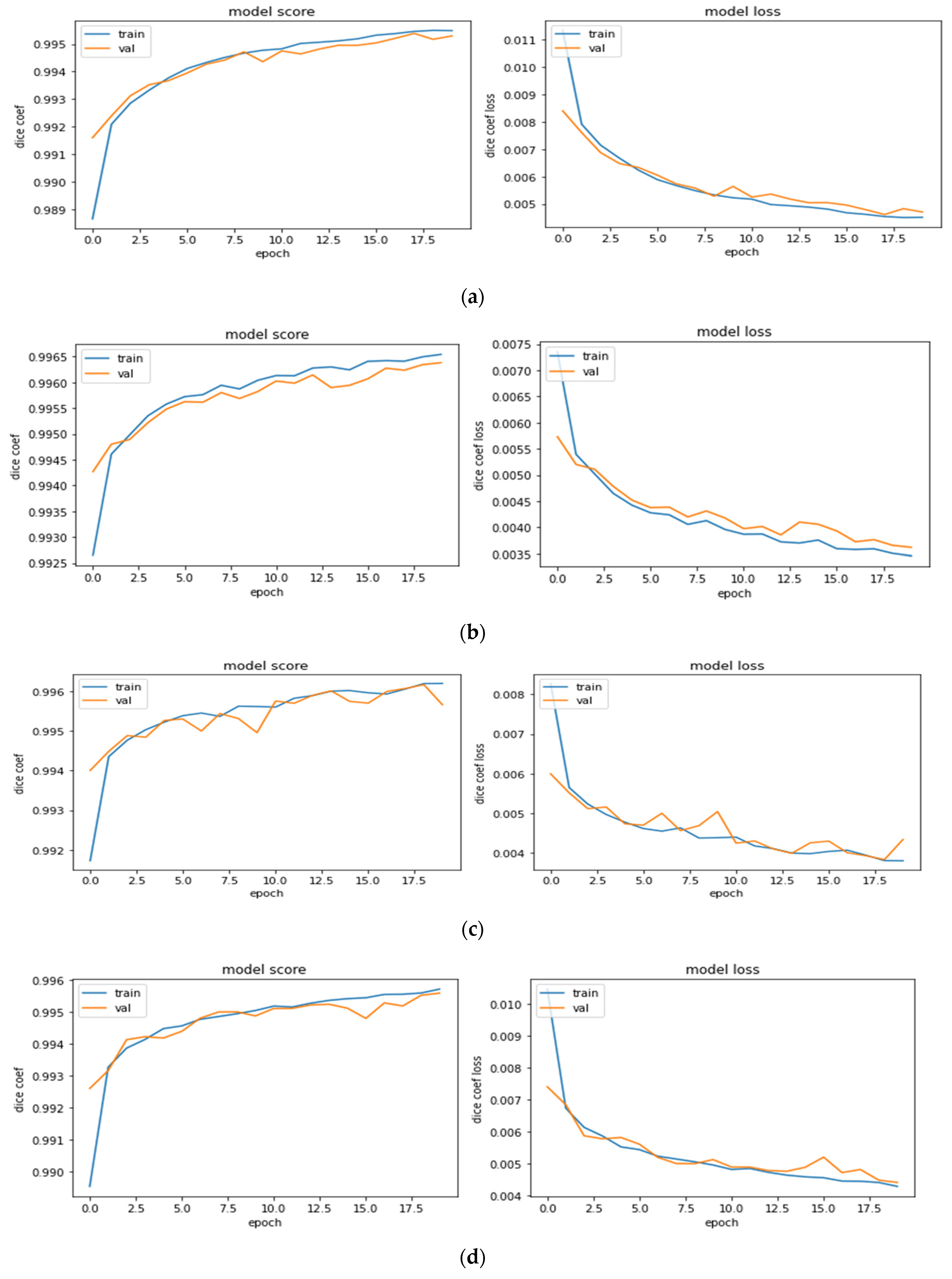
5.2. Model Outcome Observations and Refinement
5.3. U-Net Deep Learning Model Outcome for Brain Tumor Detection
6. Conclusions
- Developing a reliable system with an easy-to-use user interface for the proposed model. The interface would allow doctors to upload an image and get results on the location of the tumor and its class.
- The model can be enhanced to predict the survivability of patients suffering from a brain tumor.
- Explore a more vigorous system for the huge database of clinical images which could be noisy, be affected by external factors, and have reduced quality.
- Implement the model for the discovery and segmentation of tumors in different parts of the body.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Riries, R.; Ain, K. Edge detection for brain tumor pattern recognition. In Proceedings of the International Conference on Instrumentation, Communication, Information Technology, and Biomedical Engineering, Bandung, Indonesia, 23–25 November 2009; pp. 1–3. [Google Scholar]
- Khare, S.; Bhandari, A.; Singh, S.; Arora, A. ECG Arrhythmia Classification Using Spearman Rank Correlation and Support Vector Machine. Adv. Intell. Soft Comput. 2012, 131, 591–598. [Google Scholar] [CrossRef]
- Urva, L.; Shahid, A.R.; Raza, B.; Ziauddin, S.; Khan, M.A. An end-to-end brain tumor segmentation system using multi-inception-UNET. Int. J. Imaging Syst. Technol. 2021. [Google Scholar] [CrossRef]
- Mora, B.L.; Vilaplana, V. MRI brain tumor segmentation and uncertainty estimation using 3D-UNet architectures. arXiv 2020, arXiv:2012.15294. [Google Scholar]
- Jurdi, E.; Petitjean, R.C.; Honeine, P.; Abdallah, F. Bb-unet: U-net with bounding box prior. IEEE J. Sel. Top. Signal Process. 2020, 14, 1189–1198. [Google Scholar] [CrossRef]
- Mohammadreza, S.; Yang, G.; Lambrou, T.; Allinson, N.; Jones, T.L.; Barrick, T.R.; Howe, F.A.; Ye, X. Automated brain tumour detection and segmentation using superpixel-based extremely randomized trees in FLAIR MRI. Int. J. Comput. Assist. Radiol. Surg. 2017, 12, 183–203. [Google Scholar]
- Abdelrahman, E.; Hussein, A.; AlNaggar, E.; Zidan, M.; Zaki, M.; Ismail, M.A.; Ghanem, N.M. Brain tumor segmantation using random forest trained on iteratively selected patients. In International Workshop on Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries; Springer: Cham, Switzerland, 2016; pp. 129–137. [Google Scholar]
- Toktam, H.; Hamghalam, M.; Reyhani-Galangashi, O.; Mirzakuchaki, S. A machine learning approach to brain tumors segmentation using adaptive random forest algorithm. In Proceedings of the 2019 5th Conference on Knowledge Based Engineering and Innovation (KBEI), Tehran, Iran, 28 February–1 March 2019; pp. 76–82. [Google Scholar]
- Malathi, M.; Sinthia, P. BrainTumour Segmentation Using Convolutional Neural Network with Tensor Flow. Asian Pac. J. Cancer Prev. 2019, 20, 2095–2101. [Google Scholar] [CrossRef] [Green Version]
- Bangalore, Y.C.G.; Wagner, B.; Nalawade, S.S.; Murugesan, G.K.; Pinho, M.C.; Fei, B.; Madhuranthakam, A.J.; Maldjian, J.A. Fully automated brain tumor segmentation and survival prediction of gliomas using deep learning and mri. In International MICCAI Brainlesion Workshop; Springer: Cham, Switzerland, 2019; pp. 99–112. [Google Scholar]
- Nitish, Z.; Pawar, V. GLCM textural features for brain tumor classification. Int. J. Comput. Sci. Issues 2012, 9, 354. [Google Scholar]
- Tun, Z.H.; Maneerat, N.; Win, K.Y. Brain tumor detection based on Naïve Bayes Classification. In Proceedings of the 2019 5th International Conference on Engineering, Applied Sciences and Technology (ICEAST), Luang Prabang, Laos, 2–5 July 2019; pp. 1–4. [Google Scholar]
- Gadpayleand, P.; Mahajani, P.S. Detection and classification of brain tumor in MRI images. Int. Conf. Adv. Comput. Commun. Syst. 2013, 2320–9569. Available online: https://www.semanticscholar.org/paper/Detection-and-Classification-of-Brain-Tumor-in-MRI-Mahajani/f7faba638847a526c77d75f38f2278224aab363e (accessed on 18 October 2021).
- Bhaskarrao, B.N.; Ray, A.K.; Thethi, H.P. Image analysis for MRI based brain tumor detection and feature extraction using biologically inspired BWT and SVM. Int. J. Biomed. Imaging 2017, 2017, 9749108. [Google Scholar]
- Nabil, I.; Rahman, M.S. MultiRes UNet: Rethinking the U-Net Architecture for Multimodal Biomedical Image Segmentation. Neural Netw. Off. J. Int. Neural Netw. Soc. 2019, 121, 74–87. [Google Scholar]
- Minz, A.; Mahobiya, C. MR image classification using adaboost for brain tumor type. In Proceedings of the 2017 IEEE 7th International Advance Computing Conference (IACC), Hyderabad, India, 5–7 January 2017; pp. 701–705. [Google Scholar]
- Samjith, R.C.P.; Shreeja, R. Automatic brain tumor tissue detection in T-1 weighted MRI. In Proceedings of the 2017 International Conference on Innovations in Information, Embedded and Communication Systems (ICIIECS), Coimbatore, India, 17–18 March 2017; pp. 1–4. [Google Scholar]
- Rosy, K. SVM classification an approach on detecting abnormality in brain MRI images. Int. J. Eng. Res. Appl. 2013, 3, 1686–1690. [Google Scholar]
- Telrandhe, S.R.; Pimpalkar, A.; Kendhe, A. Detection of brain tumor from MRI images by using segmentation & SVM. In Proceedings of the 2016 World Conference on Futuristic Trends in Research and Innovation for Social Welfare (Startup Conclave), Coimbatore, India, 29 February–1 March 2016; pp. 1–6. [Google Scholar]
- Keerthana, T.K.; Xavier, S. An intelligent system for early assessment and classification of brain tumor. In Proceedings of the 2018 Second International Conference on Inventive Communication and Computational Technologies (ICICCT), Thondamuthur, India, 20–21 April 2018; pp. 1265–1268. [Google Scholar]
- Devkota, B.; Alsadoon, A.; Prasad, P.W.C.; Singh, A.K.; Elchouemi, A. Image segmentation for early stage brain tumor detection using mathematical morphological reconstruction. Procedia Comput. Sci. 2018, 125, 115–123. [Google Scholar] [CrossRef]
- Nadir, G.D.; Jehlol, H.B.; Oleiwi, A.S.A. Brain tumor detection using shape features and machine learning algorithms. Int. J. Adv. Res. Comput. Sci. Softw. Eng. 2015, 5, 454–459. [Google Scholar]
- Chavan, V.N.; Jadhav, B.D.; Patil, P.M. Detection and classification of brain tumors. Int. J. Comput. Appl. 2015, 112, 8. [Google Scholar]
- Shahariar, A.M.; Rahman, M.M.; Hossain, M.A.; Islam, M.K.; Ahmed, K.M.; Ahmed, K.T.; Singh, B.C.; Miah, M.S. Automatic human brain tumor detection in MRI image using template-based K means and improved fuzzy C means clustering algorithm. Big Data Cogn. Comput. 2019, 3, 27. [Google Scholar]
- Zoltán, K.; Lefkovits, L.; Szilágyi, L. Automatic detection and segmentation of brain tumor using random forest approach. In International Conference on Modeling Decisions for Artificial Intelligence; Springer: Cham, Switzerland, 2016; pp. 301–312. [Google Scholar]
- Chang, P.D. Fully convolutional neural networks with hyperlocal features for brain tumor segmentation. In MICCAI-BRATS Workshop; 2016; pp. 4–9. Available online: https://www.researchgate.net/publication/315920622_Fully_Convolutional_Deep_Residual_Neural_Networks_for_Brain_Tumor_Segmentation (accessed on 18 October 2021).
- Toraman, S.; Tuncer, S.A.; Balgetir, F. Is it possible to detect cerebral dominance via EEG signals by using deep learning? Med. Hypotheses 2019, 131, 109315. [Google Scholar] [CrossRef]
- Baranwal, S.; Arora, A.; Khandelwal, S. Detecting diseases in plant leaves: An optimised deep-learning convolutional neural network approach. Int. J. Environ. Sustain. Dev. 2021, 20, 166–188. [Google Scholar] [CrossRef]
- Raghav, M.; Arbel, T. 3D U-Net for brain tumour segmentation. In International MICCAI Brainlesion Workshop; Springer: Cham, Switzerland, 2018; pp. 254–266. [Google Scholar]
- Fridman, N. Brain Tumor Detection and Segmentation Using Deep Learning U-Net on Multi-Modal MRI. In Proceedings of the Pre-Conference Proceedings of the 7th MICCAI BraTS Challenge, Granada, Spain, 16 September 2018; pp. 135–143. [Google Scholar]
- Hao, D.; Yang, G.; Liu, F.; Mo, Y.; Guo, Y. Automatic brain tumor detection and segmentation using U-Net based fully convolutional networks. In Annual Conference on Medical Image Understanding and Analysis; Springer: Cham, Switzerland, 2017; pp. 506–517. [Google Scholar]
- Li, S.; Zhang, S.; Chen, H.; Luo, L. Brain tumor segmentation and survival prediction using multimodal MRI scans with deep learning. Front. Neurosci. 2019, 13, 810. [Google Scholar]
- Tuan, T.A. Brain tumor segmentation using bit-plane and unet. In International MICCAI Brainlesion Workshop; Springer: Cham, Switzerland, 2018; pp. 466–475. [Google Scholar]
- Wu, S.; Li, H.; Guan, Y. Multimodal Brain Tumor Segmentation Using U-Net. In Proceedings of the Pre-Conference Proceedings of the 7th MICCAI BraTS Challenge, Granada, Spain, 16 September 2018; pp. 508–515. [Google Scholar]
- Wei, C.; Liu, B.; Peng, S.; Sun, J.; Qiao, X. S3D-UNet: Separable 3D U-Net for brain tumor segmentation. In International MICCAI Brainlesion Workshop; Springer: Cham, Switzerland, 2018; pp. 358–368. [Google Scholar]
- Cahall, D.E.; Rasool, G.; Bouaynaya, N.C.; Fathallah-Shaykh, H.M. Inception modules enhance brain tumor segmentation. Front. Comput. Neurosci. 2019, 13, 44. [Google Scholar] [CrossRef] [Green Version]
- Kamrul, H.S.M.; Linte, C.A. A modified U-Net convolutional network featuring a Nearest-neighbor Re-sampling-based Elastic-Transformation for brain tissue characterization and segmentation. In Proceedings of the 2018 IEEE Western New York Image and Signal Processing Workshop (WNYISPW), Rochester, NY, USA, 5 October 2018; pp. 1–5. [Google Scholar]
- Çinar, A.; Yildirim, M. Detection of tumors on brain MRI images using the hybrid convolutional neural network architecture. Med. Hypotheses 2020, 139, 109684. [Google Scholar] [CrossRef]
- Kermi, A.; Mahmoudi, I.; Khadir, M.T. Deep convolutional neural networks using U-Net for automatic brain tumor segmentation in multimodal MRI volumes. In International MICCAI Brainlesion Workshop; Springer: Cham, Switzerland, 2015; pp. 37–48. [Google Scholar]
- Padmakant, D.; Phegade, M.R.; Shah, S.K. Watershed segmentation brain tumor detection. In Proceedings of the 2015 International Conference on Pervasive Computing (ICPC), Pune, India, 8–10 January 2015; pp. 1–5. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Shamir Reuben, R.; Duchin, Y.; Kim, J.; Sapiro, G.; Harel, N. Continuous dice coefficient: A method for evaluating probabilistic segmentations. arXiv 2019, arXiv:1906.11031. [Google Scholar]
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–26 October 2016; pp. 565–571. [Google Scholar]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans. Med Imaging 2015, 34, 1993–2024. [Google Scholar] [CrossRef]
- Bakas, S.; Akbari, H.; Sotiras, A.; Bilello, M.; Rozycki, M.; Kirby, J.S.; Freymann, J.B.; Farahani, K.; Davatzikos, C. Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Nat. Sci. Data 2017, 4, 170117. [Google Scholar] [CrossRef] [Green Version]
- Bakas, S.; Reyes, M.; Jakab, A.; Bauer, S.; Rempfler, M.; Crimi, A.; Shinohara, R.T.; Berger, C.; Ha, S.M.; Rozycki, M.; et al. Identifying the Best Machine Learning Algorithms for Brain Tumor Segmentation, Progression Assessment, and Overall Survival Prediction in the BRATS Challenge. arXiv 2018, arXiv:1811.02629. [Google Scholar]

| Dataset | Used in Research Paper |
|---|---|
| BraTS 2012, 2015, 2016, 2018, 2019, 2020 | [6,7,8,9,10] |
| Whole Brain Atlas (WBA) | [11] |
| Rembrandt database | [12] |
| MRI Image database, Pioneer diagnostic center | [13] |
| BrainWeb: Simulated Brain Database | [14] |
| CVC-ClinicDB | [15] |
| ISIC-2017 | [15] |
| Author | Method Used | Results |
|---|---|---|
| Mehta et al., 2018 [29] |
| ET: 0.78, WT: 0.90, CT: 0.82 |
| Fridman et al., 2018 [30] |
| No accuracy metric was presented. |
| Tuan et al., 2019 [33] |
| WT:0.82, ET: 0.68, CT:0.70 |
| Shaocheng et al., 2018 [34] |
| Training phase: WT: 0.90, CT: 0.81, ET: 0.76 Validation set: WT: 0.91, CT: 0.83, ET: 0.79 |
| Wei et al., 2018 [35] |
| ET: 0.69, WT: 0.84, CT: 0.78 |
| Nabil et al., 2019 [15] |
| Average accuracy: 80.3%, 82%, 91.65%, 88%, 78.2% |
| Cahall et al., 2019 [36] |
| Intra-tumoral: WT: 0.925, CT: 0.95, ET: 0.95 Glioma subregions: WT: 0.92, CT: 0.95, ET: 0.95 |
| Malathi et al., 2019 [9] |
| Average Dice co-efficient: 0.73 Sensitivity: 0.82 |
| Yogananda et al., 2019 [10] |
| Accuracy: 89% WT: 0.95, CT: 0.92. ET:0.90 Survival Prediction: Accuracy: 44.8% |
| Hasan et al., 2018 [37] | Preprocessing: Image scaling, translation, rotation, and shear. Classification: Proposed NNRET U-net deep convolution neural network. DataSet: BraTS 2018 | Dice coefficient: 0.87 |
| Input Image Size | 240*240*155 | |
|---|---|---|
| Training | # of HGG Images | 210 |
| # of LGG Images | 75 | |
| # of Test Dataset | 191 | |
| # of Validation Dataset | 66 | |
| Test Case | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| Image |  |  |  |  |  |
| Ground Truth |  |  |  |  |  |
| Segmented Region |  |  |  |  |  |
| Result | Fail | Pass | Fail | Pass | Pass |
| Image | Ground Truth | Segmented Region | |
|---|---|---|---|
| Test Case1 |  |  |  |
| Test Case2 |  |  |  |
| Set | Training | Validation | Test |
|---|---|---|---|
| HGG-1 | 0.9955 | 0.9953 | 0.9815 |
| HGG-2 | 0.9965 | 0.9964 | 0.9844 |
| HGG-3 | 0.9962 | 0.9957 | 0.9804 |
| LGG-1 | 0.9954 | 0.9951 | 0.9854 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arora, A.; Jayal, A.; Gupta, M.; Mittal, P.; Satapathy, S.C. Brain Tumor Segmentation of MRI Images Using Processed Image Driven U-Net Architecture. Computers 2021, 10, 139. https://doi.org/10.3390/computers10110139
Arora A, Jayal A, Gupta M, Mittal P, Satapathy SC. Brain Tumor Segmentation of MRI Images Using Processed Image Driven U-Net Architecture. Computers. 2021; 10(11):139. https://doi.org/10.3390/computers10110139
Chicago/Turabian StyleArora, Anuja, Ambikesh Jayal, Mayank Gupta, Prakhar Mittal, and Suresh Chandra Satapathy. 2021. "Brain Tumor Segmentation of MRI Images Using Processed Image Driven U-Net Architecture" Computers 10, no. 11: 139. https://doi.org/10.3390/computers10110139
APA StyleArora, A., Jayal, A., Gupta, M., Mittal, P., & Satapathy, S. C. (2021). Brain Tumor Segmentation of MRI Images Using Processed Image Driven U-Net Architecture. Computers, 10(11), 139. https://doi.org/10.3390/computers10110139