Abstract
An acidic α-galactosidase designated as TMG was purified from the fruiting bodies The purification protocol entailed ion exchange chromatography on Q-Sepharose and of Tricholoma matsutake with 136-fold purification and a specific activity of 909 units/mg. Mono-Q and fast protein liquid chromatography on Superdex 75. TMG is a monomeric protein exhibiting a molecular mass of 47 kDa in SDS-PAGE and gel filtration. The purified enzyme was identified by LC-MS/MS and three inner amino acid sequences were obtained. The optimum pH and temperature for TMG with pNPGal as substrate were pH 4.5 and 55 °C, respectively. The α-galactosidase activity was strongly inhibited by K+, Ca2+, Cd2+, Hg2+, Ag+ and Zn2+ ions. The enzyme activity was inhibited by the chemical modification agent N-bromosuccinimide (NBS), indicating the importance of tryptophan residue(s) at or near the active site. Besides hydrolyzing pNPGal, TMG also efficaciously catalyzed the degradation of natural substrates such as stachyose, raffinose, and melibiose. Thus TMG can be exploited commercially for improving the nutritional value of soy milk by degradation of indigestible oligosaccharides.
1. Introduction
α-Galactosidases (EC 3.2.1.22) are exo-glycosidases that catalyze the hydrolysis of the terminal α-linked galactoside residues from different substrates including melibiose, raffinose, stachyose, verbascose, as well as their derivatives [1]. Raffinose-family oligosaccharides, which are widely distributed in soy products, cannot be absorbed by human and other monogastirc animals leading to flatus formation [2]. Treatment with α-galactosidase for reducing stachyose and raffinose content in soybean flour has been investigated. The carbohydrase has been used in many industrial applications, mainly in the food and feed industries [3,4,5]. There are several industrial applications of α-galactosidases, mainly in the sugar industry, where they improve the crystallization of sucrose by hydrolyzing the raffinose in beet sugar syrups [6].
α-Galactosidases are widely distributed in microorganisms, plants, and animals. In recent years, many α-galactosidases have been extracted and purified from various sources. In plants, α-galactosidases are maximally distributed in seeds, fruits and leaves [7,8,9,10]. In microorganisms, α-galactosidases have been purified from the bacterium Flavobacterium sp. TN17 [11], fungi such as Aspergillus terreus [12], and the mushroom Ganoderma lucidum [13]. Some thermophilic enzymes have been discovered, such as a thermophilic α-galactosidase from Neosarotrya fischeri P1 [14], and thermostable α-galactosidases from Lenzites elegans [15], Thielavia terrestris NRRL8126 [16], and Talaromyces emersonii [17]. The thermostability makes them suitable candidates for use in feed industries.
Tricholoma matsutake, an ectomycorrhizal fungus, produces commercially important mushrooms that have iconic significance in the Far East and is highly valued as a delicacy and medicine. T. matsutake not only exhibits a delicate flavor, but also has diverse biological activities, such as immunomodulatory, anti-tumor and anti-oxidant activities [18,19,20]. Many bioactive substances have been purified from T. matsutake. A laccase from T. matsutake demonstrated activity in decolorization of azo dyes without a mediator [21]. A nuclease [22] and a polysaccharide [23] have been isolated from the fruiting bodies of T. matsutake. The enzyme and the polysaccharide are useful in the protection of the environment and human health. Now we detected high α-galactosidase activity in T. matsutake.
In the present study we reported, for the first time, the isolation and characterization of an α-galactosidase from T. matsutake named TMG. We also noted the effect of TMG on the degradation of raffinose oligosaccharides. This work would provide new evidence for further exploring the possibility of using TMG in industrial applications and advocating T. matsutake as a functional food.
2. Results and Discussion
2.1. Purification of TMG
As summarized in Table 1, the α-galactosidase from T. matsutake was purified 136-fold to homogeneity resulting in a final specific activity of 909 U/mg. The specific activity was 1.6-fold of that (561 U/mg) of α-galactosidase from a fan-shaped mushroom Coriolus versicolor [24]. The purification protocol for TMG consisted of ion exchange chromatography on Q-Sepharose and Mono-Q and gel filtration on Superdex 75 HR. Activity was located successively in peaks Q2, MonoQ1 and SU1.

Table 1.
Purification of Tricholoma matsutake α-galactosidase.
| Chromatographic Fraction | Total Protein (mg/400 g) | Total Activity (U) a | Specific Activity (U/mg) b | Yield (%) | Purification Fold c |
|---|---|---|---|---|---|
| Crude extract | 17,427 | 116,204 | 6.7 | 100 | 1 |
| Q2 | 311.7 | 102,037 | 327.3 | 87.9 | 49.1 |
| MonoQ1 | 95.6 | 34,490 | 360.8 | 29.7 | 54.1 |
| SU2 | 9.6 | 8727 | 909.1 | 7.5 | 136.3 |
a Total activity: α-galactosidase activity (U/mL) in each step × Volume (mL); b Specific activity: total activity/Total protein; c Purification fold: specific activity of each step/specific activity of the first step.
2.2. Determination of Molecular Weight and Amino Acid Sequence of TMG
Purified TMG was enriched in a symmetrical peak SU2 (with a molecular mass of 47 kDa, Figure 1a) and appeared as a single band with a molecular mass of 47 kDa in SDS-PAGE (Figure 1b). Hence the molecular mass of TMG was judged to be 47 kDa. Previously only a few α-galactosidases were purified from mushrooms, including Coriolus versicolor, Ganoderma lucidum, Lenzites elegans and Pleurotus florida and their molecular masses are spread over a wide range from 70 kDa to 249 kDa [13,15,24,25]. Hence α-galactosidases from different sources display different molecular masses and subunit properties. Although the molecular mass of TMG was lower than counterparts purified from other mushrooms, it was similar to PGG purified from Phaseolus coccineus (43 kDa) [10] and the α-galactosidase from Cicer arietinum (45 kDa) [26].
The inner amino acid sequences of TMG obtained by LC-MS/MS and database search using BLAST indicated that the peptides showed some similarity with α-galactosidases from other sources. Peptide GNVMVSLG exhibited 88% identity to α-galactosidases from Bipolaris maydis C5 (accession number EMD92465.1), Bipolaris victoriae F13 (accession number EUN27106.1), Bipolaris oryzae ATCC 44560 (accession number XP_007687294) and Bipolaris sorokiniana ND90Pr (accession number XP_007697049.1), which all belong to the glycosyl hydrolase 27 family. Peptide LLNMND showed 83% identity with the α-galactosidase from Lactobacillus otakiensis (accession number GAD17169.1) and 75% identity with α-galactosidase from Kutzneria sp. 744 (accession number WP_043715631.1). Peptide INVNDS manifested 100% identity with Bacteroides nordii α-galactosidase (accession number WP_025867713).
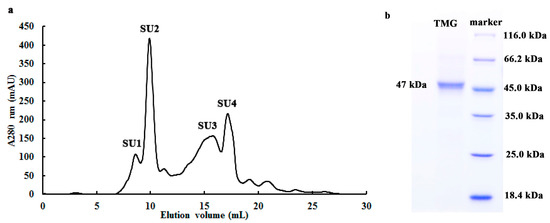
Figure 1.
(a) FPLC-gel filtration on Superdex 75 10/300 GL column and (b) SDS-PAGE of fraction SU2. (a) Eluent: 10 mM NaOAC-HOAC buffer (pH 5.2). Fraction size: 0.8 mL; Flow rate: 0.5 mL·min−1. Fraction SU1 represents purified Tricholoma matsutake α-galactosidase; (b) SDS-PAGE of fraction SU2.
Figure 1.
(a) FPLC-gel filtration on Superdex 75 10/300 GL column and (b) SDS-PAGE of fraction SU2. (a) Eluent: 10 mM NaOAC-HOAC buffer (pH 5.2). Fraction size: 0.8 mL; Flow rate: 0.5 mL·min−1. Fraction SU1 represents purified Tricholoma matsutake α-galactosidase; (b) SDS-PAGE of fraction SU2.
2.3. Biochemical Properties of TMG
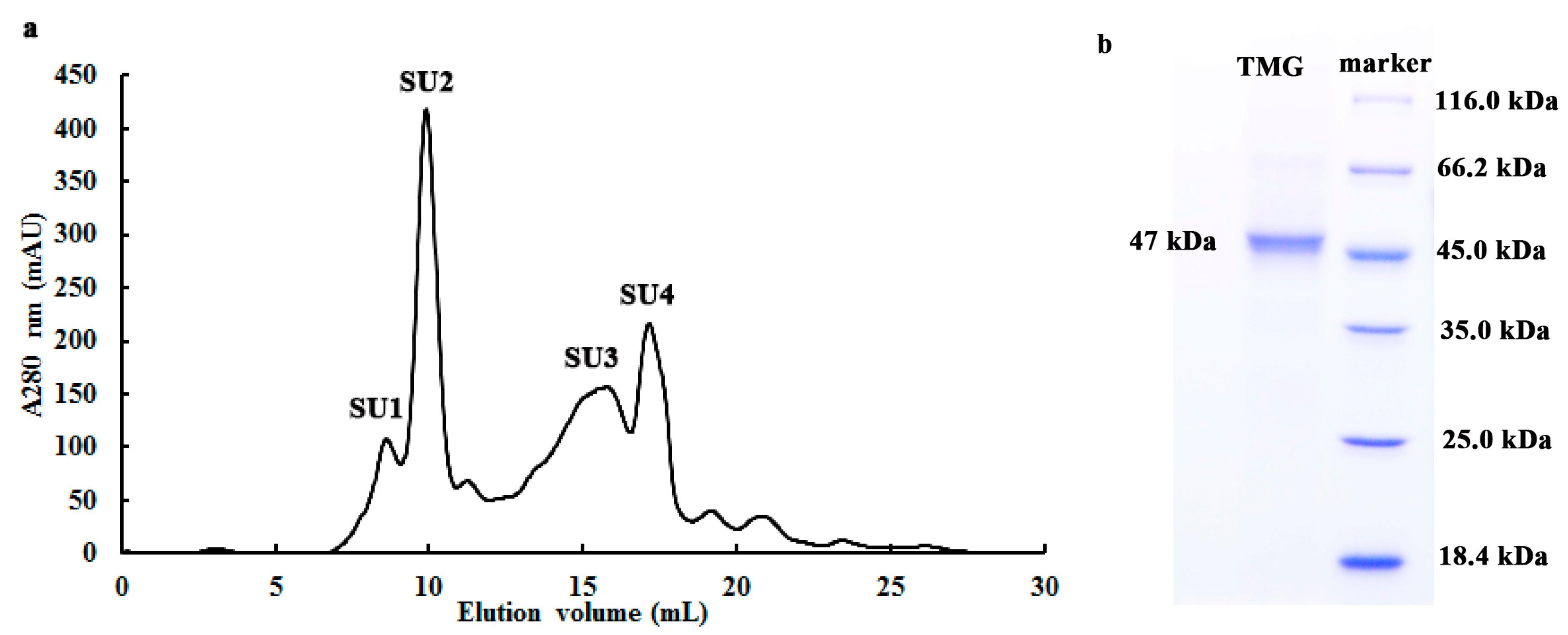
With pNPGal as substrate, the optimal pH value of TMG was observed at pH 4.5 (Figure 2a), which was identical with the optimal pH for α-galactosidases from Neosartorya fischeri [14]. Lenzites elegans [15] and Talaromyces emersonii [17]. The optimal pH values for α-galactosidase from Aspergillus terreus and an acidophilic fungus Bispora sp. MEY-1 were pH 5.5 and pH 3.5, respectively [12,27]. Although most of the optimal pH values for the α-galactosidases were an acidic pH, the optimal pH of α-galactosidase purified from Bacillus megaterium was 7.5 [28]. As shown in Figure 2a, when the pH was increased to 6, only 40.6% of the activity of TMG remained. When the pH was outside the range of 3.0 to 7.0, hardly any α-galactosidase activity could be detected.
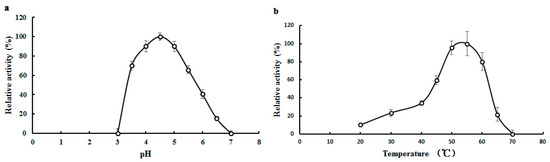
Figure 2.
Effects of pH and temperature on the activity of Tricholoma matsutake α-galactosidase. (a) Effect of pH on TMG. Buffer: 0.1 M Na2HPO4-citric acid buffers. Results represent mean ± SD (n = 3); (b) Effect of temperature on TMG. Results represent mean ± SD (n = 3).
Figure 2.
Effects of pH and temperature on the activity of Tricholoma matsutake α-galactosidase. (a) Effect of pH on TMG. Buffer: 0.1 M Na2HPO4-citric acid buffers. Results represent mean ± SD (n = 3); (b) Effect of temperature on TMG. Results represent mean ± SD (n = 3).
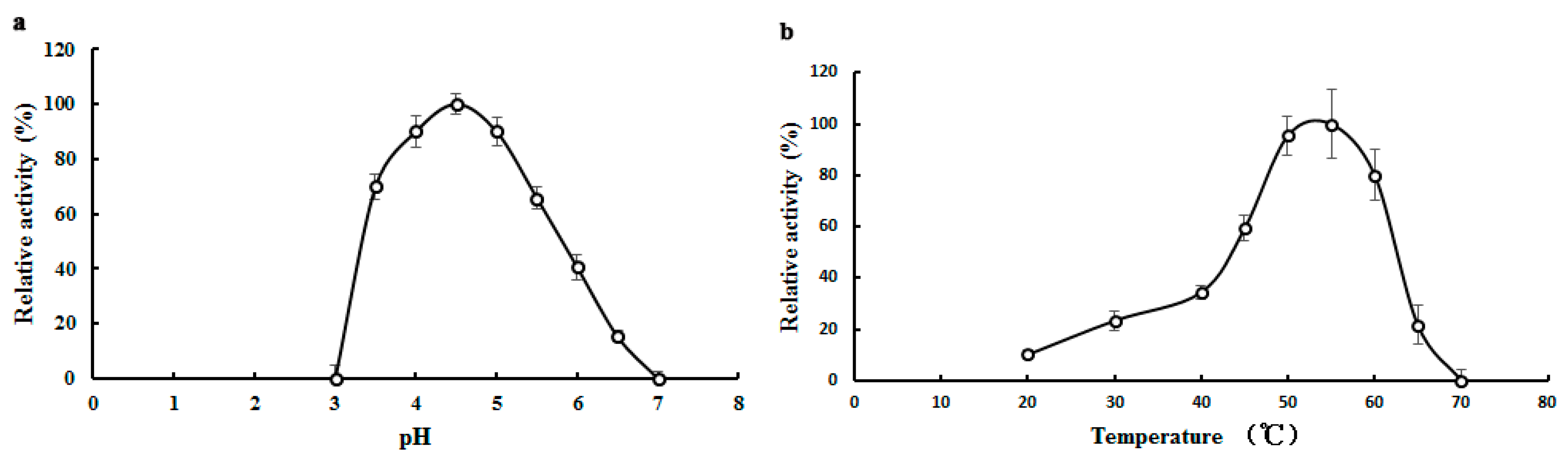
α-Galactosidases purified from different sources exhibited good enzymatic activity over a wide range of temperatures. The enzyme from Lenzites elegans [15] showed a high level of activity at temperatures ranging from 60 to 80 °C. Maximal activity of TMG was observed at 55 °C (Figure 2b) which was in good agreement with the enzyme from Bacillus megaterium [28]. When treated at 40 °C for 2 h, the activity of TMG was completely preserved (data not shown). However, TMG showed poor thermostability at higher temperatures and was completely inactivated after incubation at 50 °C for 30 min.
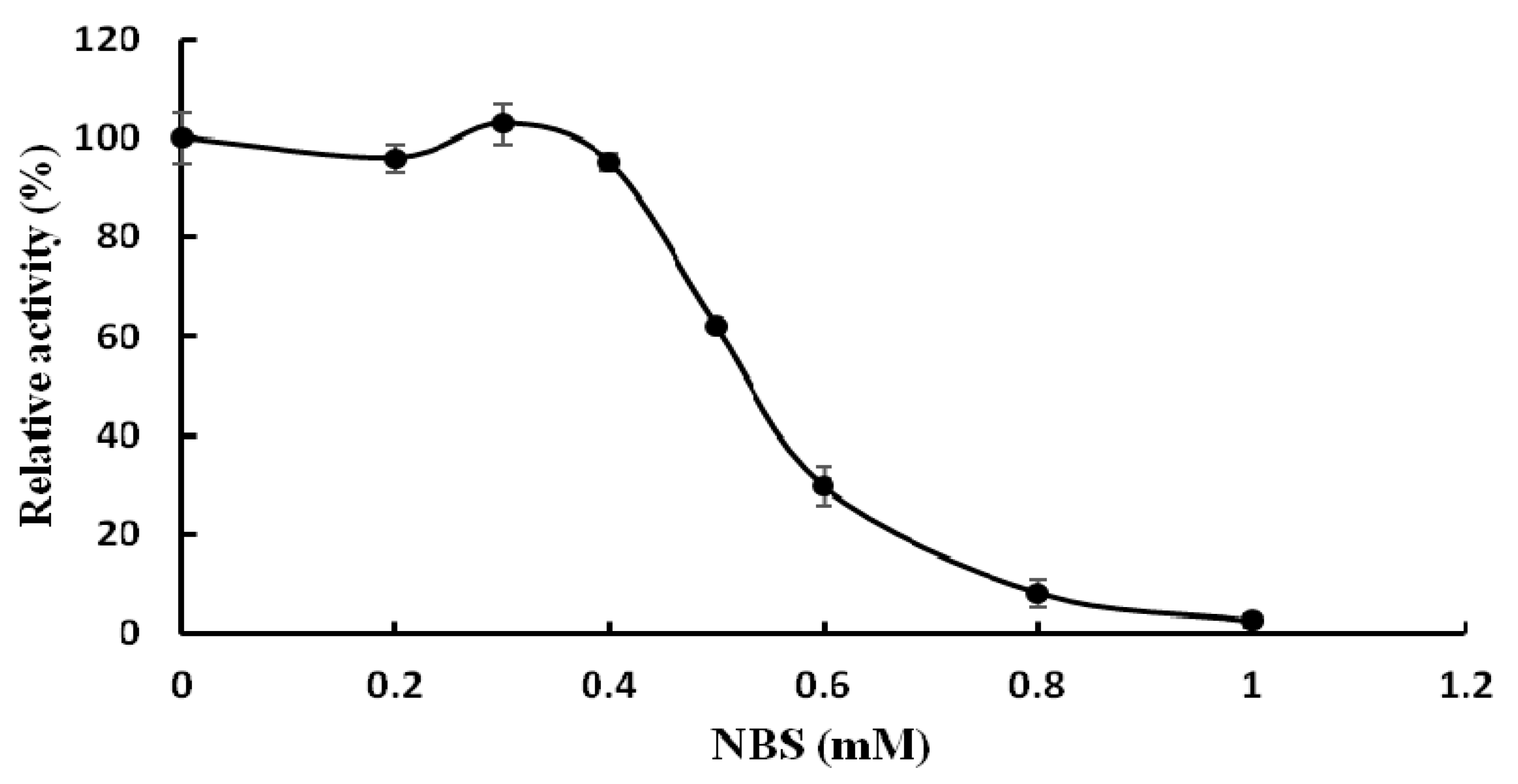
The effects of various metal ions and other chemicals on TMG activity were tested and TMG was found to be very sensitive to metal ions. The activity of TMG almost disappeared in the presence of K+, Ca2+, Cd2+, Hg2+, Ag+, and Zn2+ ions (1.25 mM–10 mM) and was strongly affected by Fe3+ ions (1.25 mM–10 mM) and Cu2+ ions (2.5 mM–10 mM). The total inhibition of α-galactosidase activity by Hg2+ and Ag+ ions was also seen in α-galactosidases from other sources, for example, the enzymes from Cicer arietinum [26] and Tachigali multijuga seeds [29]. It was reported by Singh [26] that the total inhibition by Ag+ ions was probably due to reaction of Ag+ ions with carboxyl and/or histidine residues. Hg2+ ions can react with thiol groups, amino and imidazolium groups of histidine and with peptide linkages, which was the reason why TMG was completely inhibited by Hg2+ ions [30]. Increasing the concentration of Mn2+, Mg2+, Pb2+, Al3+ ions (1.25 mM–10 mM) had little or no effect on TMG activity (Table 2). The activity of TMG was not inhibited by EDTA, implying that the enzyme is not a metalloenzyme or does not require divalent cations for activity. Similar to the present findings, EDTA had no effect on the activity of Bacillus megaterium and Ganoderma lucidum α-galactosidases [13,28]. While EDTA slightly inhibited the α-galactosidase activity from Lenzites elegans [15]. The chemical reagents modified different amino acid functional groups. The amino group of tryptophan was chemically modified by NBS while the histidine residues were modified by diethylpyrocarbonate (DEPC) [31]. The carboxyl groups were modified by carbodiimide (EDC) [32]. The disulfide bond and the arginine residues were chemically modified by dithiothreitol (DTT) [33] and diacetyl (DIC), respectively. The chemical modification reagent NBS drastically inhibited the activity of TMG. Incubation of TMG with 1 mM NBS for 30 min resulted in complete loss of α-galactosidase activity (Figure 3), which indicated the pivotal role of tryptophan at or near the active site. This finding was in keeping with the results reported for the α-galactosidases purified from Coriolus versicolor [24] and Phaseolus coccineus seeds [10]. Other chemical modification reagents (DEPC, EDC, DIC, DTT) had virtually no effect on TMG (data not shown), which indicated that the groups of tryptophan, histidine, carboxyl, arginine residues and disulfide bonds were not present at or in the proximity of the active site.

Table 2.
Effects of different metal ions on activity of Tricholoma matsutake α-galactosidase a.
| Metal Ion Concentration | Relative—Galactosidase Activity (%) | |||
|---|---|---|---|---|
| 10 mM | 5 mM | 2.5 mM | 1.25 mM | |
| Mg2+ | 30.6 ± 0.03 | 80.90.01 | 82.6 ± 0.02 | 93.0 ± 0.04 |
| Mn2+ | 43.3 ± 0.03 | 69.1 ± 0.07 | 101.6 ± 0.02 | 100.4 ± 0.02 |
| Pb2+ | 9.6 ± 0.02 | 80.0 ± 0.12 | 88.3 ± 0.09 | 95.3 ± 0.05 |
| Al3+ | 72.1 ± 0.01 | 80.7 ± 0.03 | 73.1 ± 0.05 | 82.5 ± 0.02 |
| Cu2+ | ND | ND | 31.8 ± 0.01 | 80.9 ± 0.05 |
| Fe3+ | ND | ND | ND | 31.4 ± 0.02 |
| EDTA | 116.4 ± 0.65 | 96.3 ± 0.84 | 101.3 ± 0.61 | 112.5 ± 0.45 |
a The activity of α-galactosidase without incubation with metal ions was set as 100%. Results represent mean ± SD, n = 3. The detailed data for each metal ion (Mg2+, Mn2+, Pb2+, Al3+, Cu2+ and Fe3+) are shown in Tables S1–S6 in the supplementary file. “ND” means not detected.
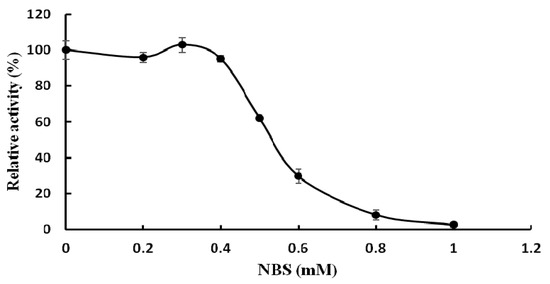
Figure 3.
Effect of the chemical modification reagent N-bromosuccinimide (NBS) on activity of Tricholoma matsutake α-galactosidase (results represent mean ± SD, n = 3).
Figure 3.
Effect of the chemical modification reagent N-bromosuccinimide (NBS) on activity of Tricholoma matsutake α-galactosidase (results represent mean ± SD, n = 3).
2.4. Substrate Specificity of TMG and Determination of Kinetic Parameters
The purified TMG showed different hydrolytic activities towards various natural and synthetic substrates (Table 3). The highest activity was demonstrated toward pNPGal, which was much higher than the activities toward other nitrophenyl derivatives (oPNPGal, 4-nitrophenyl β-d-glucuronide). TMG had hardly any activity on synthetic substrates oPNPGal, and 4-nitrophenyl β-d-glucuronide compared with pNPGal. This suggests that probably the para configuration of the substrate pNPGal facilitated its access to the active site of the enzyme. In this study, TMG exhibited higher activity toward natural substrates (stachyose, raffinose and melibiose) compared to synthetic nitrophenyl derivatives (oPNPGal, 4-nitrophenyl β-d-glucuronide). The rate of hydrolysis of natural substrates was in the order raffinose (84%) > stachyose (67%) > melibiose (17%). In particular, the relative activity of TMG for raffinose attained 84%. TMG showed meager activity on the polysaccharides, locust bean gum and guar gum, with the rate of hydrolysis being 12% and 13% respectively.

Table 3.
Hydrolysis of different substrates by Tricholoma matsutake α-galactosidase.
| Substrate | Concentration (mM) | Relative Activity (%) a |
|---|---|---|
| 4-Nitrophenyl α-d-galactopyranoside (pNPG) | 10 | 100 ± 0.77 |
| 2-Nitrophenyl β-d-galactopyranoside (oNPG) | 10 | 0.9 ± 0.03 |
| 4-Nitrophenyl β- d-glucuronide | 10 | 4 ± 0.01 |
| Stachyose | 50 | 67 ± 0.02 |
| Raffinose | 50 | 84 ± 0.01 |
| Melibiose | 50 | 17 ± 0.11 |
| Locust bean gum | 1% | 12 ± 0.07 |
| Guar gum | 1% | 13 ± 0.05 |
a Relative activities were calculated in comparison with pNPGal activity, which was considered as 100%. Results represent mean ± SD, n = 3.
The Km values of TMG for hydrolysis of pNPGal, raffinose and stachyose were 0.99 mM, 3.7 mM and 3.5 mM, respectively. This was in keeping with the above results showing that TMG displayed a higher affinity (lower Km) toward pNPGal than natural oligosaccharides as substrates. The calculated Km of TMG was lower than that of rGalQ17 (Km = 2.25 mM) from Flavobacterium ap. TN17a [11], and a little higher than that of the α-galactosidase (Km = 0.7 mM) from Cicer arietinum [26], but much higher than that of the α-galactosidase from Coriolus versicolor [24]. The catalytic efficiency expressed by Kcat/Km showed that the substrate pNPGal was used most efficiently by the enzyme (Table 4).

Table 4.
Kinetic parameters for hydrolysis of pNPGal, raffinose and stachyose by Tricholoma matsutake α-galactosidase.
| Substrate | Km (Mm) | Kcat a (s−1) | Kcat/Km (s−1·mM−1) |
|---|---|---|---|
| pPNGal | 0.99 | 9.17 | 9.3 |
| Raffinose | 3.7 | 6.4 | 1.73 |
| Stachyose | 3.5 | 6.7 | 1.9 |
a Turnover number (Kcat) is defined here as the number of mmoles of product s−1·mmol−1 of α-galactosidase, given by the relationship Vmax/[E].
Stachyose and raffinose were digested by TMG for 0.3, 1, 2, 4, 8 and 24 h, respectively. The content of reducing sugar increased gradually after treatment with TMG in the time-course of hydrolysis, which indicated that stachyose and raffinose were hydrolyzed effectively to yield galactose and sucrose (Figure 4).
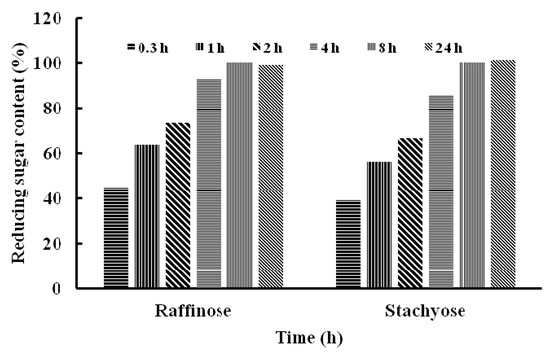
Figure 4.
The content of reducing sugar after hydrolysis of raffinose and stachyose by Tricholoma matsutake α-galactosidase.
Figure 4.
The content of reducing sugar after hydrolysis of raffinose and stachyose by Tricholoma matsutake α-galactosidase.
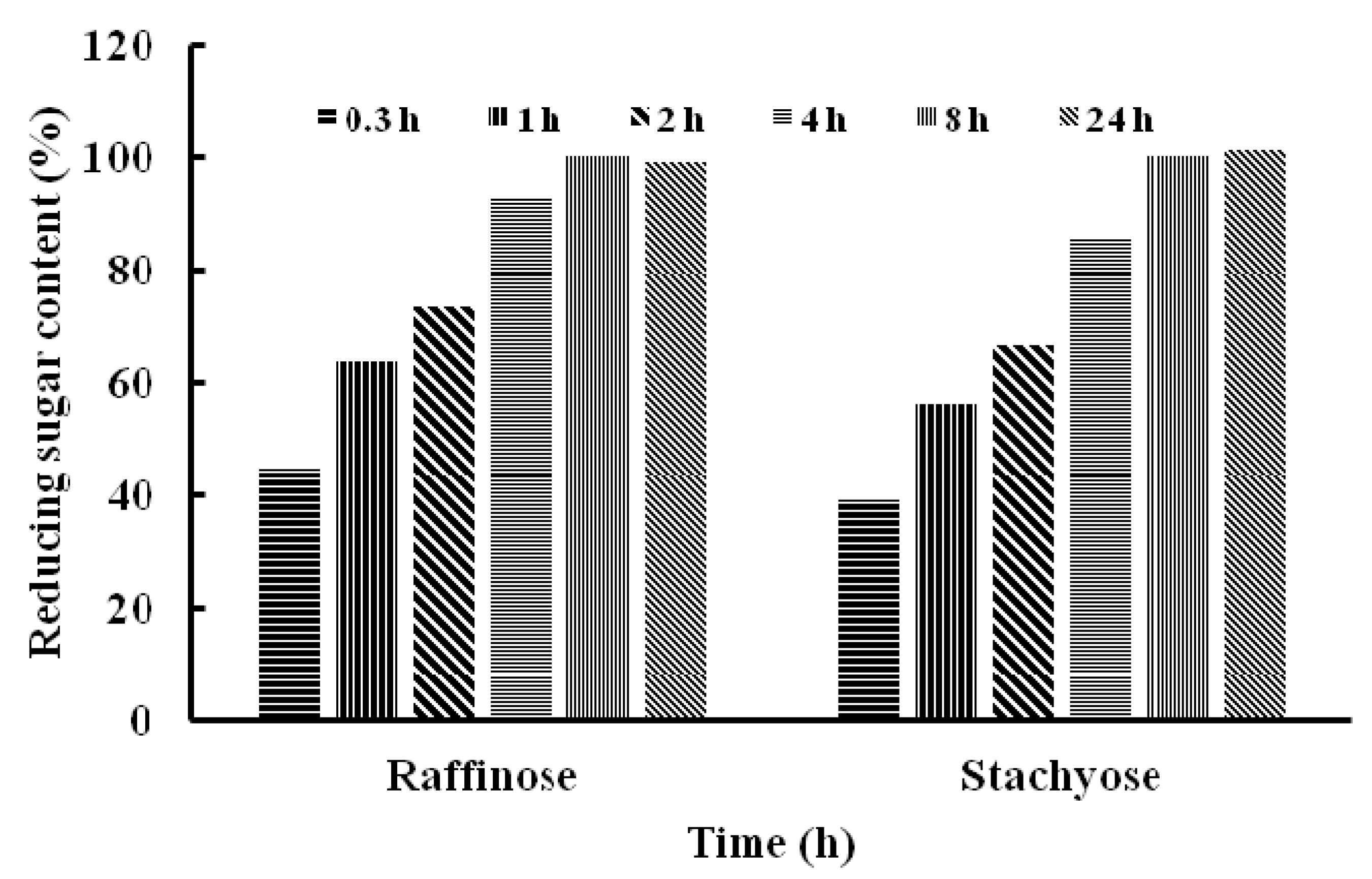
After 24 h, the content of reducing sugar released by the two oligosaccharides did not show a further increase compared with that at 8 h. The difference in efficiency of hydrolysis of the two oligosaccharides catalyzed by TMG was not remarkable, which was consistent with the Km of TMG to stachyose (Km = 3.5 mM) and raffinose (Km = 3.7 mM). Because humans and monogastric animals lack the enzyme α-galactosidase, raffinose family oligosaccharides, mainly stachyose and raffinose, cannot be absorbed and accumulate resulting in their microbial fermentation and flatus formation [2]. It is possible to improve the nutritional value of soymilk by reducing the content of RFOs and exogenous enzyme treated with RFOs has been proven to be a more effective and reliable method [10]. The ability of TMG to hydrolyze RFOs make it a good candidate as an additive for biotechnological applications. Besides, T. matsutake is a well-known wild edible mushroom and this finding can prompt further exploitations of the functions and activities of T. matsutake. There are also reports on α-galactosidases from other sources which degrade RFOs. Similarly, an α-galactosidase from symbiotic Flavobacterium sp. TN17 efficiently hydrolyzed many natural substrates including stachyose, raffinose and melibiose [11]. Wang et al., revealed that after treatment with one of α-galactosidases from Neosartorya fischeri P1 at 50 °C for 3 h, the raffinose and stachyose contents were decreased by 69.9% and 94.5%, respectively [14].To recapitulate, we have reported the purification and characterization of an α-galactosidase from T. matsutake. The good capacity of TMG in the degradation of raffinose oligosaccharides was noteworthy. Though the wild edible mushroom T. matsutake is highly valued as a delicacy and medicine, it is wild and cannot be cultivated. In order to have better productivity and facilitate the enzyme purification, it is necessary to clone the enzyme in the near future. Many α-galactosidases were cloned and a high-level expression of α-galactosidase from Rhizomucor miehei [34], the recombinant enzyme from Flavobacterium sp TN17 [11] and an α-galactosidase from Neosartorya fischeri P1 with significant hydrolysis ability of milk [14] could easily be obtained, TMG showed poor thermostability, it can be improved by immobilizing it onto functionalized graphene or onto chitosan and Amberlite, which were based on the immobilization of Cicer α-galactosidases [35,36]. The three immobilized Cicer α-galactosidases had higher thermal stability than the soluble enzymes and effective hydrolysis of RFOs increases the prospects of application of Cicer α-galactosidases in food processing industries. The TMG can also be non-covalently immobilized on a reversibly soluble-insoluble polymer [37] or immobilized on Sepabesd EC-EA and Sepabead EC-HA [38].
In future investigations, combing the cloning of the enzyme to produce higher enzyme amounts easier to be purified [11,14,34] and a suitable enzyme immobilization to improve the enzyme features (stability, activity, selectivity) [39,40,41] may convert this promising enzyme into a suitable product with potential applications in industry.
3. Experimental Section
3.1. Materials
Tricholoma matsutake fruiting bodies were collected from Yunnan Province (China). Q-Sepharose, Mono-Q, Superdex 75 HR 10/30 and AKTA Purifier were purchased from GE Healthcare (Uppsala, Sweden). The substrates, 4-nitrophenyl-α-D-galactophyranoside (pNPGal), 2-nitrophenyl β-d-galactopyranoside (oNPGal), 4-nitrophenyl β-d-glucuronide, locust bean gum, guar gum, melibiose, stachyose and raffinose were purchased from Sigma Chemical Company (St. Louis, MO, USA). All other chemicals used were of analytical grade unless otherwise stated.
3.2. Enzyme Activity Assay
The activity of α-galactosidase was assayed with the pNPGal method as described [42] with modification. Diluted enzyme (50 μL) was incubated with 50 μL 10 mM pNPGal (pH 4.6) at 50 °C for 10 min. The reaction was terminated by adding 400 μL 0.5 M Na2CO3 and the released p-nitrophenol was determined spectrophotometrically at 405 nm. One unit of α-galactosidase activity was defined as the amount of enzyme required to release 1 μmol of p-nitrophenol per min at 50 °C and pH 4.6.
3.3. Purification of α-Galactosidase from Tricholoma Matsutake
Fractionation of an extract of Tricholoma matsutake fresh fruiting bodies (400 g) was carried out by ion exchange chromatography using a 10 cm × 100 cm column of Q-Sepharose. The extract was homogenized in distilled water (1:2, w:v) with a Waring blender followed by extraction overnight at 4 °C. Then the homogenate was centrifuged at 9000 rpm for 20 min at 4 °C. The supernatant was collected and applied to a Q-Sepharose column in 10 mM NaOAc-HOAc buffer (pH 5.2). After removal of the unadsorbed fraction Q1, adsorbed proteins were eluted stepwise with 100 mM NaCl in NaAc-HAc buffer to yield fraction Q2 and then eluted with 1.0 M NaCl in NaOAc-HOAc buffer to yield fraction Q3. α-Galactosidase activity was located in fraction Q2. After dialysis against distilled water, fraction Q2 was subjected to ion exchange chromatography on Mono-Q. After the unadsorbed fraction MQ1 was eluted, the column was further eluted with a linear 0–1.0 M NaCl gradient in the same NaOAc-HOAc buffer (pH 5.6). The active fraction MQ1 was further purified by gel filtraction on a fast protein liquid chromatography Superdex 75 10/300 GL column in 10 mM NaOAc-HOAc buffer (pH 5.2).
3.4. Determination of Molecular Weight and Amino Acid Sequence
The purified α-galactosidase from Tricholoma matsutake was subjected to SDS-PAGE [43] to assesss protein homogeneity and to determine molecular mass. The native molecular mass of the α-galactosidase was determined with size exclusion chromatography using a Superdex 75 column. The α-galactosidase band obtained after SDS-PAGE was destained, digested with trypsin, and then dissolved in 0.1% formic acid and 2% acetonitrile for liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis using an LTQ—Orbitrap mass spectrometer (Thermo Electron, Bremen, Germany).
3.5. Biochemical Properties of the Enzyme
In the assay for optimum pH, a solution of 10 mM pNPGal, which was used as substrate, was freshly prepared in the pH range from pH 2.0–8.0 using 100 mM Na2HPO4-citric acid buffer, and the activity of the α-galactosidase from Tricholoma matsutake was recorded at each pH value to ascertain the pH associated with the maximal activity. For determining the optimum temperature, the activity of the α-galactosidase toward the substrate pNPGal was assayed over the temperature range of 20–70 °C at the optimum pH. The effects of various metal ions and chemical reagents(Na+, K+, Al3+, Zn2+, Mg2+, Mn2+, Fe3+, Ca2+, Cu2+, Cd2+, Hg2+, Pb2+, EDTA, NBS, DTT, DIC, EDC, DEPC) on the activity of purified enzyme were examined. The enzyme was incubated with a series of concentrations of metal ions and chemical reagents at 4 °C for 2 h and the remaining activity toward pNPGal was assayed as described above.
3.6. Substrate Specificity and Kinetic Parameters
In order to determine the substrate specificity of the purified α-galactosidase, various synthetic and natural substrates were used in the assay. The activity against synthetic substrates, pNPGal, oNPGal, 4-nitrophenyl β-d-glucuronide (10 mM), was determined under standard conditions as described for pNPGal. The ability to hydrolyze natural substrates (locust bean gum, guar gum, melibiose, stachyose and raffinose) was determined by measuring the reducing sugar released using 3,5-dinitrosalicylic acid as described by Miller [44]with slight modification. The reaction mixture consisting of appropriately diluted enzyme and 10 mM sugar in 100 mM NaAc-HAc buffer (pH 4.6) was incubated at 40 °C for 30 min. One unit of α-galactosidase activity was defined as the amount of enzyme required to release 1 μmol of reducing sugar equivalent to galactose per minute under the assay conditions. While the glucose released by the substrate melibiose was measured with a glucose-oxidase kit (Beijing BHKT Clinical Reagent Co., Ltd. Beijing, China). One unit of the enzyme activity was defined as the amount of enzyme required to release 1μmol of glucose per minute at 37 °C. The kinetic parameter Km of the purified α-galactosidase on different substrates was determined. pNPGal (1–10 mM), stachyose(1–10 mM) and raffinose(1–10 mM) were used as substrates, and the reactions were performed in the same conditions as before. The apparent Michaelis constant Km was calculated using the Grafit program.
Supplementary Materials
Supplementary materials can be accessed at: http://www.mdpi.com/ 1420-3049/20/08/13550/s1.
Acknowledgments
This work was financially supported by China Agriculture Research System (CARS24) and Special Fund for Agro-scientific Research in the Public Interest (No. 201303080).
Author Contributions
X.G. performed the experiments. G.T. collected and analysed the data. Y.Z. and L.Z. provided the materials and reagents. H.W. conceived and designed the study. T.B.N. wrote the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Katrolia, P.; Jia, H.; Yan, Q.; Song, S.; Jiang, Z.; Xu, H. Characterization of a protease-resistant alpha-galactosidase from the thermophilic fungus Rhizomucor miehei and its application in removal of raffinose family oligosaccharides. Bioresour. Technol. 2012, 110, 578–586. [Google Scholar] [CrossRef] [PubMed]
- Steggerda, F.R. Gastrointestinal gas following food consumption. Ann. N. Y. Acad. Sci. 1968, 150, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Kayastha, A.M. A novel application of Cicer alpha-galactosidase in reduction of raffinose family oligosaccharides in soybean folur. J. Plant Biochem. Biotechnol. 2013, 22, 353–356. [Google Scholar] [CrossRef]
- Ghazi, S.; Rooke, J.A.; Galbraith, H. Improvement of the nutritive value of soybean meal by protease and alpha-galactosidase treatment in broiler cockerels and broiler chicks. Br. Poult. Sci. 2003, 44, 410–418. [Google Scholar] [CrossRef] [PubMed]
- Kotiguda, G.; Kapnoor, S.S.; Kulkarni, D.; Mulimani, V.H. Degradation of raffinose oligosaccharides in soymilk by immobilized alpha-galactosidase of Aspergillus oryzae. J. Microbiol. Biotechnol. 2007, 17, 1430–1436. [Google Scholar] [PubMed]
- Kobayashi, H.; Suzuki, H. Studies on the decomposition of raffinose by α-galactosidase of mold. J. Fermet. Technol. 1972, 50, 625–632. [Google Scholar]
- Mittal, Y.; Sharma, C.B. Development of α-galactosidase isoenzymes in chickpea seeds. Plant Sci. 1991, 77, 185–190. [Google Scholar] [CrossRef]
- Gao, Z.; Schaffer, A.A. A novel alkaline alpha-galactosidase from melon fruit with a substrate preference for raffinose. Plant Physiol. 1999, 119, 979–988. [Google Scholar] [CrossRef] [PubMed]
- Smart, E.L.; Pharr, D.M. Characterization of alpha-Galactosidase from Cucumber Leaves. Plant Physiol. 1980, 66, 731–734. [Google Scholar] [CrossRef] [PubMed]
- Du, F.; Zhu, M.; Wang, H.; Ng, T. Purification and characterization of an alpha-galactosidase from Phaseolus coccineus seeds showing degrading capability on raffinose family oligosaccharides. Plant Physiol. Biochem. 2013, 69, 49–53. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Shi, P.; Huang, H.; Cao, Y.; Meng, K.; Yang, P.; Zhang, R.; Chen, X.; Yao, B. A new alpha-galactosidase from symbiotic Flavobacterium sp. TN17 reveals four residues essential for alpha-galactosidase activity of gastrointestinal bacteria. Appl. Microbiol. Biotechnol. 2010, 88, 1297–1309. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, J.G.; Reis, A.P.; Guimaraes, V.M.; Falkoski, D.L.; da Silva Fialho, L.; de Rezende, S.T. Purification and characterization of Aspergillus terreus alpha-galactosidases and their use for hydrolysis of soymilk oligosaccharides. Appl. Biochem. Biotechnol. 2011, 164, 1111–1125. [Google Scholar] [CrossRef] [PubMed]
- Sripuan, T.; Aoki, K.; Yamamoto, K.; Tongkao, D.; Kumagai, H. Purification and characterization of thermostable alpha-galactosidase from Ganoderma lucidum. Biosci. Biotechnol. Biochem. 2003, 67, 1485–1491. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Shi, P.; Luo, H.; Huang, H.; Yang, P.; Yao, B. A thermophilic alpha-galactosidase from Neosartorya fischeri P1 with high specific activity, broad substrate specificity and significant hydrolysis ability of soymilk. Bioresour. Technol. 2014, 153, 361–364. [Google Scholar] [CrossRef] [PubMed]
- Sampietro, D.; Quiroga, E.; Sgariglia, M.; Soberon, J.; Vattuone, M.A. A thermostable alpha-galactosidase from Lenzites elegans (Spreng.) ex Pat. MB445947: Purification and properties. Antonie Leeuwenhoek 2012, 102, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Saad, R.R.; Fawzi, E.M. Purification and characterization of a thermostable alpha-galactosidase from Thielavia terrestris NRRL 8126 in solid state fermentation. Acta Biol. Hung. 2012, 63, 138–150. [Google Scholar] [CrossRef] [PubMed]
- Simila, J.; Gernig, A.; Murray, P.; Fernandes, S.; Tuohy, M.G. Cloning and expression of a thermostable alpha-galactosidase from the thermophilic fungus Talaromyces emersonii in the methylotrophic yeast Pichia pastoris. J. Microbiol. Biotechnol. 2010, 20, 1653–1663. [Google Scholar] [PubMed]
- Hoshi, H.; Yagi, Y.; Iijima, H.; Matsunaga, K.; Ishihara, Y.; Yasuhara, T. Isolation and characterization of a novel immunomodulatory alpha-glucan-protein complex from the mycelium of Tricholoma matsutake in basidiomycetes. J. Agric. Food Chem. 2005, 53, 8948–8956. [Google Scholar] [CrossRef] [PubMed]
- Ikekawa, T.; Uehara, N.; Maeda, Y.; Nakanishi, M.; Fukuoka, F. Antitumor activity of aqueous extracts of edible mushrooms. Cancer Res. 1969, 29, 734–735. [Google Scholar] [PubMed]
- Mau, J.L.; Lin, H.C.; Song, S.F. Antioxidant properties of several specialty mushrooms. Food Res. Int. 2002, 35, 519–526. [Google Scholar] [CrossRef]
- Xu, L.; Zhu, M.; Chen, X.; Wang, H.; Zhang, G. A novel laccase from fresh fruiting bodies of the wild medicinal mushroom Tricholoma matsutake. Acta Biochim. Pol. 2015, 62, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, T.; Mizuno, K.; Kurosawa, S. Purification and properties of a nuclease from the fruit body of Tricholoma matsutake. Biosci. Biotechnol. Biochem. 2010, 74, 206–208. [Google Scholar] [CrossRef] [PubMed]
- Tong, H.; Liu, X.; Tian, D.; Sun, X. Purification, chemical characterization and radical scavenging activities of alkali-extracted polysaccharide fractions isolated from the fruit bodies of Tricholoma matsutake. World J. Microbiol. Biotechnol. 2013, 29, 775–780. [Google Scholar] [CrossRef] [PubMed]
- Du, F.; Liu, Q.; Wang, H.; Ng, T. Purification an alpha-galactosidase from Coriolus versicolor with acid-resistant and good degradation ability on raffinose family oligosaccharides. World J. Microbiol. Biotechnol 2014, 30, 1261–1267. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam; Saraswathy, N.; Sadasivam, S.; Subha, K.; Poorani, N. Purification and properties of alpha-galactosidase from white-rot fungus Pleurotus florida. Indian J. Biochem. Biophys. 2007, 44, 76–81. [Google Scholar] [PubMed]
- Singh, N.; Kayastha, A.M. Purification and characterization of alpha-galactosidase from white chickpea (Cicer arietinum). J. Agric. Food chem. 2012, 60, 3253–3259. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Luo, H.; Li, J.; Bai, Y.; Huang, H.; Shi, P.; Fan, Y.; Yao, B. An alpha-galactosidase from an acidophilic Bispora sp. MEY-1 strain acts synergistically with beta-mannanase. Bioresour. Technol. 2010, 101, 8376–8382. [Google Scholar] [CrossRef] [PubMed]
- Patil, A.G.G. α-Galactosidase from bacillus megaterium VHM1 and its application in removal of flatulence-causing factors from soymilk. J. Microbiol. Biotechnol. 2010, 20, 1546–1554. [Google Scholar] [CrossRef] [PubMed]
- Da Silva Fialho, L; Guimaraes, V.M.; Callegari, C.M.; Reis, A.P.; Barbosa, D.S.; de Lima Borgesb, E.E.; Moreira, M.A.; de Rezende, S.T. Characterization and biotechnological application of an acid alpha-galactosidase from Tachigali multijuga Benth. seeds. Phytochemistry 2008, 69, 2579–2585. [Google Scholar]
- Dey, P.M.; Pridham, J.B. Biochemistry of α-galactosidases. Adv. Enzymol. Relat. Areas Mol. Biol. 1972, 36, 114–118. [Google Scholar]
- Gote, M.M.; Khan, M.I.; Khire, J.M. Active site directed chemical modification of alpha-galactosidase from Bacillus stearothermophilus (NCIM 5146): Involvement of lysine, tryptophan and carboxylate residues in catalytic site. Enzyme Microb. Technol. 2007, 40, 1312–1320. [Google Scholar] [CrossRef]
- Mathew, C.D.; Balasubramaniam, K. Chemical modification of alpha-galactosidase from coconut. Phytochemistry 1986, 25, 2439–2443. [Google Scholar] [CrossRef]
- Huang, K.; Chen, S.Z.; Yang, K.Y. Crystallization and chemical modification of disulfide bond of calf chymosin. Chin. J. Biotechnol. 1991, 7, 83–92. [Google Scholar] [PubMed]
- Chen, Z.; Yan, Q.; Jiang, Z.; Liu, Y.; Li, Y. High-level expression of a novel α-galactosidase gene from Rhizomucor miehei in Pichia pastoris and characterization of the recombinant enyzme. Protein Expr. Purif. 2015, 110, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Srivastava, G.; Talat, M.; Raghubanshi, H.; Srivastava, O.N.; Kayastha, A.M. Cicer alpha-galactosidase immobilization onto functionalized graphene nanosheets using response surface method and its applications. Food Chem. 2014, 142, 430–438. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Kayastha, A.M. Cicer alpha-galactosidase immobilization onto chitosan and Amberlite MB-150: Optimization, characterization, and its applications. Carbohydr. Res. 2012, 358, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Park, I.; Cho, J. Immobilization of the antarctic Bacillus sp. LX-1 alpha-galactosidase on eudragit L-100 for the production of a functional feed additive. Asian Australas. J. Anim. Sci. 2013, 26, 552–557. [Google Scholar] [CrossRef] [PubMed]
- Bayraktar, H.; Serilmez, M.; Karkas, T.; Celem, E.B.; Onal, S. Immobilization and stabilization of alpha-galactosidase on Sepabeads EC-EA and EC-HA. Intl. J. Biol. Macromol. 2011, 49, 855–860. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, O.; Ortiz, C.; Berenguer-Murcia, A.; Torres, R.; Rodrigues, R.C.; Fernandez-Lafuente, R. Strategies for the one-step immobilization-purification of enzymes as industrial biocatalysts. Biotechnol. Adv. 2015, 33, 435–456. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, R.C.; Ortiz, C.; Berenguer-Murcia, A.; Torres, R.; Fernandez-Lafuente, R. Modifying enzyme activity and selectivity by immobilization. Chem. Soc. Rev. 2013, 42, 6290–6307. [Google Scholar] [CrossRef] [PubMed]
- Husain, Q. Beta galactosidases and their potential applications: A review. Crit. Rev. Biotechnol. 2010, 30, 41–62. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Wang, Y.; Luo, H.; Shi, P.; Meng, K.; Zhou, Z.; Zhang, Z.; Yao, B. Molecular cloning and expression of a novel protease-resistant GH-36 alpha-galactosidase from Rhizopus sp. F78 ACCC 30795. J. Microbiol. Biotechnol. 2009, 19, 1295–1300. [Google Scholar] [PubMed]
- Laemmli, U.K.; Favre, M. Maturation of the head of bacteriophage T4. I. DNA packaging events. J. Mol. Biol. 1973, 80, 575–599. [Google Scholar] [CrossRef]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing suGAR. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Sample Availability: Samples of the compounds are available from the authors.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
