Cu(II) Complexes of 4-[(1E)-N-{2-[(Z)-Benzylidene-amino]ethyl}ethanimidoyl]benzene-1,3-diol Schiff Base: Synthesis, Spectroscopic, In-Vitro Antioxidant, Antifungal and Antibacterial Studies
Abstract
:1. Introduction
2. Results and Discussion
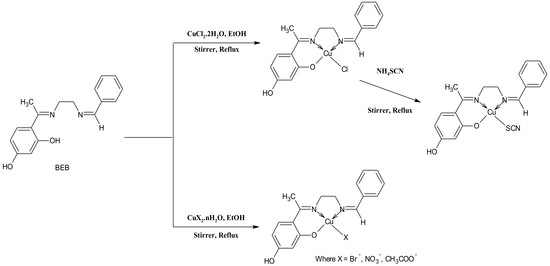
2.1. Synthesis and Characterization of the Compounds
2.2. Molar Conductivity Measurements
2.3. Infrared Absorption Spectra
2.4. 1H-NMR Spectral Studies
2.5. Electronic Spectral Measurements
2.6. Powder X-ray Diffraction Spectroscopy
2.7. Thermal Decomposition of the Complexes
2.8. Antioxidant Assays
2.8.1. DPPH Radical Scavenging Assay
2.8.2. 2,2′-Azino-bis(3-ethylbenzothiazoline-6-sulfonic Acid) Radical Scavenging Activity
2.9. In Vitro Antimicrobial Activity
3. Materials and Methods
3.1. Materials
3.2. Physical Measurements
3.3. Synthesis of the Schiff Base Ligand: 4-[(1E)-N-{2-[(Z)-Benzylideneamino]ethyl}ethanimidoyl]benzene-1,3-diol (BEB)
3.4. General Procedure for the Synthesis of Cu(II) Complexes
3.5. Biological Evaluations
3.5.1. Antioxidant Assay
3.5.2. In Vitro Antimicrobial Studies
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Karaliota, A.; Kresti, O.; Tzougraki, C. Synthesis and characterization of a binuclear coumarin-3-carboxylatecopper(II) complex. J. Inorg. Biochem. 2001, 84, 33–37. [Google Scholar] [CrossRef]
- Alaghaz, A.M.A.; Bayoumi, H.A.; Ammar, Y.A.; Aldhlmani, S.A. Synthesis, characterization, and antipathogenic studies of some transition metal complexes with N, O-chelating Schiff’s base ligand incorporating azo and sulfonamide Moieties. J. Mol. Struct. 2013, 1035, 383–399. [Google Scholar] [CrossRef]
- Scozzafava, A.; Mastrolorenzo, A.; Supuran, C.T. Antimycobacterial activity of 9-sulfonylated/sulfenylated-6-mercaptopurine derivatives. Bioorg. Med. Chem. Lett. 2001, 11, 1675–1678. [Google Scholar] [CrossRef]
- Aderoju, A.O.; Ingo, O.; Oladunni, M.O. Synthesis, spectroscopic, anticancer, and antimicrobial properties of some metal(II) complexes of (substituted) nitrophenol Schiff base. Int. J. Inorg. Chem. 2012, 2012, 206417. [Google Scholar] [CrossRef]
- Annapoorani, S.; Krishnan, C. Synthesis and spectroscopic studies of trinuclear N4 Schiff base complexes international. J. ChemTech Res. 2013, 5, 180–185. [Google Scholar]
- Alias, M.; Kassum, H.; Shakir, C. Synthesis, physical characterization and biological evaluation of Schiff base M(II) complexes. JAAUBAS 2014, 15, 28–34. [Google Scholar] [CrossRef]
- Gürbüz, D.; Çinarli, A.; Tavman, A.; Tan, A.S.B. Synthesis, characterization and antimicrobial activity of some transition metal complexes of N-(5-chloro-2-hydroxyphenyl)-3-methoxy-salicylaldimine. Bull. Chem. Soc. Ethiop. 2015, 29, 63–74. [Google Scholar] [CrossRef]
- Iftikhar, B.; Javed, K.; Khan, M.S.U.; Akhter, Z.; Mirza, B.; Mckee, V. Synthesis, characterization and biological assay of salicylaldehyde Schiff base Cu(II) complexes and their precursors. J. Mol. Struct. 2018, 1155, 337–348. [Google Scholar] [CrossRef]
- Ejidike, I.P.; Ajibade, P.A. Synthesis and in vitro anticancer, antibacterial, and antioxidant studies of unsymmetrical Schiff base derivatives of 4-[(1E)-N-(2-aminoethyl) ethanimidoyl]benzene-1,3-diol. Res. Chem. Intermed. 2016, 42, 6543–6555. [Google Scholar] [CrossRef]
- Ejidike, I.P.; Ajibade, P.A. Synthesis, characterization and biological studies of metal(II) complexes of (3E)-3-[(2-{(E)-[1-(2,4-Dihydroxyphenyl)ethylidene]amino}ethyl)imino]-1-phenylbutan-1-one Schiff base. Molecules 2015, 20, 9788–9802. [Google Scholar] [CrossRef] [PubMed]
- Issa, R.M.; Gaber, M.; Al-Wakiel, N.A.; Fathalla, S.K. Synthesis, spectral, thermal and biological studies of Mn(II), Co(II), Ni(II) and Cu(II) complexes with 1-(((5-Mercapto-1H-1,2,4-triazol-3-yl)imino)-methyl)naphthalene-2-ol. Chin. J. Chem. 2012, 30, 547–556. [Google Scholar] [CrossRef]
- Creaven, B.S.; Duff, B.; Egan, D.A. Anticancer and antifungal activity of copper(II) complexes of quinolin-2(1H)-one-derived Schiff bases. Inorg. Chim. Acta 2010, 363, 4048–4058. [Google Scholar] [CrossRef]
- Prasad, K.S.; Kumar, L.S.; Chandan, S.; Vijaya, B.; Revanasiddappa, H.D. Synthesis, characterization and DNA interaction studies of Copper(II) complex of 4(3H)-quinazolinone-derived Schiff base. An. Univ. Bucuresti Chim. 2011, 20, 7–13. [Google Scholar]
- Ejidike, I.P.; Ajibade, P.A. Ruthenium(III) complexes of heterocyclic tridentate (ONN) Schiff base: Synthesis, characterization and its biological properties as an antiradical and antiproliferative agent. Int. J. Mol. Sci. 2016, 17, 60. [Google Scholar] [CrossRef] [PubMed]
- Ammar, R.A.A.; Alaghaz, A.M.A. Synthesis, spectroscopic characterization and potentiometric studies of a tetradentate [N2O2] Schiff base, N,N′-bis(2-hydroxybenzylidene)-1,1-diaminoethane and Its Co(II), Ni(II), Cu(II) and Zn(II) Complexes. Int. J. Electrochem. Sci. 2013, 8, 8686–8699. [Google Scholar]
- Ejidike, I.P.; Ajibade, P.A. Synthesis, spectroscopic, antibacterial and free radical scavenging studies of Cu(II), Ni(II), Zn(II) and Co(II) complexes of 4,4′-{ethane-1,2-diylbis[nitrilo(1E)eth-1-yl-1-ylidene]}dibenzene-1,3-diol Schiff base. J. Pharm. Sci. Res. 2017, 9, 593–600. [Google Scholar]
- Geary, W.J. The use of conductivity measurements in organic solvents for the characterization of coordination compounds. Coord. Chem. Rev. 1971, 7, 81–122. [Google Scholar] [CrossRef]
- Sharaby, C.M.; Amine, M.F.; Hamed, A.A. Synthesis, structure characterization and biological activity of selected metal complexes of sulfonamide Schiff base as a primary ligand and some mixed ligand complexes with glycine as a secondary ligand. J. Mol. Struct. 2017, 1134, 208–216. [Google Scholar] [CrossRef]
- Al-Shaalan, N.H. Synthesis, characterization and biological activities of Cu(II), Co(II), Mn(II), Fe(II), and UO2(VI) complexes with a new Schiff base Hydrazone: O-Hydroxyacetophenone-7-chloro-4-quinoline Hydrazone. Molecules 2011, 16, 8629–8645. [Google Scholar] [CrossRef] [PubMed]
- Siddappa, K.; Mayana, N.S. Synthesis, spectroscopic characterization, and biological evaluation studies of 5-Bromo-3-(((hydroxy-2-methylquinolin-7-yl)methylene)hydrazono) indolin-2-one and its metal(II) complexes. Bioinorg. Chem. Appl. 2014, 2014, 483282. [Google Scholar] [CrossRef] [PubMed]
- Dikio, C.W.; Ejidike, I.P.; Mtunzi, F.M.; Klink, M.J.; Dikio, E.D. Hydrazide Schiff bases of acetylacetonate metal complexes: Synthesis, spectroscopic and biological studies. Int. J. Pharm. Pharm. Sci. 2017, 9, 257–267. [Google Scholar] [CrossRef]
- Abdel-Rahman, L.H.; Ismail, N.M.; Ismael, M.; Abu-Dief, A.M.; Ahmed, E.A. Synthesis, characterization, DFT calculations and biological studies of Mn(II), Fe(II), Co(II) and Cd(II) complexes based on a tetradentate ONNO donor Schiff base ligand. J. Mol. Struct. 2017, 1134, 851–862. [Google Scholar] [CrossRef]
- Neelakantan, M.A.; Marriappan, S.S.; Dharmaraja, J.; Jeyakumar, T.; Muthukumaran, K. Spectral, XRD, SEM and biological activities of transition metal complexes of polydentate ligands containing thiazole moiety. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2008, 71, 628–635. [Google Scholar] [CrossRef] [PubMed]
- Ejidike, I.P.; Ajibade, P.A. Synthesis, characterization, anticancer, and antioxidant studies of Ru(III) complexes of monobasic tridentate Schiff bases. Bioinorg. Chem. Appl. 2016, 2016, 9672451. [Google Scholar] [CrossRef] [PubMed]
- Philippopoulos, A.I.; Terzis, A.; Raptopoulou, C.P.; Catalanon, V.J.; Felaras, P. Synthesis, characterization, and sensitizing properties of heteroleptic RuII complexes based on 2,6-bis(1-pyrazolyl)pyridine and 2,2′-Bipyridine-4,4′-dicarboxylic acid ligands. Eur. J. Inorg. Chem. 2007, 5633–5644. [Google Scholar] [CrossRef]
- Alaghaz, A.M.A.; Zayed, M.E.; Alharbi, S.A. Synthesis, spectral characterization, molecular modeling and antimicrobial studies of tridentate azo-dye Schiff base metal complexes. J. Mol. Struct. 2015, 1084, 36–45. [Google Scholar] [CrossRef]
- Thangadurai, T.D.; Ihm, S.-K. Ruthenium(II) complexes derived from substituted cyclobutane and substituted thiazole Schiff base ligands: Synthetic, spectral, catalytic and antimicrobial studies. Synth. React. Inorg. Met. Org. Chem. 2005, 35, 499–507. [Google Scholar] [CrossRef]
- Souaya, E.R.; Hanna, W.G.; Ismail, E.H.; Milad, N.E. Studies on some acid divalent-metal nitrilotriacetate complexes. Molecules 2000, 5, 1121–1129. [Google Scholar] [CrossRef]
- Joseyphus, R.S.; Nair, M.S. Synthesis, characterization and biological studies of some Co(II), Ni(II) and Cu(II) complexes derived from indole-3-carboxaldehyde and glycylglycine as Schiff base ligand. Arab. J. Chem. 2010, 3, 195–204. [Google Scholar] [CrossRef]
- Elisha, I.L.; Botha, F.S.; McGaw, L.J.; Eloff, J.N. The antibacterial activity of extracts of nine plant species with good activity against Escherichia coli against five other bacteria and cytotoxicity of extracts. BMC Complement. Altern. Med. 2017, 17, 133. [Google Scholar] [CrossRef] [PubMed]
- Nastasă, C.; Vodnar, D.C.; Ionuţ, I.; Stana, A.; Benedec, D.; Tamaian, R.; Oniga, O.; Tiperciuc, B. Antibacterial evaluation and virtual screening of new Thiazolyl-Triazole Schiff bases as potential DNA-Gyrase inhibitors. Int. J. Mol. Sci. 2018, 19, 222. [Google Scholar] [CrossRef] [PubMed]
- Masoko, P.; Picard, J.; Eloff, J.N. Antifungal activities of six South African Terminalia species (Combretaceae). J. Ethnopharmacol. 2005, 99, 301–308. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds ...... are available from the authors. |









| Compounds | Empirical Formula | F. Wt (Gram) | Colour | Yield (%) | % Found (Calcd.) | Decomp. Temp, °C | Conductance (µS cm−1) | ||
|---|---|---|---|---|---|---|---|---|---|
| BEB | C17H18N2O2 | 282.34 | Dark-yellow | 71.50 | 72.17 (72.32) | 6.31 (6.43) | 9.86 (9.92) | 233–234 | - |
| [Cu(BEB)Cl]·2H2O | C17H21N2O4ClCu | 416.36 | Greyish | 69.84 | 50.39 (49.04) | 4.69 (5.08) | 7.38 (6.73) | 181–182 | 13.83 |
| [Cu(BEB)Br]·H2O | C17H19N2O3BrCu | 442.79 | Dark-brow | 75.49 | 47.87 (46.11) | 5.13 (4.32) | 6.82 (6.33) | 175–176 | 13.15 |
| [Cu(BEB)SCN]·2H2O | C18H21N3O4SCu | 438.99 | Dark-brown | 60.72 | 48.21 (49.25) | 5.29 (4.82) | 10.29 (9.57) | 178–179 | 15.09 |
| [Cu(BEB)NO3]·3H2O | C17H23N3O8Cu | 460.93 | Dark-blackish | 81.64 | 45.08 (44.30) | 5.67 (5.03) | 9.43 (9.12) | 173–174 | 22.70 |
| [Cu(BEB)CH3COO]·2H2O | C19H24N2O6Cu | 439.95 | Darkish-brown | 65.09 | 50.94 (51.87) | 5.01 (5.50) | 6.89 (6.37) | 171–172 | 10.71 |
| Compound | ν(OH)/(H2O) | ν(CH3/CH2) | ν(SCN) | ν(C=N) | ν(C=C) | ν(NO3/OAc) | ν(C−O) | ν(M−N) | ν(M−O) |
|---|---|---|---|---|---|---|---|---|---|
| asy&(sy) | asy&(sy) | ||||||||
| BEB | 3404mb | 3166w, 3057w | 1613vs | 1538m, 1436m | 1266s | ||||
| [Cu(BEB)Cl]·2H2O | 3384mb | 3158w, 3067w | 1588s | 1538m, 1436m | 1237s | 594m | 452m | ||
| [Cu(BEB)Br]·H2O | 3392mb | 3167w, 3064w | 1589vs | 1539m, 1435m | 1237s | 526m | 455m | ||
| [Cu(BEB)SCN]·2H2O | 3392mb | 3157w, 3059w | 2161m | 1589vs | 1539s, 1435w | 1241s | 567m | 455m | |
| (2096m) | |||||||||
| [Cu(BEB)NO3]·3H2O | 3378mb | 3160w, 3062w | 1590vs | 1539m, 1436w | 1417w | 1240m | 593m | 457m | |
| (1297m) | |||||||||
| [Cu(BEB)CH3COO]·2H2O | 3396mb | 3162w, 3062w | 1589s | 1542w, 1435w | 1538m | 1240m | 597m | 462m | |
| (1340m) |
| Compounds | Absorption Transition, λmax (nm, DMF) | Band Assignments | |
|---|---|---|---|
| BEB | C18H20N2O4 | 277, 309, 381 | π-π*, π-π*, n-π* |
| [Cu(BEB)Cl]·2H2O | C18H20N2O5Cu | 325, 345, 375, 545 | π-π*, L → M (LMCT), 2B1g → 2A1g |
| [Cu(BEB)Br]·H2O | C18H22N2O6Cu | 335, 395, 565 | π-π*, L → M (LMCT), 2B1g → 2A1g |
| [Cu(BEB)SCN]·2H2O | C18H22N2O6Cu | 330, 380, 547 | π-π*, L → M (LMCT), 2B1g → 2A1g |
| [Cu(BEB)NO3]·3H2O | C18H22N2O6Cu | 335, 395, 570 | π-π*, L → M (LMCT), 2B1g → 2A1g |
| Compounds | DPPH Radical Scavenging Activity | ABTS Radical Scavenging Activity | ||
|---|---|---|---|---|
| IC50 (µM) | R2 | IC50 (µM) | R2 | |
| BEB | 3.49 ± 1.57 | 0.913 | 2.77 ± 1.44 | 0.781 |
| [Cu(BEB)Cl] | 3.43 ± 0.82 | 0.965 | 3.02 ± 0.47 | 0.852 |
| [Cu(BEB)Br] | 3.40 ± 1.51 | 0.962 | 3.73 ± 0.77 | 0.739 |
| [Cu(BEB)SCN] | 2.60 ± 1.46 | 0.904 | 3.42 ± 0.41 | 0.917 |
| [Cu(BEB)NO3] | 2.01 ± 1.49 | 0.995 | 4.56 ± 1.99 | 0.766 |
| [Cu(BEB)CH3COO] | 2.88 ± 0.66 | 0.969 | 4.33 ± 1.62 | 0.778 |
| Vit. C * | 1.74 ± 1.19 | 0.976 | - | - |
| Rutin * | 2.49 ± 1.27 | 0.835 | 2.86 ± 0.92 | 0.931 |
| Gallic acid * | 1.44 ± 1.11 | 0.890 | 1.22 ± 1.08 | 0.949 |
| BHT * | - | - | 2.31 ± 1.30 | 0.971 |
| Compounds | Gram (+) bacteria | Gram (−) bacteria | Fungi | |||
|---|---|---|---|---|---|---|
| S. aureus | E. faecalis | K. pneumoniae | P. aeruginosa | C. albicans | C. neoformans | |
| BEB | 1563 | 3125 | 1563 | 1563 | 781.3 | 195.3 |
| [Cu(BEB)Cl] | 48.83 | 390.6 | 781.3 | 390.6 | 48.83 | 48.83 |
| [Cu(BEB)Br] | 97.66 | 781.3 | 781.3 | 390.6 | 48.83 | 48.83 |
| [Cu(BEB)SCN] | 48.83 | 195.3 | 781.3 | 390.6 | 48.83 | 48.83 |
| [Co(BEB)NO3] | 97.66 | 1563 | 1563 | 1563 | 48.83 | 48.83 |
| [Cu(BEB)CH3COO] | 390.6 | 3125 | 1563 | 1563 | 195.3 | 195.3 |
| Neomycin a | 1.953 | 3.906 | 1.953 | 3.906 | - | - |
| Amphotericin B a | - | - | - | - | 48.83 | 48.83 |
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ejidike, I.P. Cu(II) Complexes of 4-[(1E)-N-{2-[(Z)-Benzylidene-amino]ethyl}ethanimidoyl]benzene-1,3-diol Schiff Base: Synthesis, Spectroscopic, In-Vitro Antioxidant, Antifungal and Antibacterial Studies. Molecules 2018, 23, 1581. https://doi.org/10.3390/molecules23071581
Ejidike IP. Cu(II) Complexes of 4-[(1E)-N-{2-[(Z)-Benzylidene-amino]ethyl}ethanimidoyl]benzene-1,3-diol Schiff Base: Synthesis, Spectroscopic, In-Vitro Antioxidant, Antifungal and Antibacterial Studies. Molecules. 2018; 23(7):1581. https://doi.org/10.3390/molecules23071581
Chicago/Turabian StyleEjidike, Ikechukwu P. 2018. "Cu(II) Complexes of 4-[(1E)-N-{2-[(Z)-Benzylidene-amino]ethyl}ethanimidoyl]benzene-1,3-diol Schiff Base: Synthesis, Spectroscopic, In-Vitro Antioxidant, Antifungal and Antibacterial Studies" Molecules 23, no. 7: 1581. https://doi.org/10.3390/molecules23071581
APA StyleEjidike, I. P. (2018). Cu(II) Complexes of 4-[(1E)-N-{2-[(Z)-Benzylidene-amino]ethyl}ethanimidoyl]benzene-1,3-diol Schiff Base: Synthesis, Spectroscopic, In-Vitro Antioxidant, Antifungal and Antibacterial Studies. Molecules, 23(7), 1581. https://doi.org/10.3390/molecules23071581