Oral Cancer Discrimination and Novel Oral Epithelial Dysplasia Stratification Using FTIR Imaging and Machine Learning
Abstract
:1. Introduction
2. Materials and Methods
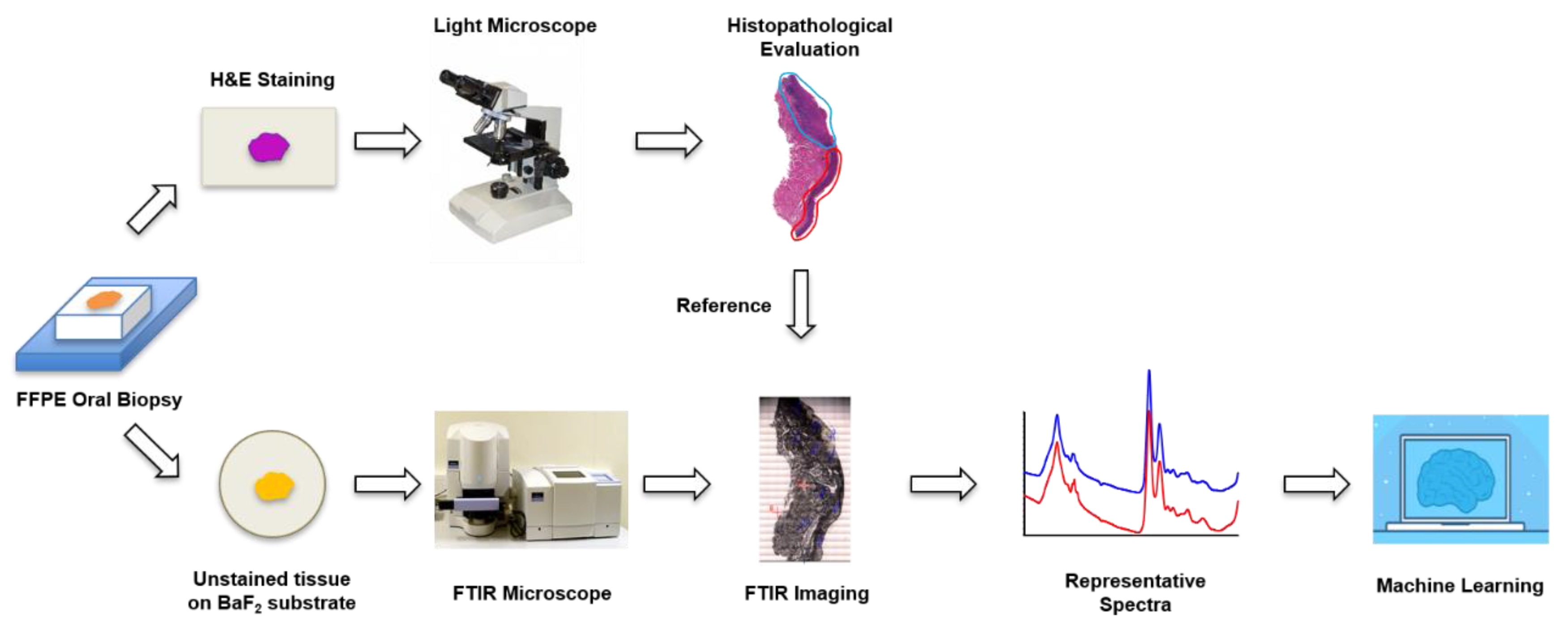
2.1. Sample Preparation
2.2. FTIR Imaging
2.3. Data Analysis
2.3.1. Spectral Preprocessing
2.3.2. Unsupervised Exploratory Analysis Using PCA and HCA
2.3.3. Supervised Discrimination between HK and OSCC Samples
2.3.4. A novel Strategy for OED Classification
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA: A Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Ali, J.; Sabiha, B.; Jan, H.U.; Haider, S.A.; Khan, A.A.; Ali, S.S. Genetic etiology of oral cancer. Oral Oncol. 2017, 70, 23–28. [Google Scholar] [CrossRef]
- Irani, S. New Insights into Oral Cancer-Risk Factors and Prevention: A Review of Literature. Int. J. Prev. Med. 2020, 11, 202. [Google Scholar] [CrossRef]
- Shah, F.D.; Begum, R.; Vajaria, B.N.; Patel, K.R.; Patel, J.B.; Shukla, S.N.; Patel, P.S. A Review on Salivary Genomics and Proteomics Biomarkers in Oral Cancer. Indian J. Clin. Biochem. 2011, 26, 326–334. [Google Scholar] [CrossRef] [Green Version]
- Van der Waal, I. Potentially malignant disorders of the oral and oropharyngeal mucosa; terminology, classification and present concepts of management. Oral Oncol. 2009, 45, 317–323. [Google Scholar] [CrossRef]
- Warnakulasuriya, S. Oral potentially malignant disorders: A comprehensive review on clinical aspects and management. Oral Oncol. 2020, 102, 104550. [Google Scholar] [CrossRef] [PubMed]
- Wenig, B.M. Squamous cell carcinoma of the upper aerodigestive tract: Precursors and problematic variants. Mod. Pathol. Off. J. United States Can. Acad. Pathol. Inc. 2002, 15, 229–254. [Google Scholar] [CrossRef] [Green Version]
- Shirani, S.; Kargahi, N.; Razavi, S.M.; Homayoni, S. Epithelial dysplasia in oral cavity. Iran. J. Med. Sci. 2014, 39, 406–417. [Google Scholar] [PubMed]
- Warnakulasuriya, S.; Reibel, J.; Bouquot, J.; Dabelsteen, E. Oral epithelial dysplasia classification systems: Predictive value, utility, weaknesses and scope for improvement. J. Oral Pathol. Med. 2008, 37, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Van der Waal, I. Oral potentially malignant disorders: Is malignant transformation predictable and preventable? Med. Oral Patol. Oral Y Cir. Bucal 2014, 19, e386–e390. [Google Scholar] [CrossRef]
- Reibel, J. Prognosis of Oral Pre-malignant Lesions: Significance of Clinical, Histopathological, and Molecular Biological Characteristics. Crit. Rev. Oral Biol. Med. 2003, 14, 47–62. [Google Scholar] [CrossRef]
- Ranganathan, K.; Kavitha, L. Oral epithelial dysplasia: Classifications and clinical relevance in risk assessment of oral potentially malignant disorders. J. Oral Maxillofac. Pathol. 2019, 23, 19–27. [Google Scholar] [PubMed]
- Celentano, A.; Glurich, I.; Borgnakke, W.S.; Farah, C.S. World Workshop on Oral Medicine VII: Prognostic biomarkers in oral leukoplakia and proliferative verrucous leukoplakia—A systematic review of retrospective studies. Oral Dis. 2020, 27, 848–880. [Google Scholar] [CrossRef]
- Talari, A.C.S.; Martinez, M.A.G.; Movasaghi, Z.; Rehman, S.; Rehman, I.U. Advances in Fourier transform infrared (FTIR) spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2017, 52, 456–506. [Google Scholar] [CrossRef]
- Fabian, H.; Naumann, D. Methods to study protein folding by stopped-flow FT-IR. Methods 2004, 34, 28–40. [Google Scholar] [CrossRef]
- Ghimire, H.; Garlapati, C.; Janssen, E.A.M.; Krishnamurti, U.; Qin, G.; Aneja, R.; Perera, A.G.U. Protein conformational changes in breast cancer sera using infrared spectroscopic analysis. Cancers 2020, 12, 1708. [Google Scholar] [CrossRef]
- Kelly, J.G.; Martin-Hirsch, P.L.; Martin, F.L. Discrimination of base differences in oligonucleotides using mid-infrared spectroscopy and multivariate analysis. Anal. Chem. 2009, 81, 5314–5319. [Google Scholar] [CrossRef]
- Petibois, C.; Déléris, G. Evidence that erythrocytes are highly susceptible to exercise oxidative stress: FT-IR spectrometric studies at the molecular level. Cell Biol. Int. 2005, 29, 709–716. [Google Scholar] [CrossRef] [PubMed]
- Benseny-Cases, N.; Klementieva, O.; Cotte, M.; Ferrer, I.; Cladera, J. Microspectroscopy (μFTIR) reveals co-localization of lipid oxidation and amyloid plaques in human Alzheimer disease brains. Anal. Chem. 2014, 86, 12047–12054. [Google Scholar] [CrossRef] [PubMed]
- Bogomolny, E.; Huleihel, M.; Suproun, Y.; Sahu, R.K.; Mordechai, S. Early spectral changes of cellular malignant transformation using Fourier transform infrared microspectroscopy. J. Biomed. Opt. 2007, 12, 024003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papamarkakis, K.; Bird, B.; Schubert, J.M.; Miljković, M.; Wein, R.; Bedrossian, K.; Laver, N.; Diem, M. Cytopathology by optical methods: Spectral cytopathology of the oral mucosa. Lab. Investig. 2010, 90, 589–598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Srinivasan, A.; Nikolajeff, F. Role of Infrared Spectroscopy and Imaging in Cancer Diagnosis. Curr. Med. Chem. 2018, 25, 1055–1072. [Google Scholar] [CrossRef]
- Beć, K.; Grabska, J.; Huck, C. Biomolecular and bioanalytical applications of infrared spectroscopy—A review. Anal. Chim. Acta 2020, 1133, 150–177. [Google Scholar] [CrossRef]
- Pallua, J.D.; Brunner, A.; Zelger, B.; Stalder, R.; Unterberger, S.H.; Schirmer, M.; Tappert, M. Clinical infrared microscopic imaging: An overview. Pathol. Res. Pract. 2018, 214, 1532–1538. [Google Scholar] [CrossRef]
- Bel’skaya, L. Use of IR Spectroscopy in Cancer Diagnosis. A Review. J. Appl. Spectrosc. 2019, 86, 187–205. [Google Scholar] [CrossRef]
- Wang, R.; Wang, Y. Fourier Transform Infrared Spectroscopy in Oral Cancer Diagnosis. Int. J. Mol. Sci. 2021, 22, 1206. [Google Scholar] [CrossRef]
- Bro, R.; Smilde, A.K. Principal component analysis. Anal. Methods 2014, 6, 2812–2831. [Google Scholar] [CrossRef] [Green Version]
- Morais, C.L.M.; Lima, K.M.G.; Singh, M.; Martin, F.L. Tutorial: Multivariate classification for vibrational spectroscopy in biological samples. Nat. Protoc. 2020, 15, 2143–2162. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Perez, D.; Guan, H.; Madhivanan, P.; Mathee, K.; Narasimhan, G. So you think you can PLS-DA? BMC Bioinform. 2020, 21, 2. [Google Scholar] [CrossRef]
- Song, W.; Wang, H.; Maguire, P.; Nibouche, O. Collaborative representation based classifier with partial least squares regression for the classification of spectral data. Chemom. Intell. Lab. Syst. 2018, 182, 79–86. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Piryonesi, S.M.; El-Diraby, T.E. Data Analytics in Asset Management: Cost-Effective Prediction of the Pavement Condition Index. J. Infrastruct. Syst. 2020, 26, 04019036. [Google Scholar] [CrossRef]
- Guang, P.; Huang, W.; Guo, L.; Yang, X.; Huang, F.; Yang, M.; Wen, W.; Li, L. Blood-based FTIR-ATR spectroscopy coupled with extreme gradient boosting for the diagnosis of type 2 diabetes: A STARD compliant diagnosis research. Medicine 2020, 99, e19657. [Google Scholar] [CrossRef]
- Šimundić, A.-M. Measures of Diagnostic Accuracy: Basic Definitions. EJIFCC 2009, 19, 203–211. [Google Scholar]
- Barth, A. Infrared spectroscopy of proteins. Biochim. Et Biophys. Acta 2007, 1767, 1073–1101. [Google Scholar] [CrossRef] [Green Version]
- Casal, H.L.; Mantsch, H.H. Polymorphic phase behaviour of phospholipid membranes studied by infrared spectroscopy. Biochim. Et Biophys. Acta (BBA)—Rev. Biomembr. 1984, 779, 381–401. [Google Scholar] [CrossRef]
- Baker, M.J.; Trevisan, J.; Bassan, P.; Bhargava, R.; Butler, H.J.; Dorling, K.M.; Fielden, P.R.; Fogarty, S.W.; Fullwood, N.J.; Heys, K.A.; et al. Using Fourier transform IR spectroscopy to analyze biological materials. Nat. Protoc. 2014, 9, 1771–1791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiercigroch, E.; Szafraniec, E.; Czamara, K.; Pacia, M.Z.; Majzner, K.; Kochan, K.; Kaczor, A.; Baranska, M.; Malek, K. Raman and infrared spectroscopy of carbohydrates: A review. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017, 185, 317–335. [Google Scholar] [CrossRef] [PubMed]
- Bellisola, G.; Sorio, C. Infrared spectroscopy and microscopy in cancer research and diagnosis. Am. J. Cancer Res. 2012, 2, 1–21. [Google Scholar] [PubMed]
- Chiang, T.-E.; Lin, Y.-C.; Wu, C.-T.; Yang, C.-Y.; Wu, S.-T.; Chen, Y.-W. Comparison of the accuracy of diagnoses of oral potentially malignant disorders with dysplasia by a general dental clinician and a specialist using the Taiwanese Nationwide Oral Mucosal Screening Program. PLoS ONE 2021, 16, e0244740. [Google Scholar] [CrossRef] [PubMed]
- Tilakaratne, W.M.; Jayasooriya, P.R.; Jayasuriya, N.S.; De Silva, R.K. Oral epithelial dysplasia: Causes, quantification, prognosis, and management challenges. Periodontology 2000 2019, 80, 126–147. [Google Scholar] [CrossRef] [PubMed]
- Schultz, C.P.; Mantsch, H.H. Biochemical imaging and 2D classification of keratin pearl structures in oral squamous cell carcinoma. Cell. Mol. Biol. (Noisy-Le-Grand Fr.) 1998, 44, 203–210. [Google Scholar]
- Schultz, C.P.; Liu, K.Z.; Kerr, P.D.; Mantsch, H.H. In situ infrared histopathology of keratinization in human oral/oropharyngeal squamous cell carcinoma. Oncol. Res. 1998, 10, 277–286. [Google Scholar]
- Fukuyama, Y.; Yoshida, S.; Yanagisawa, S.; Shimizu, M. A study on the differences between oral squamous cell carcinomas and normal oral mucosas measured by Fourier transform infrared spectroscopy. Biospectroscopy 1999, 5, 117–126. [Google Scholar] [CrossRef]
- Bruni, P.; Conti, C.; Giorgini, E.; Pisani, M.; Rubini, C.; Tosi, G. Histological and microscopy FT-IR imaging study on the proliferative activity and angiogenesis in head and neck tumours. Faraday Discuss. 2004, 126, 19–26, discussion 77–92. [Google Scholar] [CrossRef]
- Pallua, J.D.; Pezzei, C.; Zelger, B.; Schaefer, G.; Bittner, L.K.; Huck-Pezzei, V.A.; Schoenbichler, S.A.; Hahn, H.; Kloss-Brandstaetter, A.; Kloss, F.; et al. Fourier transform infrared imaging analysis in discrimination studies of squamous cell carcinoma. Analyst 2012, 137, 3965–3974. [Google Scholar] [CrossRef]
- Wong, P.T.; Wong, R.K.; Caputo, T.A.; Godwin, T.A.; Rigas, B. Infrared spectroscopy of exfoliated human cervical cells: Evidence of extensive structural changes during carcinogenesis. Proc. Natl. Acad. Sci. USA 1991, 88, 10988–10992. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, S.; Pal, M.; Chakrabarty, J.; Petibois, C.; Paul, R.R.; Giri, A.; Chatterjee, J. Fourier-transform-infrared-spectroscopy based spectral-biomarker selection towards optimum diagnostic differentiation of oral leukoplakia and cancer. Anal. Bioanal. Chem. 2015, 407, 7935–7943. [Google Scholar] [CrossRef]
- Naurecka, M.L.; Sierakowski, B.M.; Kasprzycka, W.; Dojs, A.; Dojs, M.; Suszyński, Z.; Kwaśny, M. FTIR-ATR and FT-Raman Spectroscopy for Biochemical Changes in Oral Tissue. Am. J. Anal. Chem. 2017, 8, 180–188. [Google Scholar] [CrossRef] [Green Version]
- Rai, V.; Mukherjee, R.; Routray, A.; Ghosh, A.K.; Roy, S.; Ghosh, B.P.; Mandal, P.B.; Bose, S.; Chakraborty, C. Serum-based diagnostic prediction of oral submucous fibrosis using FTIR spectrometry. Spectrochim. Acta—Part A: Mol. Biomol. Spectrosc. 2018, 189, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Sabbatini, S.; Conti, C.; Rubini, C.; Librando, V.; Tosi, G.; Giorgini, E. Infrared microspectroscopy of Oral Squamous Cell Carcinoma: Spectral signatures of cancer grading. Vib. Spectrosc. 2013, 68, 196–203. [Google Scholar] [CrossRef]
- Bunaciu, A.A.; Aboul-Enein, H.; Fleschin, Ş. Vibrational Spectroscopy in Clinical Analysis. Appl. Spectrosc. Rev. 2015, 50, 176–191. [Google Scholar] [CrossRef]
- Kimber, J.A.; Kazarian, S.G. Spectroscopic imaging of biomaterials and biological systems with FTIR microscopy or with quantum cascade lasers. Anal. Bioanal. Chem. 2017, 409, 5813–5820. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| Wavenumber (cm−1) | Vibrational Modes and Biochemical Assignments |
|---|---|
| 1704 | Ester carbonyl C=O stretching, fatty acid esters, lipids |
| 1670 | Amide I, secondary structure of proteins |
| 1660 | Amide I, secondary structure of proteins |
| 1654 | C=O stretching of amide I, secondary structure of proteins, |
| 1640 | Amide I, secondary structure of proteins |
| 1548 | C-N and CN-H stretching of amide II, secondary structure of proteins |
| 1516 | Amide II, secondary structure of proteins |
| 1482 | deformation vibrations of –CH3, lipid |
| 1238 | Asymmetric phosphodiester stretching νas (–PO2−), lipid, nuclei acid, amide III (C-N stretching, N-H bending) proteins |
| 1082 | Symmetric phosphodiester stretching νs (–PO2−), protein phosphorylation, phospholipids, collagen, DNA |
| 1026 | Vibrational frequency of -CH2OH groups of carbohydrates (e.g., glucose, glycogen, etc.) C-O stretching, C-O stretching coupled with C-O bending of the C-OH groups of carbohydrates |
| 966 | C-O stretching of the phosphodiester, deoxyribose, C-C of DNA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, R.; Naidu, A.; Wang, Y. Oral Cancer Discrimination and Novel Oral Epithelial Dysplasia Stratification Using FTIR Imaging and Machine Learning. Diagnostics 2021, 11, 2133. https://doi.org/10.3390/diagnostics11112133
Wang R, Naidu A, Wang Y. Oral Cancer Discrimination and Novel Oral Epithelial Dysplasia Stratification Using FTIR Imaging and Machine Learning. Diagnostics. 2021; 11(11):2133. https://doi.org/10.3390/diagnostics11112133
Chicago/Turabian StyleWang, Rong, Aparna Naidu, and Yong Wang. 2021. "Oral Cancer Discrimination and Novel Oral Epithelial Dysplasia Stratification Using FTIR Imaging and Machine Learning" Diagnostics 11, no. 11: 2133. https://doi.org/10.3390/diagnostics11112133






