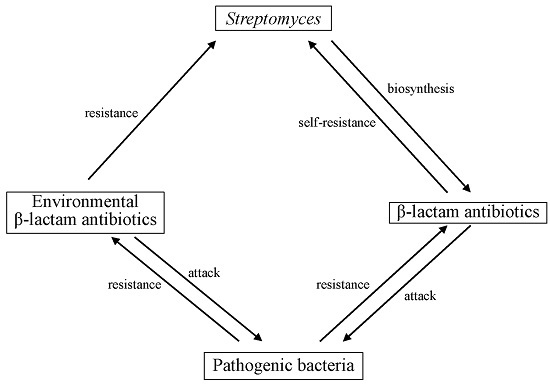
Self-resistance in Streptomyces, with Special Reference to ?-Lactam Antibiotics
Abstract
:1. Introduction
2. Self-resistance mechanisms
2.1. β-Lactamases
2.1.1. Class A β-lactamases
2.1.2. Class B β-lactamases
2.1.3. Class C β-lactamases
2.2. Penicillin-Binding Proteins (PBPs)
2.2.1. Class A PBPs
2.2.2. Class B PBPs
3. PASTA Domains: Connector between Penicillin-Binding Proteins and Protein Kinases
4. Relationship between β-Lactam Biosynthetic Gene, β-Lactamase and PBP: Additional Comments
5. Conclusions
Supplementary Materials
Conflicts of Interest
References
- Fleming, A. On the antibacterial action of cultures of penicillium, with special reference to their use in the isolation of B. influenzae. Br. J. Exp. Pathol. 1929, 10, 226–236. [Google Scholar] [CrossRef]
- Chain, E.; Florey, H.W.; Gardner, A.D.; Heatley, N.G.; Jennings, M.A.; Orr-Ewing, J.; Sanders, A.G. Penicillin as a chemotherapeutic agent. Lancet 1940, 236, 226–228. [Google Scholar] [CrossRef]
- Abraham, E.P.; Chain, E.; Fletcher, C.M.; Florey, H.W.; Gardner, A.D.; Heatley, N.G.; Jennings, M.A. Further observations in penicillin. Lancet 1941, 238, 177–189. [Google Scholar] [CrossRef]
- Abraham, E.P.; Chain, E. An enzyme from bacteria able to destroy penicillin. Rev. Infect. Dis. 1940, 10, 677–678. [Google Scholar] [CrossRef]
- Plough, H.H. Penicillin resistance of Staphylococcus aureus and its clinical implications. Am. J. Clin. Pathol. 1945, 15, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Bondi, A., Jr.; Dietz, C.C. Penicillin resistant staphylococci. Proc. Soc. Exp. Biol. Med. 1945, 60, 55–58. [Google Scholar] [CrossRef] [PubMed]
- Eriksen, K.R. Studies on induced resistance to penicillin in pneumococcus type I. Acta Pathol. Microbiol. Scand. 1945, 22, 398–405. [Google Scholar] [CrossRef] [PubMed]
- Gots, J.S. Production of extracellular penicillin-inactivating substances associated with penicillin resistance in Staphylococcus aureus. Proc. Soc. Exp. Biol. Med. 1945, 60, 165–168. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.P.; Bohnhoff, M. Studies on the action of penicillin; development of penicillin resistance by gonococcus. Proc. Soc. Exp. Biol. Med. 1945, 60, 354–356. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.P.; Bohnhoff, M. Studies on action of penicillin; virulence of penicillin resistant strains of meningococcus. Proc. Soc. Exp. Biol. Med. 1945, 60, 356–362. [Google Scholar] [CrossRef] [PubMed]
- Shwartzman, G. Studies on the nature of resistance of Gram-negative bacilli to penicillin. J. Exp. Med. 1946, 83, 65–88. [Google Scholar] [CrossRef] [PubMed]
- Waksman, S.A.; Reilly, H.C.; Schatz, A. Strain specificity and production of antibiotic substances. V. Strain resistance of bacteria to antibiotic substances, especially streptomycin. Proc. Natl. Acad. Sci. USA 1945, 31, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Ogawara, H.; Horikawa, S.; Shimada-Miyoshi, S.; Yasuzawa, K. Production and property of beta-lactamases in Streptomyces: Comparison of the strains isolated newly and thirty years ago. Antimicrob. Agents Chemother. 1978, 13, 865–870. [Google Scholar] [CrossRef] [PubMed]
- D’Costa, V.M.; King, C.E.; Kalan, L.; Morar, M.; Sung, W.W.; Schwarz, C.; Froese, D.; Zazula, G.; Calmels, F.; Debruyne, R.; et al. Antibiotic resistance is ancient. Nature 2011, 477, 457–461. [Google Scholar] [CrossRef] [PubMed]
- About antimicrobial resistance. Available online: http://www.cdc.gov/drugresistance/ (accessed on 20 March 2016).
- Liu, B.; Pop, M. ARDB—Antibiotic resistance genes database. Nucleic Acids Res. 2009, 37, D443–D447. Available online: http://ardb.cbcb.umd.edu/ (accessed on 20 March 2016). [Google Scholar] [CrossRef] [PubMed]
- Walker, M.S.; Walker, J.B. Streptomycin biosynthesis and metabolism. Enzymatic phosphorylation of dihydrostreptobiosamine moieties of dihydrostreptomycin-(streptidino) phosphate and dihydrostreptomycin by Streptomyces extracts. J. Biol. Chem. 1970, 245, 6683–6689. [Google Scholar] [PubMed]
- Benveniste, R.; Davies, J. Aminoglycoside antibiotic-inactivating enzymes in actinomycetes similar to those present in clinical isolates of antibiotic-resistant bacteria. Proc. Natl. Acad. Sci. USA 1973, 70, 2276–2280. [Google Scholar] [CrossRef] [PubMed]
- Ogawara, H. Antibiotic resistance in pathogenic and producing bacteria, with special reference to β-lactam antibiotics. Microbiol. Rev. 1981, 45, 591–619. [Google Scholar] [PubMed]
- Dantas, G.; Sommer, M.O.; Oluwasegun, R.D.; Church, G.M. Bacteria subsisting on antibiotics. Science 2008, 320, 100–103. [Google Scholar] [CrossRef] [PubMed]
- Riesenfeld, C.S.; Goodman, R.M.; Handelsman, J. Uncultured soil bacteria are a reservoir of new antibiotic resistance genes. Environ. Microbiol. 2004, 6, 981–989. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.D. The antibiotic resistome: The nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 2007, 5, 175–186. [Google Scholar] [CrossRef] [PubMed]
- D’Costa, V.M.; McGrann, K.M.; Hughes, D.W.; Wright, G.D. Sampling the antibiotic resistome. Science 2006, 311, 374–377. [Google Scholar] [CrossRef] [PubMed]
- Forsberg, K.J.; Reyes, A.; Wang, B.; Selleck, E.M.; Sommer, M.O.; Dantas, G. The shared antibiotic resistome of soil bacteria and human pathogens. Science 2012, 337, 1107–1111. [Google Scholar] [CrossRef] [PubMed]
- Mole, B. MRSA: Farming up trouble. Nature 2013, 499, 398–400. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.L. Antibiotics and antibiotic resistance genes in natural environments. Science 2008, 321, 365–367. [Google Scholar] [CrossRef] [PubMed]
- Aminov, R.I. Horizontal gene exchange in environmental microbiota. Front. Microl. 2011, 2. [Google Scholar] [CrossRef] [PubMed]
- Perry, J.A.; Wright, G.D. The antibiotic resistance “mobilome”: Searching for the link between environment and clinic. Front. Microbiol. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Francino, M.P. Antibiotics and the human gut microbiome: Dysbioses and accumulation of resistance. Front. Micribiol. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Gupta, R.S. Phylogenetic framework and molecular signatures for the main clades of the phylum Actinobacteria. Microbiol. Mol. Biol. Rev. 2012, 76, 66–112. [Google Scholar] [CrossRef] [PubMed]
- Ventura, M.; Canchaya, C.; Tauch, A.; Chandra, G.; Fitzgerald, G.F.; Chater, K.F.; van Sinderen, D. Genomics of Actinobacteria: Tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 2007, 71, 495–548. [Google Scholar] [CrossRef] [PubMed]
- Chater, K.F.; Biró, S.; Lee, K.J.; Palmer, T.; Schrempf, H. The complex extracellular biology of Streptomyces. FEMS Microbiol. Rev. 2010, 34, 171–198. [Google Scholar] [CrossRef] [PubMed]
- Liras, P.; Martin, J.F. Gene clusters for β-lactam antibiotics and control of their expression: Why have clusters evolved, and from where did they originate? Int. Microbiol. 2006, 9, 9–19. [Google Scholar] [PubMed]
- Liu, G.; Chater, K.F.; Chandra, G.; Niu, G.; Tan, H. Molecular regulation of antibiotic biosynthesis in streptomyces. Microbiol. Mol. Biol. Rev. 2013, 77, 112–143. [Google Scholar] [CrossRef] [PubMed]
- Kahan, J.S.; Kahan, F.M.; Goegelman, R.; Currie, S.A.; Jackson, M.; Stapley, E.; Miller, T.W.; Miller, A.K.; Hendlin, D.; Mochales, S.; et al. Thienamycin, a new beta-lactam antibiotic. I. Discovery, taxonomy, isolation and physical properties. J. Antibiot. 1979, 32, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Reading, C.; Cole, M. Clavulanic acid: A beta-lactamase-inhiting beta-lactam from Streptomyces clavuligerus. Antimicrob. Agents Chemother. 1977, 11, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Koch, A.L. Penicillin binding proteins, β-lactams, and lactamases: Offensives, attacks, and defensive countermeasures. Crit. Rev. Microbiol. 2000, 26, 205–220. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.D.; Maas, W.K. Analysis of the biochemical mechanism of drug resistance in certain bacterial mutants. Proc. Natl. Acad. Sci. USA 1952, 38, 775–785. [Google Scholar] [CrossRef] [PubMed]
- The Comprehensive Antibiotic Resistance Database. Available online: http://arpcard.mcmaster.ca/ (accessed on 20 March 2016).
- King, D.T.; Sobhanifar, S.; Strynadka, N.C. One ring to rule them all: Current trends in combating bacterial resistance to the β-lactams. Protein Sci. 2016. [Google Scholar] [CrossRef] [PubMed]
- Cundliffe, E.; Demain, A.L. Avoidance of suicide in antibiotic-producing microbes. J. Ind. Microbiol. Biotechnol. 2010, 37, 643–672. [Google Scholar] [CrossRef] [PubMed]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef] [PubMed]
- Gibson, M.K.; Forsberg, K.J.; Dantas, G. Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. ISME J. 2015, 9, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Richmond, M.H.; Sykes, R.B. The β-lactamases of gram-negative bacteria and their possible physiological role. Adv. Microb. Physiol. 1973, 9, 31–88. [Google Scholar] [PubMed]
- Ogawara, H. Production and property of β-lactamases in Streptomyces. Antimicrob. Agents Chemother. 1975, 8, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Abraham, E.P.; Waley, S.G. Beta-Lactamases; Hamilton-Miller, J.M.T., Smith, J.T., Eds.; Academic Press: London, UK, 1979; p. 338. [Google Scholar]
- Kushner, D.J.; Breuil, C. Penicillinase (β-lactamase) formation by blue-green algae. Arch. Microbiol. 1977, 112, 219–223. [Google Scholar] [CrossRef] [PubMed]
- Urbach, C.; Fastrez, J.; Soumillion, P. A new family of cyanobacterial penicillin-binding proteins. A missing link in the evolution of class A β-lactamases. J. Biol. Chem. 2008, 283, 32516–32526. [Google Scholar] [CrossRef] [PubMed]
- Podzelinska, K.; He, S.-M.; Wathier, M.; Yakunin, A.; Proudfoot, M.; Hove-Jensen, B.; Zechel, D.L.; Jia, Z. Structure of PhnP, a phosphodiesterase of the carbon-phosphorus lyase pathway for phosphonate degradation. J. Biol. Chem. 2009, 284, 17216–17226. [Google Scholar] [CrossRef] [PubMed]
- Sattler, S.A.; Wang, X.; Lewis, K.M.; DeHan, P.J.; Park, C.M.; Xin, Y.; Liu, H.; Xian, M.; Xun, L.; Kang, C. Characterizations of two bacterial persulfide dioxygenases of the metallo-β-lactamase superfamily. J. Biol. Chem. 2015, 290, 18914–18923. [Google Scholar] [CrossRef] [PubMed]
- Daiyasu, H.; Osaka, K.; Ishino, Y.; Toh, H. Expansion of the zinc metallo-hydrolase family of the β-lactamase fold. FEBS Lett. 2001, 503, 1–6. [Google Scholar] [CrossRef]
- Dominski, Z. Nucleases of the metallo-β-lactamase family and their role in DNA and RNA metabolism. Crit. Rev. Biochem. Mol. Biol. 2007, 42, 67–93. [Google Scholar] [CrossRef] [PubMed]
- Vogel, A.; Schilling, O.; Späth, B.; Marchfelder, A. The tRNase Z family of proteins: Physiological functions, substrate specificity and structural properties. Biol. Chem. 2005, 386, 1253–1264. [Google Scholar] [CrossRef] [PubMed]
- Lapidus, A.; Clum, A.; Labutti, K.; Kaluzhnaya, M.G.; Lim, S.; Beck, D.A.; del Glavina Rio, T.; Nolan, M.; Mavromatis, K.; Huntemann, M.; et al. Genomes of three methylotrophs from a single niche reveal the genetic and metabolic divergence of the Methylophilaceae. J. Bacteriol. 2011, 193, 3757–3764. [Google Scholar] [CrossRef] [PubMed]
- Limphong, P.; Nimako, G.; Thomas, P.W.; Fast, W.; Makaroff, C.A.; Crowder, M.W. Arabidopsis thaliana mitochondrial glyoxalase 2-1 exhibits β-lactamase activity. Biochemistry 2009, 48, 8491–8493. [Google Scholar] [CrossRef] [PubMed]
- Florea, L.; Di Francesco, V.; Miller, J.; Turner, R.; Yao, A.; Harris, M.; Walenz, B.; Mobarry, C.; Merkulov, G.V.; Charlab, R.; et al. Gene and alternative splicing annotation with AIR. Genome Res. 2005, 15, 54–66. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.S.; Southan, C.; Ellington, K.; Campbell, D.; Tew, D.G.; Debouck, C. Identification, genomic organization, and mRNA expression of LACTB, encoding a serine β-lactamase-like protein with an amino-terminal transmembrane domain. Genomics 2001, 78, 12–14. [Google Scholar] [CrossRef] [PubMed]
- Toth, M.; Smith, C.; Frase, H.; Mobashery, S.; Vakulenko, S. An antibiotic-resistance enzyme from a deep-sea bacterium. J. Am. Chem. Soc. 2010, 132, 816–823. [Google Scholar] [CrossRef] [PubMed]
- Michaux, C.; Massant, J.; Kerff, F.; Frère, J.-M.; Docquier, J.D.; Vandenberghe, I.; Samyn, B.; Pierrard, A.; Feller, G.; Charlier, P.; et al. Crystal structure of a cold-adapted class C β-lactamase. FEBS J. 2008, 275, 1687–1697. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-B.; Joo, S.; Yoon, S.; Kim, S.; Moon, J.; Ryu, Y.; Kim, K.K.; Kim, T.D. Purification, crystallization and preliminary crystallographic analysis of Est-Y29: A novel oligomeric β-lactamase. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2009, 65, 310–312. [Google Scholar] [CrossRef] [PubMed]
- Allen, H.K.; Moe, L.; Rodbumrer, J.; Gaarder, A.; Handelsman, J. Functional metagenomics reveals diverse β-lactamases in a remote Alaskan soil. ISME J. 2009, 3, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Donato, J.J.; Moe, L.A.; Converse, B.J.; Smart, K.D.; Berklein, F.; McManus, P.S.; Handelsman, J. Metagenomic analysis of apple orchard soil reveals antibiotic resistance genes encoding predicted bifunctional proteins. Appl. Environ. Microbiol. 2010, 76, 4396–4401. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhang, L.; Zhang, H.; He, Z.G. Characterization of a bifunctional β-lactamase/ribonuclease and its interaction with a chaperone-like protein in the pathogen Mycobacterium tuberculosis H37Rv. Biochemistry 2011, 76, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Kumar, A.; Arya, S.; Gupta, U.D.; Singh, K.; Kaur, J. Characterization of a novel esterase Rv1497 of Mycobacterium tuberculosis H37Rv demonstrating β-lactamase activity. Enzyme Microb. Technol. 2016, 82, 180–190. [Google Scholar] [CrossRef] [PubMed]
- Ambler, R.P. The structure of beta-lactamases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1980, 289, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Bush, K.; Jacoby, G.A. Updated functional classification of β-lactamases. Antimicrob. Agents Chemother. 2010, 54, 969–976. [Google Scholar] [CrossRef] [PubMed]
- Jaurin, B.; Grundström, T. ampC cephalosporinase of Escherichia coli K-12 has a different evolutionary origin from that of beta-lactamases of the penicillinase type. Proc. Natl. Acad. Sci. USA 1981, 78, 4897–4901. [Google Scholar] [CrossRef] [PubMed]
- Ouellette, M.; Bissonnette, L.; Roy, P.H. Precise insertion of antibiotic resistance determinants into Tn21-like transposons: Nucleotide sequence of the OXA-1 β-lactamase gene. Proc. Natl. Acad. Sci. USA 1987, 84, 7378–7382. [Google Scholar] [CrossRef] [PubMed]
- Palzkill, T. Metallo-β-lactamase structure and function. Ann. N. Y. Acad. Sci. 2013, 1277, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Frère, J.M.; Galleni, M.; Bush, K.; Dideberg, O. Is it necessary to change the classification of β-lactamases? J. Antimicrob. Chemother. 2005, 55, 1051–1053. [Google Scholar] [CrossRef] [PubMed]
- Bush, K.; Jacoby, G.A.; Medeiros, A.A. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 1995, 39, 1211–1233. [Google Scholar] [CrossRef] [PubMed]
- Bradford, P.A. Extended-spectrum β-lactamases in the 21st century: Characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 2001, 14, 933–951. [Google Scholar] [CrossRef] [PubMed]
- Pettinati, I.; Brem, J.; Lee, S.Y.; McHugh, P.J.; Schofield, C.J. The chemical biology of human metallo-β-lactamase fold proteins. Trends Biochem. Sci. 2016, 41, 338–355. [Google Scholar] [CrossRef] [PubMed]
- Bebrone, C. Metallo-β-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem. Pharmacol. 2007, 74, 1686–1701. [Google Scholar] [CrossRef] [PubMed]
- Garau, G.; Di Guilmi, A.M.; Hall, B.G. Structure-based phylogeny of the metallo-β-lactamases. Antimicrob. Agents Chemother. 2005, 49, 2778–2784. [Google Scholar] [CrossRef] [PubMed]
- Walsh, T.R.; Toleman, M.A.; Poirel, L.; Nordmann, P. Metallo-β-lactamases: The quiet before the storm? Clin. Microbiol. Rev. 2005, 18, 306–325. [Google Scholar] [CrossRef] [PubMed]
- Ogawara, H. Phylogenetic tree and sequence similarity of β-lactamases. Mol. Phylogenet. Evol. 1993, 2, 97–111. [Google Scholar] [CrossRef] [PubMed]
- Ogawara, H. Self-resistance to β-lactam antibiotics in Streptomyces (in Japanese). Bull. Meiji Pharmaceut. Univ. 2014, 43, 1–20. [Google Scholar]
- Ogawara, H. Molecular phylogenetics of β-lactamases in Actinobacteria. Bull. Meiji Pharmaceut. Univ. 2013, 42, 1–18. [Google Scholar]
- BioProject. Available online: http://www.ncbi.nlm.nih.gov/guide/all/ (accessed on 10 September 2011).
- Stackebrandt, E.; Rainey, F.A.; Ward-Rainey, N.L. Proposal for a New Hierarchic Classification System, Actinobacteria classis nov. Int. J. Syst. Bacteriol. 1997, 47, 479–491. [Google Scholar] [CrossRef]
- Ogawara, H.; Kawamura, N.; Kudo, T.; Suzuki, K.I.; Nakase, T. Distribution of β-lactamases in actinomycetes. Antimicrob. Agents Chemother. 1999, 43, 3014–3017. [Google Scholar] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Notredame, C.; Higgins, D.G.; Heringa, J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000, 302, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Galleni, M.; Lamotte-Brasseur, J.; Rossolini, G.M.; Spencer, J.; Dideberg, O.; Frère, J.-M. Metallo-β-lactamases Working Group. Standard numbering scheme for class B beta-lactamases. Antimicrob. Agents Chemother. 2001, 45, 660–663. [Google Scholar] [CrossRef] [PubMed]
- Lobkovsky, E.; Moews, P.C.; Liu, H.; Zhao, H.; Frere, J.-M.; Knox, J.R. Evolution of an enzyme activity: Crystallographic structure at 2-A resolution of cephalosporinase from the ampC gene of Enterobacter cloacae P99 and comparison with a class A penicillinase. Proc. Natl. Acad. Sci. USA 1993, 90, 11257–11261. [Google Scholar] [CrossRef] [PubMed]
- Goffin, C.; Ghuysen, J.M. Biochemistry and comparative genomics of SxxK superfamily acyltransferases offer a clue to the mycobacterial paradox: Presence of penicillin-susceptible target proteins versus lack of efficiency of penicillin as therapeutic agent. Microbiol. Mol. Biol. Rev. 2002, 66, 702–738. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- β-Lactamase Classification and Amino Acid Sequences for TEM, SHV and OXA Extended-Spectrum and Inhibitor Resistant Enzymes. Available online: http://www.lahey.org/studies/ (accessed on 20 March 2016).
- Protdist-Program to Compute Distance Matrix from Protein Sequences. Available online: http://evolution.genetics.washington.edu/phylip/doc/protdist.html (accessed on 20 March 2016).
- Urabe, H.; Toyama, K.; Ogawara, H. Cloning from Streptomyces cellulosae of the gene encoding beta-lactamase, a blue-dextran binding protein. J. Antibiot. 1990, 43, 1483–1488. [Google Scholar] [CrossRef] [PubMed]
- Ogawara, H. Sequence of a gene encoding β-lactamase from Streptomyces cellulosae. Gene 1993, 124, 111–114. [Google Scholar] [CrossRef]
- Jensen, S.E.; Paradkar, A.S.; Mosher, R.H.; Anders, C.; Beatty, P.H.; Brumlik, M.J.; Griffin, A.; Barton, B. Five additional genes are involved in clavulanic acid biosynthesis in Streptomyces clavuligerus. Antimicrob. Agents Chemother. 2004, 48, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Khaleeli, N.; Townsend, C.A. Expansion of the clavulanic acid gene cluster: Identification and in vivo functional analysis of three new genes required for biosynthesis of clavulanic acid by Streptomyces clavuligerus. J. Bacteriol. 2000, 182, 4087–4095. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, G.A. AmpC β-lactamases. Clin. Microbiol. Rev. 2009, 22, 161–182. [Google Scholar] [CrossRef] [PubMed]
- Macheboeuf, P.; Contreras-Martel, C.; Job, V.; Dideberg, O.; Dessen, A. Penicillin binding proteins: Key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol. Rev. 2006, 30, 673–691. [Google Scholar] [CrossRef] [PubMed]
- Typas, A.; Banzhaf, M.; Gross, C.A.; Vollmer, W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 2012, 10, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Van Heijenoort, J. Lipid intermediates in the biosynthesis of bacterial peptidoglycan. Microbiol. Mol. Biol. Rev. 2007, 71, 620–635. [Google Scholar] [CrossRef] [PubMed]
- Suginaka, H.; Blumberg, P.M.; Strominger, J.L. Multiple penicillin-binding components in Bacillus subtilis, Bacillus cereus, Staphylococcus aureus, and Escherichia coli. J. Biol. Chem. 1972, 247, 5279–5288. [Google Scholar] [PubMed]
- Spratt, B.G. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc. Natl. Acad. Sci. USA 1975, 72, 2999–3003. [Google Scholar] [CrossRef] [PubMed]
- Pratt, R.F. Substrate specificity of bacterial DD-peptidases (penicillin-binding proteins). Cell. Mol. Life Sci. 2008, 65, 2138–2155. [Google Scholar] [CrossRef] [PubMed]
- Sauvage, E.; Kerff, F.; Terrak, M.; Ayala, J.A.; Charlier, P. The penicillin-binding proteins: Structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 2008, 32, 234–258. [Google Scholar] [CrossRef] [PubMed]
- Ogawara, H. Penicillin-binding proteins in Actinobacteria. J. Antib. 2015, 68, 223–245. [Google Scholar] [CrossRef] [PubMed]
- Goffin, C.; Ghuysen, J.M. Multimodular penicillin-binding proteins: An enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 1998, 62, 1079–1093. [Google Scholar] [PubMed]
- Ishida, K.; Hung, T.V.; Liou, K.; Lee, H.C.; Shin, C.H.; Sohng, J.K. Characterization of pbpA and pbp2 encoding penicillin-binding proteins located on the downstream of clavulanic acid gene cluster in Streptomyces clavuligerus. Biotechnol. Lett. 2006, 28, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Ogawara, H.; Horikawa, S. Penicillin-binding proteins of Streptomyces cacaoi, Streptomyces olivaceus, and Streptomyces clavuligerus. Antimicrob. Agents Chemother. 1980, 17, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Paradkar, A.S.; Aidoo, K.A.; Wong, A.; Jensen, S.E. Molecular analysis of a β-lactam resistance gene encoded within the cephamycin gene cluster of Streptomyces clavuligerus. J. Bacteriol. 1996, 178, 6266–6274. [Google Scholar] [PubMed]
- Hanks, S.K.; Hunter, T. Protein kinases 6. The eukaryotic protein kinase superfamily: Kinase (catalytic) domain structure and classification. FASEB J. 1995, 9, 576–596. [Google Scholar] [PubMed]
- Hunter, T. Tyrosine phosphorylation: Thirty years and counting. Curr Opin. Cell Biol. 2009, 21, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Dorado, J.; Inouye, S.; Inouye, M. A gene encoding a protein serine/threonine kinase is required for normal development of M. xanthus, a gram-negative bacterium. Cell 1991, 67, 995–1006. [Google Scholar] [CrossRef]
- Pereira, S.F.; Goss, L.; Dworkin, J. Eukaryote-like serine/threonine kinases and phosphatases in bacteria. Microbiol. Mol. Biol. Rev. 2011, 75, 192–212. [Google Scholar] [CrossRef] [PubMed]
- Manuse, S.; Fleurie, A.; Zucchini, L.; Lesterlin, C.; Grangeasse, C. Role of eukaryotic-like serine/threonine kinases in bacterial cell division and morphogenesis. FEMS Microbiol. Rev. 2016, 40, 41–56. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, L.E.; Koonin, E.V.; Zhulin, I.B. One-component systems dominate signal transduction in prokaryotes. Trends. Microbiol. 2005, 13, 52–56. [Google Scholar] [CrossRef] [PubMed]
- Yeats, C.; Finn, R.D.; Bateman, A. The PASTA domain: A β-lactam-binding domain. Trends Biochem. Sci. 2002, 27, 438–440. [Google Scholar] [CrossRef]
- Gordon, E.; Mouz, N.; Duée, E.; Dideberg, O. The crystal structure of the penicillin-binding protein 2x from Streptococcus pneumoniae and its acyl-enzyme form: Implication in drug resistance. J. Mol. Biol. 2000, 299, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Calvanese, L.; Falcigno, L.; Maglione, C.; Marasco, D.; Ruggiero, A.; Squeglia, F.; Berisio, R.; D’Auria, G. Structural and binding properties of the PASTA domain of PonA2, a key penicillin binding protein from Mycobacterium tuberculosis. Biopolymers 2014, 101, 712–719. [Google Scholar] [CrossRef] [PubMed]
- Ruggiero, A.; De Simone, P.; Smaldone, G.; Squeglia, F.; Berisio, R. Bacterial cell division regulation by Ser/Thr kinases: A structural perspective. Curr. Protein Pept. Sci. 2012, 13, 756–766. [Google Scholar] [CrossRef] [PubMed]
- Peters, K.; Schweizer, I.; Beilharz, K.; Stahlmann, C.; Veening, J.W.; Hakenbeck, R.; Denapaite, D. Streptococcus pneumoniae PBP2x mid-cell localization requires the C-terminal PASTA domains and is essential for cell shape maintenance. Mol. Microbiol. 2014, 92, 733–755. [Google Scholar] [CrossRef] [PubMed]
- Ogawara, H. Distribution of PASTA domains in penicillin-binding proteins and serine/threonine kinases of Actinobacteria. J. Antibiot. 2016. [Google Scholar] [CrossRef] [PubMed]
- McCormick, J.R.; Flärdh, K. Signals and regulators that govern Streptomyces development. FEMS Micribiol. Rev. 2012, 36, 206–231. [Google Scholar] [CrossRef] [PubMed]
- Ladwig, N.; Franz-Wachtel, M.; Hezel, F.; Soufi, B.; Macek, B.; Wohlleben, W.; Muth, G. Control of morphological differentiation of Streptomyces coelicolor A3(2) by phosphorylation of MreC and PBP2. PLoS ONE 2015, 10, e0125425. [Google Scholar] [CrossRef] [PubMed]
- Jones, G.; Del Sol, R.; Dudley, E.; Dyson, P. Forkhead-associated proteins genetically linked to the serine/threonine kinase PknB regulate carbon flux towards antibiotic biosynthesis in Streptomyces coelicolor. Microb. Biotechnol. 2011, 4, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Llarena, F.; Martín, J.F.; Galleni, M.; Coque, J.J.; Fuente, J.L.; Frère, J.M.; Liras, P. The bla gene of the cephamycin cluster of Streptomyces clavuligerus encodes a class A β-lactamase of low enzymatic activity. J. Bacteriol. 1997, 179, 6028–6034. [Google Scholar]
- Streptomyces clavuligerus ATCC 27064 chromosome, whole genome shotgun sequence. Available online: http://www.ncbi.nlm.nih.gov/nuccore/294323433?report=graph (accessed on 20 March 2016).
- Streptomyces cattleya NRRL 8057 = DSM 46488, complete genome. Available online: http://www.ncbi.nlm.nih.gov/nuccore/357397620?report=graph (accessed on 20 March 2016).
- Jensen, S.E. Biosynthesis of clavam metabolites. J. Ind. Microbiol. Biotechnol. 2012, 39, 1407–1419. [Google Scholar] [CrossRef] [PubMed]
- Paradkar, A. Clavulanic acid production by Streptomyces clavuligerus: Biogenesis, regulation and strain improvement. J. Antibiot. 2013, 66, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Mellado, E.; Lorenzana, L.M.; Rodríguez-Sáiz, M.; Díez, B.; Liras, P.; Barredo, J.L. The clavulanic acid biosynthetic cluster of Streptomyces clavuligerus: Genetic organization of the region upstream of the car gene. Microbiology 2002, 148, 1427–1438. [Google Scholar]
- Coque, J.J.; Liras, P.; Martín, J.F. Genes for a β-lactamase, a penicillin-binding protein and a transmembrane protein are clustered with the cephamycin biosynthetic genes in Nocardia lactamdurans. EMBO J. 1993, 12, 631–639. [Google Scholar] [PubMed]
- Kimura, H.; Izawa, M.; Sumino, Y. Molecular analysis of the gene cluster involved in cephalosporin biosynthesis from Lysobacter lactamgenus YK90. Appl. Microbiol. Biotechnol. 1996, 44, 589–596. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.F. Clusters of genes for the biosynthesis of antibiotics: Regulatory genes and overproduction of pharmaceuticals. J. Ind. Microbiol. 1992, 9, 73–90. [Google Scholar] [CrossRef] [PubMed]
- Brakhage, A.A.; Al-Abdallah, Q.; Tüncher, A.; Spröte, P. Evolution of β-lactam biosynthesis genes and recruitment of trans-acting factors. Phytochemistry 2005, 66, 1200–1210. [Google Scholar] [CrossRef] [PubMed]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Klenk, H.P.; Clément, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, Physiology, and Natural Products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80. [Google Scholar] [CrossRef] [PubMed]
- Alanis, A.J. Resistance to antibiotics: Are we in the post-antibiotic era? Arch. Med. Res. 2005, 36, 697–705. [Google Scholar] [CrossRef] [PubMed]
- Cooper, M.A.; Shlaes, D. Fix the antibiotics pipeline. Nature 2014, 472. [Google Scholar] [CrossRef] [PubMed]
- Rokem, J.S.; Lantz, A.E.; Nielsen, J. Systems biology of antibiotic production by microorganisms. Nat. Prod. Rep. 2007, 24, 1262–1287. [Google Scholar] [CrossRef] [PubMed]
- Bull, A.T.; Stach, J.E. Marine actinobacteria: New opportunities for natural product search and discovery. Trends Microbiol. 2007, 15, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Demain, A.L. Importance of microbial natural products and the need to revitalize their discovery. J. Ind. Microbiol. Biotechnol. 2014, 41, 185–201. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, B.O.; Van Lanen, S.G.; Baltz, R.H. Microbial genome mining for accelerated natural products discovery: Is a renaissance in the making? J. Ind. Microbiol. Biotechnol. 2014, 41, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Katz, L.; Baltz, R.H. Natural product discovery: Past, present, and future. J. Ind. Microbiol. Biotechnol. 2016, 43, 155–176. [Google Scholar] [CrossRef] [PubMed]
- Baltz, R.H. Genetic manipulation of secondary metabolite biosynthesis for improved production in Streptomyces and other actinomycetes. J. Ind. Microbiol. Biotechnol. 2016, 43, 343–370. [Google Scholar] [CrossRef] [PubMed]





| Bacteria | Prefix | Class A | Class B | Class C | Class D |
|---|---|---|---|---|---|
| Streptomyces albulus CCRC11814 | K530 | 01315, 35835 * | 18626, 25018, 25240 | 02767, 10843, 40281 | none |
| Streptomyces cattleya NRRL805 | SCAT | 0807, 1418, 4581, 5692 | 0258, 1598, 4145, 4517, 4822 | 1363, 1488, 5541, 5637 | none |
| Streptomyces clavuligerus ATCC27064 | SSCG | 00130, 00160, 01467 | 00563, 00791, 02181, 04638, 05032, 05130, 05295, 05571 | 01647, 03303, 03668, 04090, 04120, 04299 | none |
| Streptomyces collinus Tu365 | B446 | 02285, 33010 | 02000, 02210, 05525, 13210, 29385, 33085, 33290 | 01315, 03940, 11745, 11965, 12445, 13750, 19000, 22405, 33975 | none |
| Streptomyces davawensis JCM4913 | BN159 | 5790 | 0265, 0293, 4295, 5380, 5740, 6071, 7079,7978, 8291 | 1677, 1822, 3452, 3645, 4558, 5112, 5953, 5970, 6145, 7932, 8163 | none |
| Streptomyces hygroscopicus subsp. jinggangensis 5008 | SHJG | 8264, 8266 | 0444, 1156, 1422, 2523, 3996, 4046, 8335 | 0734, 0750, 1734, 2053, 2219, 2828, 3769, 3873, 4357, 4919, 5160, 8639 | none |
| Streptomyces rimosus subsp. rimosus ATCC10970 | SRIM | 07618, 10531 | 08043, 16115, 16845, 19739, 21549, 23716, 26422 | 00410, 02396, 04151, 04583, 10671, 11591, 15850, 24376, 28551, 31085, 32256, 37671 | none |
| Streptomyces scabiei 87.22 | SCAB | 38731 | 25181 | 26931, 46171, 52151, 55861 | none |
| Streptomyces sp. PAMC26508 | F750 | 2387, 6336, 6823 | 0075, 2374 | 0558, 2251, 3609, 5886 | none |
| Streptomyces tsukubaensis NRRL18488 | STSU | 15167, 15177, 32330 | 07338, 08188, 24671, 31620 | 00260, 00675, 00895, 15744, 17713, 20672, 23056, 33145, | none |
| Streptomyces venezuelae ATCC 10712 | SVEN | 0089 | 0710, 1591, 2335, 2660, 7291 | 0306, 0457, 1972, 2198, 3165, 3520, 3691, 5322, 6030, 6060, 7092 | none |
| Streptomyces viridochromogenes DSM 40736 | SSQG | 01127 | 02569, 02955, 05776, 05938, 06409 | 00225, 00387, 00417, 02214, 02353, 02491, 02679, 03888, 04096, 04848, 06609 | none |
| β-Lactamase | SXXK73 | SDN132 | RW/YE166 | D179 | KT/SG236 |
|---|---|---|---|---|---|
| YP_004910945_SCAT_1418 | X | X | X | ○ | X |
| YP_004914072_SCAT_4581 | X | X | X | ○ | X |
| ZP_05002833_SSCG_00160 | ○ | ○ | △ | ○ | ○ |
| YP_007863245_F750_6336 | ○ | ○ | △ | ○ | ○ |
| YP_006249403_SHJG_8264 | ○ | ○ | X | E | ○ |
| YP_008384100_B446_02285 | ○ | ○ | ○ | X | ○ |
| YP_008390217_B446_33010 | ○ | ○ | ○ | X | ○ |
| EIF91547_STSU_15177 | ○ | ○ | △ | X | ○ |
| YP_004910346_SCAT_0807 | ○ | ○ | △ | X | ○ |
| YP_004915182_SCAT_5692 | ○ | ○ | HYE | ○ | ○ |
| ZP_05002803_SSCG_00130 | ○ | ○ | QYE | ○ | ○ |
| WP_037636700_K530_RS35835 | ○ | ○ | ○ | ○ | ○ |
| EIF91545_STSU_15167 | ○ | ○ | ○ | ○ | ○ |
| ELQ83985_SRIM_07318 | X | X | ○ | ○ | ○ |
| WP_020929156_K530_RS01315 | ○ | ○ | ○ | ○ | ○ |
| YP_006249405_SHJG_8266 | ○ | ○ | ○ | ○ | ○ |
| ELQ83341_SRIM_10531 | ○ | ○ | ○ | ○ | ○ |
| YP_003489505_SCAB_38731 | ○ | ○ | ○ | ○ | ○ |
| YP_007524296_BN159_5790 | ○ | ○ | ○ | ○ | ○ |
| ZP_05003887_SSCG_01467 | ○ | ○ | ○ | ○ | ○ |
| YP_006875636_SVEN_0089 | ○ | ○ | ○ | ○ | ○ |
| ZP_07302240_SSQG_01127 | ○ | ○ | ○ | ○ | ○ |
| YP_007859362_F750_2387 | X | X | ○ | E | X |
| EIF88144_STSU_32330 | ○ | ○ | X | X | ○ |
| YP_007863731_F750_6823 | ○ | ○ | ○ | ○ | ○ |
| Class A β-lactamases | S70XXK73 | S130DN132 | R164W/YE166 | D179 | K234T/SG236 | |
|---|---|---|---|---|---|---|
| Class B β-lactamases: | H116XHXD120 | G123 | H196 | H263 | ||
| Class C β-lactamases | S64XXK67 | Y150XN152 | K315TG317S/A318 | |||
| Class D β-lactamases | S67TFK70 | S115XV | Y142G | W154 | K205TG | W221XXG |
| Class A PBPs | S465XXK468 | S524XN526 | K716TG718 | |||
| Class B PBPs: | S307XXK310 | S356XN358 | S356XN358 |
| β-Lactamase | HXHXD120 | G123 | H196 | H263 |
|---|---|---|---|---|
| EOT02442_K530_18626 | ○ | D | X | X |
| YP_006878205_SVEN_2660 | ○ | D | X | X |
| YP_007523886_BN159_5380 | ○ | D | X | X |
| ZP_07304068_SSQG_02955 | ○ | D | X | X |
| ZP_05007878_SSCG_05130 | △ | ○ | ○ | ○ |
| YP_006249473_SHJG_8335 | △ | ○ | ○ | ○ |
| YP_004913644_SCAT_4145 | △ | ○ | ○ | ○ |
| ELQ82101_SRIM_16845 | △ | ○ | ○ | ○ |
| ZP_07307522_SSQG_06409 | ○ | E | X | R |
| AAA22276_Bacillus_cereus | ○ | ○ | ○ | ○ |
| YP_008384085_B446_02210 | ○ | ○ | X | ○ |
| YP_008390232_B446_33085 | ○ | ○ | X | ○ |
| ZP_05004854_SSCG_02181 | ○ | ○ | ○ | ○ |
| YP_004909803_SCAT_0258 | ○ | ○ | ○ | ○ |
| ELQ82279_SRIM_16115 | ○ | ○ | ○ | ○ |
| ZP_05007361_SSCG_04638 | ○ | L | ○ | R |
| YP_007524577_BN159_6071 | ○ | L | ○ | ○ |
| YP_007857070_F750_0075 | ○ | Y | ○ | ○ |
| ZP_05003465_SSCG_00791 | ○ | ○ | E | ○ |
| ZP_05007967_SSCG_05295 | ○ | ○ | ○ | ○ |
| ELQ83811_SRIM_08043 | ○ | ○ | ○ | ○ |
| YP_004911125_SCAT_1598 | ○ | ○ | ○ | ○ |
| ZP_07303682_SSQG_02569 | ○ | ○ | ○ | ○ |
| YP_007524246_BN159_5740 | ○ | ○ | ○ | ○ |
| YP_008386271_B446_13210 | ○ | ○ | ○ | ○ |
| YP_006245191_SHJG_4046 | ○ | ○ | ○ | ○ |
| YP_007859349_F750_2374 | ○ | ○ | ○ | ○ |
| YP_006877880_SVEN_2335 | ○ | ○ | ○ | ○ |
| ZP_05007774_SSCG_05032 | ○ | ○ | ○ | ○ |
| EIF89665_STSU_24671 | ○ | ○ | ○ | ○ |
| YP_007518799_BN159_0293 | ○ | N | ○ | ○ |
| YP_006242571_SHJG_1422 | ○ | W | ○ | ○ |
| ELQ80751_SRIM_23716 | ○ | W | ○ | ○ |
| YP_006242306_SHJG_1156 | ○ | W | ○ | X |
| ELQ81190_SRIM_21549 | ○ | W | ○ | T |
| YP_006241594_SHJG_0444 | ○ | W | ○ | D |
| YP_006245141_SHJG_3996 | ○ | W | ○ | X |
| YP_008384043_B446_02000 | ○ | S | ○ | ○ |
| YP_008389494_B446_29385 | ○ | ○ | G | A |
| ZP_07307051_SSQG_05938 | ○ | ○ | G | A |
| YP_004914313_SCAT_4822 | ○ | ○ | G | A |
| YP_004914015_SCAT_4517 | ○ | ○ | G | T |
| YP_003488184_SCAB_25181 | ○ | A | G | S |
| ZP_07306889_SSQG_05776 | ○ | ○ | G | S |
| ELQ81507_SRIM_19739 | ○ | ○ | G | S |
| ZP_05008219_SSCG_05571 | ○ | ○ | G | S |
| EIF92931_STSU_08188 | ○ | ○ | G | S |
| YP_007526484_BN159_7978 | ○ | ○ | ○ | ○ |
| YP_007526796_BN159_8291 | ○ | ○ | ○ | ○ |
| EOT01146_K530_25240 | ○ | ○ | X | ○ |
| EIF93163_STSU_07338 | ○ | ○ | X | ○ |
| ELQ80212_SRIM_26422 | ○ | S | ○ | ○ |
| YP_008384742_B446_05525 | ○ | ○ | ○ | ○ |
| YP_006243671_SHJG_2523 | ○ | ○ | ○ | ○ |
| EIF88273_STSU_31620 | ○ | A | ○ | ○ |
| YP_006876256_SVEN_0710 | ○ | A | ○ | ○ |
| β-Lactamase | SXXK70 | YXN152 | KTG317 |
|---|---|---|---|
| YP_006249777_SHJG_8639 | ○ | X | X |
| YP_006243975_SHJG_2828 | X | X | ○ |
| YP_006242882_SHJG_1734 | ○ | ○ | X |
| AGS69568_B446_13750 | ○ | ○ | ○ |
| YP_006879065_SVEN_3520 | ○ | ○ | HTG |
| ZP_05006222_SSCG_03668 | X | X | X |
| AmpC_Escherichia_coli | ○ | ○ | ○ |
| ELQ82327_SRIM_15850 | ○ | YSS | HDG |
| ELQ79325_SRIM_31085 | X | X | X |
| YP_003491153_SCAB_55861 | ○ | YSG | HDG |
| EIF89980_STSU_23056_391 | ○ | ○ | ○ |
| YP_006878710_SVEN_3165 | ○ | X | RAG |
| YP_007520183_BN159_1677 | ○ | ○ | HTG |
| YP_007526438_BN159_7932 | X | YSH | X |
| YP_006882637_SVEN_7092 | △ | YSH | X |
| YP_003488355_SCAB_26931 | ○ | ○ | HDG |
| ELQ85060_SRIM_02396 | ○ | ○ | ○ |
| YP_006880867_SVEN_5322 | ○ | ○ | ○ |
| ZP_07305209_SSQG_04096 | ○ | ○ | ○ |
| YP_004915128_SCAT_5637 | ○ | YNG | ○ |
| YP_006241884_SHJG_0734 | ○ | YNG | ○ |
| YP_006881575_SVEN_6030 | ○ | ○ | HNG |
| YP_008384429_B446_03940 | ○ | ○ | HNG |
| YP_006243201_SHJG_2053 | ○ | ○ | HNG |
| EIF94426_STSU_00675 | ○ | ○ | HNG |
| ZP_05005976_SSCG_03303 | ○ | ○ | X |
| YP_006246060_SHJG_4919 | ○ | YRG | ○ |
| YP_006877518_SVEN_1972 | ○ | ○ | HGG |
| ELQ78022_SRIM_37671 | ○ | ○ | HGG |
| YP_008388104_B446_22405 | ○ | CSN | RSG |
| ZP_07303604_SSQG_02491 | X | ○ | HDG |
| ELQ79790_SRIM_28551 | ○ | ○ | HNG |
| YP_007522151_BN159_3645 | ○ | ○ | HSG |
| ZP_07305961_SSQG_04848 | ○ | ○ | HNG |
| ZP_05006696_SSCG_04120 | ○ | ○ | HSG |
| ZP_07301500_SSQG_00387 | ○ | ○ | KSG |
| ZP_07303792_SSQG_02679 | ○ | ○ | HGG |
| EIF87980_STSU_33145 | ○ | YHA | X |
| YP_007521958_BN159_3452 | ○ | YHS | X |
| YP_007524459_BN159_5953 | ○ | YHA | ○ |
| ELQ84672_SRIM_04151 | ○ | YHA | HGG |
| YP_007523618_BN159_5112 | ○ | YHA | HPG/RGG |
| YP_006245502_SHJG_4357 | ○ | YHG | HTG/RGG |
| ZP_05006916_SSCG_04299 | ○ | YHG | HTG/RGG |
| EIF91419_STSU_15744 | ○ | YHG | HTG/RGG |
| YP_004915032_SCAT_5541 | ○ | YSV | X |
| YP_007520328_BN159_1822 | ○ | YSV | X |
| ZP_07307722_SSQG_06609 | ○ | YSV | ○ |
| EOT05599_K530_02767 | ○ | YNV | X |
| EIF94507_STSU_00260 | ○ | YHV | X |
| EIF90453_STSU_20672 | ○ | YNT | ○ |
| YP_008383910_B446_01315 | ○ | YDT | RVG |
| YP_008390406_B446_33975 | ○ | YDT | RVG |
| YP_006241900_SHJG_0750 | ○ | YGT | RYG |
| ELQ85328_SRIM_00410 | ○ | ○ | HSG |
| ZP_05006666_SSCG_04090 | ○ | ○ | HSG |
| EIF91041_STSU_17713 | ○ | ○ | HTG |
| YP_003490218_SCAB_46171 | ○ | ○ | HGG |
| ZP_07305001_SSQG_03888 | ○ | ○ | HTG |
| YP_006246301_SHJG_5160 | ○ | ○ | HSG |
| YP_007523064_BN159_4558 | ○ | ○ | HSG |
| YP_008387425_B446_19000 | ○ | ○ | HSG |
| YP_006879236_SVEN_3691 | ○ | ○ | HTG |
| YP_007860556_F750_3609 | ○ | ○ | HSG |
| YP_003490794_SCAB_52151 | ○ | ○ | ○ |
| ELQ83369_SRIM_10671 | ○ | ○ | HNG |
| YP_008385982_B446_11745 | ○ | ○ | HDG |
| ZP_07301338_SSQG_00225 | X | ○ | HSG |
| ELQ79081_SRIM_32256 | ○ | ○ | HSG |
| YP_007862798_F750_5886 | ○ | ○ | HSG |
| ELQ80643_SRIM_24376 | ○ | ○ | X |
| YP_006876003_SVEN_0457 | ○ | YTD | HFG |
| YP_006881605_SVEN_6060 | ○ | YTD | HYG |
| YP_004911015_SCAT_1488 | ○ | ○ | HFG |
| ZP_05004320_SSCG_01647 | ○ | ○ | HFG |
| YP_006877743_SVEN_2198 | ○ | ○ | HFG |
| EOT04000_K530_10843 | ○ | ○ | HFG |
| ELQ84558_SRIM_04583 | ○ | ○ | HFG |
| YP_007859229_F750_2251 | ○ | ○ | HFG |
| YP_007524476_BN159_5970 | ○ | ○ | HFG |
| ZP_07303466_SSQG_02353 | ○ | ○ | HFG |
| YP_006245018_SHJG_3873 | ○ | ○ | HFG |
| YP_008386122_B446_12445 | ○ | ○ | HFG |
| YP_007857553_F750_0558 | ○ | YSD | HTG |
| YP_006243367_SHJG_2219 | ○ | YSD | HTG |
| YP_007526668_BN159_8163 | ○ | YSD | HTG |
| ZP_07301530_SSQG_00417 | ○ | YSD | HTG |
| EIF94367_STSU_00895 | ○ | YSD | HTG |
| YP_006875852_SVEN_0306 | ○ | YSD | HTG |
| YP_004910890_SCAT_1363 | ○ | YSD | HTG |
| ELQ83067_SRIM_11591 | ○ | YSD | HTG |
| ZP_07303327_SSQG_02214 | ○ | YSD | HTG |
| YP_007524651_BN159_6145 | ○ | YSD | HTG |
| YP_008386026_B446_11965 | ○ | YSD | HTG |
| YP_006244914_SHJG_3769 | ○ | YSD | HTG |
| Bacteria | Prefix | Genome Size (Mb) | Class A PBP | Class B PBP | PBP with PASTA Domain * | No of Protein Kinase | Protein Kinase with PASTA Domain * |
|---|---|---|---|---|---|---|---|
| Streptomyces albulus NK660 | DC74 | 9.37 | 3326, 4231, 4477, 5204, 7647 | 3128, 3135, 3598, 4185, 5459 | none | 40 | 2595(4), 4186(4) |
| Streptomyces albus J1074 | XNR | 6.84 | 1770, 2736, 2983, 4127 | 1496, 2096, 2097, 3038, 4337, 4789 | none | 25 | 3037(4), 3064(1), 4768(4) |
| Streptomyces avermitilis MA-4680 | SAV | 9.12 | 3225, 4294, 4423, 4583, 5179, 7219 | 2952, 3603, 3604, 4339, 5458, 6116, 6387 | none | 33 | 4338(4), 4371(1), 6092(4) |
| Streptomyces bingchenggensis BCW-1 | SBI | 11.94 | 03076, 04174, 05361, 05810, 06697, 09068 | 02283, 04376, 05407, 06233, 07119, 07873 | none | 67 | 05406(4), 07851(4) |
| Streptomyces cattleya NRRL8057 | SCAT | 8.09 | 1929, 2889, 3140, 3906 | 0768, 1207, 1730, 1901, 3088, 4153, 5676 | none | 18 | 1232(4), 3053(1), 3089(4) |
| Streptomyces clavuligerus ATCC27064 | SCLAV | 8.56 | 2006, 2887, 3942 | 1087, 1301, 1774, 2276, 2947, 4154, 4179, 4180, 4198 | none | 33 | 1326(4), 2946(4), 2991(1) |
| Streptomyces coelicoflavus ZG0656 | SMCF | 8.48 | 1708, 7595 | 3764, 4686, 7469, 7795, 7796, 8190, 8286, 8884 | none | 22 | 6490 (4), 8300 (1),8885 (4) |
| Streptomyces coelicolor A3(2) | SCO | 9.05 | 2897, 3580, 3901, 5039 | 1875, 2090, 2608, 3156, 3157, 3771, 3847, 4013, 5301 | none | 37 | 2110(4), 3821(1), 3848(4) |
| Streptomyces collinus Tu365 | B446 | 8.38 | 15140, 19060, 23580 | 09755, 10960, 13820, 16355, 19320, 24955 | none | 39 | 11080(4), 19315(4), 19450(1) |
| Streptomyces davawensis JCM4913 | BN159 | 9.56 | 3357, 4150, 4546, 5391 | 3075, 4478, 5121, 5122, 5684, 6352, 6632 | none | 35 | 4396(1), 4479(4), 6328(4) |
| Streptomyces ghanaensis ATCC 14672 | SSFG | 8.51 | 02387, 02608, 03635, 04479 | 02394, 03587, 04216, 04217, 04765, 05266 | none | 35 | 03552(1), 03588(4) |
| Streptomyces griseoflavus Tu4000 | SSRG | 8.05 | 02182, 02879, 03203, 03961 | 01957, 03076, 03158, 03705, 03706, 04177, 04634, 04850 | none | 35 | 03115(1), 03159(4), 04610(4) |
| Streptomyces griseus subsp. griseus NBRC 13350 | SGR | 8.55 | 2494, 3341, 3679, 4647 | 2203, 3726, 4232, 4340, 4934, 5621 | none | 30 | 3725(4), 5391(4) |
| Streptomyces hygroscopicus subsp. jinggangensis 5008 | SHJG | 10.38 | 3853, 4373, 5171, 5432, 6136 | 3336, 4100, 4627, 4628, 5219, 6411 | none | 43 | 3594(4), 5218(4), 5252(1) |
| Streptomyces lividans TK24 | SLIV | 8.35 | 13190, 18760, 20390, 23205 | 11865, 18345, 19035, 19450, 21910, 21915, 24635, 28340 | none | 35 | 19030(4), 19175(1), 27155(4) |
| Streptomyces rimosus subsp. rimosus ATCC10970 | SRIM | 9.5 | 00295, 08328, 13873, 22689 | 00065, 04191, 06646, 15770, 26297, 31850, 31885 | none | 40 | 00070(4), 07563(1), 24996(4) |
| Streptomyces scabiei 87.22 | SCAB | 10.15 | 33601, 41401, 56801, 64431 | 10101, 29591, 45551, 53611, 53621, 60051, 70631 | none | 43 | 0931(1), 45201(1), 67711(4) |
| Streptomyces sp. PAMC26508 | F750 | 7.63 | 2719, 3337, 3596, 4580 | 1743, 2434, 2998, 3546, 4834, 6320 | none | 27 | 1975(4), 3512(1), 3547(4) |
| Streptomyces sp. SirexAA-E | SACTE | 7.41 | 2371, 3027, 3329, 4291 | 1307, 1519, 2029, 2618, 2701, 3283, 4532 | none | 32 | 1542(4), 3284(3) |
| Streptomyces sviceus ATCC 29083 | SSEG | 9.31 | 01073, 07525, 03439, 04164 | 00010, 00011, 00733, 01896, 09019, 09517, | none | 40 | 02705(4), 06024(4) |
| Streptomyces venezuelae ATCC 10712 | SVEN | 8.23 | 2646, 3350, 3677, 4705 | 1522, 1745, 2386, 2985, 3631, 4995 | none | 37 | 1769(4), 3592(1), 3632(4) |
| Streptomyces violaceusniger Tu4113 | STRVI | 11.14 | 1350, 2314, 3845, 8252, 9005 | 0275, 1135, 3190, 7171, 7897, 7904 | none | 38 | 0274(4), 7194(4) |
| Streptomyces viridochromogenes DSM 40736 | SSQG | 8.65 | 02328, 02941, 03901, 04279, 05113 | 01781, 02628, 03242, 03243, 03958, 05348 | none | 37 | 02054(4), 03956(4), 03996(1) |
| Streptomyces zinciresistens K42 | SZN | 8.22 | 06389, 16730, 18682, 28493 | 02952, 10458, 13352, 17932, 18819, 22026 | none | 39 | 08009(4), 17937(4), 21616(1) |
| Gene No. | Protein Name | No. of aa | Gene No. | Protein Name | No. of aa | Gene No. | Protein Name | No. of aa |
|---|---|---|---|---|---|---|---|---|
| SCO3844 | hypothetical protein | 172aa+ * | SCLAV_2950 | hypothetical protein | 132aa− | SGR_3729 | hypothetical protein | 173aa− |
| SCO3845 | protein phosphatase | 515aa+ | SCLAV_2949 | protein phosphatase | 507aa− | SGR_3728 | protein phosphatase | 499aa− |
| SCO3846 | FtsW/RodA family protein | 479aa+ | SCLAV_2948 | FtsW/RodA family protein | 470aa− | SGR_3727 | FtsW/RodA family protein | 466aa− |
| SCO3847 | PBP | 490aa+ | SCLAV_2947 | PBP | 484aa− | SGR_3726 | PBP | 485aa− |
| SCO3848 | STPK | 673aa+ | SCLAV_2946 | STPK | 682aa− | SGR_3725 | STPK | 664aa− |
| SCO3849 | hypothetical protein | 253aa− | SCLAV_2945 | hypothetical protein | 231aa+ | SGR_3724 | hypothetical protein | 245aa+ |
| SCO3850 | hypothetical protein | 352aa− | SCLAV_2944 | hypothetical protein | 461aa+ | SGR_3723 | hypothetical protein | 474aa+ |
| SCO3851 | glutamine amidotransferase | 212aa− | SCLAV_2943 | glutamine amidotransferase | 212aa+ | SGR_3722 | glutamine amidotransferase | 212aa+ |
| SCO3852 | hypothetical protein | 69aa− | SCLAV_2942 | hypothetical protein | 68aa+ | SGR_3721 | hypothetical protein | 62aa+ |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ogawara, H. Self-resistance in Streptomyces, with Special Reference to ?-Lactam Antibiotics. Molecules 2016, 21, 605. https://doi.org/10.3390/molecules21050605
Ogawara H. Self-resistance in Streptomyces, with Special Reference to ?-Lactam Antibiotics. Molecules. 2016; 21(5):605. https://doi.org/10.3390/molecules21050605
Chicago/Turabian StyleOgawara, Hiroshi. 2016. "Self-resistance in Streptomyces, with Special Reference to ?-Lactam Antibiotics" Molecules 21, no. 5: 605. https://doi.org/10.3390/molecules21050605
APA StyleOgawara, H. (2016). Self-resistance in Streptomyces, with Special Reference to ?-Lactam Antibiotics. Molecules, 21(5), 605. https://doi.org/10.3390/molecules21050605