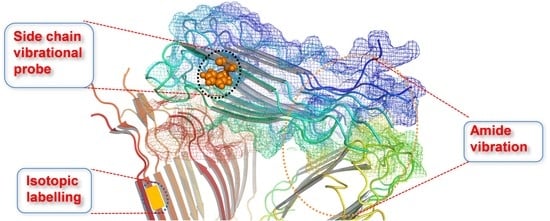
Vibrational Approach to the Dynamics and Structure of Protein Amyloids
Abstract
:1. Introduction
2. Backbone Vibrational Probe
2.1. Amyloid-β
2.2. Islet Amyloid Polypeptide
2.3. α-Synuclein
2.4. Examples of Other Disease-Associated Proteins and Model Peptides
3. Isotopic Labeling Probe
3.1. Amyloid-β
3.2. Islet Amyloid Polypeptide
3.3. Examples of Other Disease-Associated Proteins and Model Peptides
4. Side Chain Vibrational Probe
4.1. Azide Probe
4.2. Nitrile Probe
4.3. Ester Carbonyl Probe
5. Conclusions and Outlook
Funding
Conflicts of Interest
References
- Selkoe, D.J. Folding proteins in fatal ways. Nature 2003, 426, 900–904. [Google Scholar] [CrossRef] [PubMed]
- Chiti, F.; Dobson, C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006, 75, 333–366. [Google Scholar] [CrossRef] [PubMed]
- Sipe, J.D.; Cohen, A.S. Review: History of the amyloid fibril. J. Struct. Biol. 2000, 130, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Jahn, T.R.; Makin, O.S.; Morris, K.L.; Marshall, K.E.; Tian, P.; Sikorski, P.; Serpell, L.C. The common architecture of cross-β amyloid. J. Mol. Biol. 2010, 395, 717–727. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, D.; Jucker, M. The amyloid state of proteins in human diseases. Cell 2012, 148, 1188–1203. [Google Scholar] [CrossRef] [PubMed]
- Kayed, R.; Head, E.; Thompson, J.L.; McIntire, T.M.; Milton, S.C.; Cotman, C.W.; Glabe, C.G. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 2003, 300, 486–489. [Google Scholar] [CrossRef]
- Shankar, G.M.; Li, S.; Mehta, T.H.; Garcia-Munoz, A.; Shepardson, N.E.; Smith, I.; Brett, F.M.; Farrell, M.A.; Rowan, M.J.; Lemere, C.A. Amyloid-β protein dimers isolated directly from Alzheimer’s brains impair synaptic plasticity and memory. Nat. Med. 2008, 14, 837–842. [Google Scholar] [CrossRef]
- Harper, J.D.; Lansbury, P.T. Models of amyloid seeding in Alzheimier’s disease and scrapie: Mechanistic truths and physiological consequences of the time-dependent solubility of amyloid proteins. Annu. Rev. Biochem. 1997, 66, 385–407. [Google Scholar] [CrossRef]
- Naiki, H.; Hasegawa, K.; Yamaguchi, I.; Nakamura, H.; Gejyo, F.; Nakakuki, K. Apolipoprotein E and antioxidants have different mechanisms of inhibiting Alzheimer’s β-amyloid fibril formation in vitro. Biochemistry 1998, 37, 17882–17889. [Google Scholar] [CrossRef]
- Powers, E.T.; Powers, D.L. Mechanisms of protein fibril formation: Nucleated polymerization with competing off-pathway aggregation. Biophys. J. 2008, 94, 379–391. [Google Scholar] [CrossRef]
- Howie, A.J.; Brewer, D.B. Optical properties of amyloid stained by Congo red: History and mechanisms. Micron 2009, 40, 285–301. [Google Scholar] [CrossRef] [PubMed]
- Biancalana, M.; Koide, S. Molecular mechanism of Thioflavin-T binding to amyloid fibrils. Biochem. Biophys. Acta 2010, 1804, 1405–1412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mechaly, A.E.; Bellomio, A.; Gil-Carton, D.; Morante, K.; Valle, M.; Gonzalez-Manas, J.M.; Guerin, D.M. Structural insights into the oligomerization and architecture of eukaryotic membrane pore-forming toxins. Structure 2011, 19, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Mustata, M.; Capone, R.; Jang, H.; Arce, F.T.; Ramachandran, S.; Lal, R.; Nussinov, R. K3 fragment of amyloidogenic β2-microglobulin forms ion channels: Implication for dialysis related amyloidosis. J. Am. Chem. Soc. 2009, 131, 14938–14945. [Google Scholar] [CrossRef] [PubMed]
- Pires, R.H.; Saraiva, M.J.; Damas, A.M.; Kellermayer, M.S. Structure and assembly-disassembly properties of wild-type transthyretin amyloid protofibrils observed with atomic force microscopy. J. Mol. Recognit. 2011, 24, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Vestergaard, B.; Groenning, M.; Roessle, M.; Kastrup, J.S.; van de Weert, M.; Flink, J.M.; Frokjaer, S.; Gajhede, M.; Svergun, D.I. A helical structural nucleus is the primary elongating unit of insulin amyloid fibrils. PLoS Biol. 2007, 5, e134. [Google Scholar] [CrossRef] [PubMed]
- Chimon, S.; Shaibat, M.A.; Jones, C.R.; Calero, D.C.; Aizezi, B.; Ishii, Y. Evidence of fibril-like β-sheet structures in a neurotoxic amyloid intermediate of Alzheimer’s β-amyloid. Nat. Struct. Mol. Biol. 2007, 14, 1157–1164. [Google Scholar] [CrossRef] [PubMed]
- Walsh, P.; Yau, J.; Simonetti, K.; Sharpe, S. Morphology and secondary structure of stable β-oligomers formed by amyloid peptide PrP(106–126). Biochemistry 2009, 48, 5779–5781. [Google Scholar] [CrossRef]
- Patel, H.R.; Pithadia, A.S.; Brender, J.R.; Fierke, C.A.; Ramamoorthy, A. In Search of Aggregation Pathways of IAPP and Other Amyloidogenic Proteins: Finding Answers through NMR Spectroscopy. J. Phys. Chem. Lett. 2014, 5, 1864–1870. [Google Scholar] [CrossRef]
- Wälti, M.A.; Ravotti, F.; Arai, H.; Glabe, C.G.; Wall, J.S.; Böckmann, A.; Güntert, P.; Meier, B.H.; Riek, R. Atomic-resolution structure of a disease-relevant Aβ (1–42) amyloid fibril. Proc. Natl. Acad. Sci. USA 2016, 113, E4976–E4984. [Google Scholar] [CrossRef]
- Sawaya, M.R.; Sambashivan, S.; Nelson, R.; Ivanova, M.I.; Sievers, S.A.; Apostol, M.I.; Thompson, M.J.; Balbirnie, M.; Wiltzius, J.J.; McFarlane, H.T.; et al. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature 2007, 447, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Cho, M. Infrared probes for studying the structure and dynamics of biomolecules. Chem. Rev. 2013, 113, 5817–5847. [Google Scholar] [CrossRef] [PubMed]
- Moran, S.D.; Zanni, M.T. How to get insight into amyloid structure and formation from infrared spectroscopy. J. Phys. Chem. Lett. 2014, 5, 1984–1993. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Pazos, I.M.; Zhang, W.; Culik, R.M.; Gai, F. Site-specific infrared probes of proteins. Annu. Rev. Phys. Chem. 2015, 66, 357–377. [Google Scholar] [CrossRef] [PubMed]
- Schweitzer-Stenner, R. Visible and UV-resonance Raman spectroscopy of model peptides. J. Raman Spectrosc. 2001, 32, 711–732. [Google Scholar] [CrossRef]
- Barth, A. Infrared spectroscopy of proteins. Biochem. Biophys. Acta 2007, 1767, 1073–1101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDonald, R.S. Infrared spectrometry. Anal. Chem. 1986, 58, 1906–1925. [Google Scholar] [CrossRef]
- Rostron, P.; Gaber, S.; Gaber, D. Raman Spectroscopy, Review. IJETR 2016, 6, 50–64. [Google Scholar]
- Krimm, S.; Bandekar, J. Vibrational spectroscopy and conformation of peptides, polypeptides, and proteins. Adv. Prot. Chem. 1986, 38, 181–364. [Google Scholar]
- Barth, A.; Zscherp, C. What vibrations tell us about proteins. Q. Rev. Biophys. 2002, 35, 369–430. [Google Scholar] [CrossRef]
- Haris, P.I.; Severcan, F. FTIR spectroscopic characterization of protein structure in aqueous and non-aqueous media. J. Mol. Catal. B Enzym. 1999, 7, 207–221. [Google Scholar] [CrossRef]
- Moore, W.H.; Krimm, S. Transition dipole coupling in Amide I modes of βpolypeptides. Proc. Natl. Acad. Sci. USA 1975, 72, 4933–4935. [Google Scholar] [CrossRef] [PubMed]
- Woutersen, S.; Ham, S. Structure Determination of Trialanine in Water Using Polarization Sensitive Two-Dimensional Vibrational Spectroscopy. J. Phys. Chem. B 2000, 104, 11316–11320. [Google Scholar] [CrossRef]
- Lee, C.; Cho, M. Local Amide I Mode Frequencies and Coupling Constants in Multiple-Stranded Antiparallel β-Sheet Polypeptides. J. Phys. Chem. B 2004, 108, 20397–20407. [Google Scholar] [CrossRef]
- Myshakina, N.S.; Asher, S.A. Peptide bond vibrational coupling. J. Phys. Chem. B 2007, 111, 4271–4279. [Google Scholar] [CrossRef] [PubMed]
- Ganim, Z.; Chung, H.S.; Smith, A.W.; Deflores, L.P.; Jones, K.C.; Tokmakoff, A. Amide I two-dimensional infrared spectroscopy of proteins. Acc. Chem. Res. 2008, 41, 432–441. [Google Scholar] [CrossRef] [PubMed]
- Rygula, A.; Majzner, K.; Marzec, K.M.; Kaczor, A.; Pilarczyk, M.; Baranska, M. Raman spectroscopy of proteins: A review. J. Raman Specrosc. 2013, 44, 1061–1076. [Google Scholar] [CrossRef]
- Goormaghtigh, E.; Cabiaux, V.; Ruysschaert, J.-M. Determination of soluble and membrane protein structure by Fourier transform infrared spectroscopy. In Physicochemical Methods in the Study of Biomembranes; Springer: New York, NY, USA, 1994; pp. 405–450. [Google Scholar]
- Dobson, C.M. Protein folding and misfolding. Nature 2003, 426, 884–890. [Google Scholar] [CrossRef]
- Zandomeneghi, G.; Krebs, M.R.; McCammon, M.G.; Fandrich, M. FTIR reveals structural differences between native β-sheet proteins and amyloid fibrils. Protein Sci. 2004, 13, 3314–3321. [Google Scholar] [CrossRef]
- Lomont, J.P.; Ostrander, J.S.; Ho, J.J.; Petti, M.K.; Zanni, M.T. Not All β-Sheets Are the Same: Amyloid Infrared Spectra, Transition Dipole Strengths, and Couplings Investigated by 2D IR Spectroscopy. J. Phys. Chem. B 2017, 121, 8935–8945. [Google Scholar] [CrossRef] [Green Version]
- Sarroukh, R.; Goormaghtigh, E.; Ruysschaert, J.M.; Raussens, V. ATR-FTIR: A “rejuvenated” tool to investigate amyloid proteins. Biochem. Biophys. Acta 2013, 1828, 2328–2338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venyaminov, S.; Prendergast, F.G. Water (H2O and D2O) molar absorptivity in the 1000–4000 cm−1 range and quantitative infrared spectroscopy of aqueous solutions. Anal. Biochem. 1997, 248, 234–245. [Google Scholar] [CrossRef] [PubMed]
- Fayer, M.D. Dynamics of liquids, molecules, and proteins measured with ultrafast 2D IR vibrational echo chemical exchange spectroscopy. Annu. Rev. Phys. Chem. 2009, 60, 21–38. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Hochstrasser, R.M. Applications of 2D IR spectroscopy to peptides, proteins, and hydrogen-bond dynamics. J. Phys. Chem. B 2009, 113, 8231–8251. [Google Scholar] [CrossRef] [PubMed]
- Strasfeld, D.B.; Ling, Y.L.; Gupta, R.; Raleigh, D.P.; Zanni, M.T. Strategies for extracting structural information from 2D IR spectroscopy of amyloid: Application to islet amyloid polypeptide. J. Phys. Chem. B 2009, 113, 15679–15691. [Google Scholar] [CrossRef] [PubMed]
- Mattson, M.P. Pathways towards and away from Alzheimer’s disease. Nature 2004, 430, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J.; Selkoe, D.J. Medicine—The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science 2002, 297, 353–356. [Google Scholar] [CrossRef]
- Tanzi, R.E.; Bertram, L. Twenty years of the Alzheimer’s disease amyloid hypothesis: A genetic perspective. Cell 2005, 120, 545–555. [Google Scholar] [CrossRef]
- Sarroukh, R.; Cerf, E.; Derclaye, S.; Dufrene, Y.F.; Goormaghtigh, E.; Ruysschaert, J.M.; Raussens, V. Transformation of amyloid β(1–40) oligomers into fibrils is characterized by a major change in secondary structure. Cell Mol. Life Sci. 2011, 68, 1429–1438. [Google Scholar] [CrossRef]
- Klementieva, O.; Willén, K.; Martinsson, I.; Israelsson, B.; Engdahl, A.; Cladera, J.; Uvdal, P.; Gouras, G. Pre-plaque conformational changes in Alzheimer’s disease-linked Aβ and APP. Nat. Commun. 2017, 8, 14726. [Google Scholar] [CrossRef] [Green Version]
- Lomont, J.P.; Rich, K.L.; Maj, M.; Ho, J.J.; Ostrander, J.S.; Zanni, M.T. Spectroscopic Signature for Stable β-Amyloid Fibrils versus β-Sheet-Rich Oligomers. J. Phys. Chem. B 2018, 122, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Tidy, R.J.; Lam, V.; Fimognari, N.; Mamo, J.C.; Hackett, M.J. FTIR studies of the similarities between pathology induced protein aggregation in vivo and chemically induced protein aggregation ex vivo. Vib. Spec. 2017, 91, 68–76. [Google Scholar] [CrossRef]
- Nabers, A.; Ollesch, J.; Schartner, J.; Kotting, C.; Genius, J.; Hafermann, H.; Klafki, H.; Gerwert, K.; Wiltfang, J. Amyloid-β-Secondary Structure Distribution in Cerebrospinal Fluid and Blood Measured by an Immuno-Infrared-Sensor: A Biomarker Candidate for Alzheimer’s Disease. Anal. Chem. 2016, 88, 2755–2762. [Google Scholar] [CrossRef]
- Hull, R.L.; Westermark, G.T.; Westermark, P.; Kahn, S.E. Islet amyloid: A critical entity in the pathogenesis of type 2 diabetes. J. Clin. Endocrinol. Metab. 2004, 89, 3629–3643. [Google Scholar] [CrossRef] [PubMed]
- Westermark, P.; Andersson, A.; Westermark, G.T. Islet amyloid polypeptide, islet amyloid, and diabetes mellitus. Physiol. Rev. 2011, 91, 795–826. [Google Scholar] [CrossRef] [PubMed]
- Janson, J.; Ashley, R.H.; Harrison, D.; McIntyre, S.; Butler, P.C. The mechanism of islet amyloid polypeptide toxicity is membrane disruption by intermediate-sized toxic amyloid particles. Diabetes 1999, 48, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Rawat, A.; Maity, B.K.; Chandra, B.; Maiti, S. Aggregation-induced conformation changes dictate islet amyloid polypeptide (IAPP) membrane affinity. Biochim. Biophys. Acta Biomembr. 2018, 1860, 1734–1740. [Google Scholar] [CrossRef]
- Goldsbury, C.; Goldie, K.; Pellaud, J.; Seelig, J.; Frey, P.; Müller, S.; Kistler, J.; Cooper, G.; Aebi, U. Amyloid fibril formation from full-length and fragments of amylin. J. Struct. Biol. 2000, 130, 352–362. [Google Scholar] [CrossRef]
- Strasfeld, D.B.; Ling, Y.L.; Shim, S.H.; Zanni, M.T. Tracking fiber formation in human islet amyloid polypeptide with automated 2D-IR spectroscopy. J. Am. Chem. Soc. 2008, 130, 6698–6699. [Google Scholar] [CrossRef]
- Abedini, A.; Plesner, A.; Cao, P.; Ridgway, Z.; Zhang, J.; Tu, L.H.; Middleton, C.T.; Chao, B.; Sartori, D.J.; Meng, F.; et al. Time-resolved studies define the nature of toxic IAPP intermediates, providing insight for anti-amyloidosis therapeutics. Elife 2016, 5, e12977. [Google Scholar] [CrossRef]
- Shim, S.H.; Strasfeld, D.B.; Ling, Y.L.; Zanni, M.T. Automated 2D IR spectroscopy using a mid-IR pulse shaper and application of this technology to the human islet amyloid polypeptide. Proc. Natl. Acad. Sci. USA 2007, 104, 14197–14202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunkelberger, E.B.; Grechko, M.; Zanni, M.T. Transition Dipoles from 1D and 2D Infrared Spectroscopy Help Reveal the Secondary Structures of Proteins: Application to Amyloids. J. Phys. Chem. B 2015, 119, 14065–14075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishra, R.; Bulic, B.; Sellin, D.; Jha, S.; Waldmann, H.; Winter, R. Small-molecule inhibitors of islet amyloid polypeptide fibril formation. Angew. Chem. Int. Ed. 2008, 47, 4679–4682. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.; Sellin, D.; Radovan, D.; Gohlke, A.; Winter, R. Inhibiting islet amyloid polypeptide fibril formation by the red wine compound resveratrol. Chem. BioChem. 2009, 10, 445–449. [Google Scholar] [CrossRef] [PubMed]
- Sellin, D.; Yan, L.M.; Kapurniotu, A.; Winter, R. Suppression of IAPP fibrillation at anionic lipid membranes via IAPP-derived amyloid inhibitors and insulin. Biophys. Chem. 2010, 150, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Estel, K.; Seeliger, J.; Friedrich, R.P.; Dogan, S.; Wanker, E.E.; Winter, R.; Ebbinghaus, S. Modulation of human IAPP fibrillation: Cosolutes, crowders and chaperones. Phys. Chem. Chem. Phys. 2015, 17, 8338–8348. [Google Scholar] [CrossRef] [PubMed]
- Forno, L.S. Neuropathology of Parkinson’s disease. J. Neuropathol. Exp. Neuro. 1996, 55, 259–272. [Google Scholar] [CrossRef]
- Polymeropoulos, M.H.; Lavedan, C.; Leroy, E.; Ide, S.E.; Dehejia, A.; Dutra, A.; Pike, B.; Root, H.; Rubenstein, J.; Boyer, R.; et al. Mutation in the α-synuclein gene identified in families with Parkinson’s disease. Science 1997, 276, 2045–2047. [Google Scholar] [CrossRef]
- Kruger, R.; Kuhn, W.; Muller, T.; Woitalla, D.; Graeber, M.; Kosel, S.; Przuntek, H.; Epplen, J.T.; Schols, L.; Riess, O. Ala30Pro mutation in the gene encoding α-synuclein in Parkinson’s disease. Nat. Genet. 1998, 18, 106–108. [Google Scholar] [CrossRef]
- Spillantini, M.G.; Crowther, R.A.; Jakes, R.; Hasegawa, M.; Goedert, M. α-Synuclein in filamentous inclusions of Lewy bodies from Parkinson’s disease and dementia with lewy bodies. Proc. Natl. Acad. Sci. USA 1998, 95, 6469–6473. [Google Scholar] [CrossRef]
- Deas, E.; Cremades, N.; Angelova, P.R.; Ludtmann, M.H.; Yao, Z.; Chen, S.; Horrocks, M.H.; Banushi, B.; Little, D.; Devine, M.J.; et al. α-Synuclein Oligomers Interact with Metal Ions to Induce Oxidative Stress and Neuronal Death in Parkinson’s Disease. Antioxid. Redox. Signal. 2016, 24, 376–391. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Li, J.; Fink, A.L. Evidence for a partially folded intermediate in α-synuclein fibril formation. J. Biol. Chem. 2001, 276, 10737–10744. [Google Scholar] [CrossRef] [PubMed]
- Celej, M.S.; Sarroukh, R.; Goormaghtigh, E.; Fidelio, G.D.; Ruysschaert, J.-M.; Raussens, V. Toxic prefibrillar α-synuclein amyloid oligomers adopt a distinctive antiparallel β-sheet structure. Biochem. J. 2012, 443, 719–726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conway, K.A.; Harper, J.D.; Lansbury, P.T. Fibrils formed in vitro from α-synuclein and two mutant forms linked to Parkinson’s disease are typical amyloid. Biochemistry 2000, 39, 2552–2563. [Google Scholar] [CrossRef] [PubMed]
- Roeters, S.J.; Iyer, A.; Pletikapić, G.; Kogan, V.; Subramaniam, V.; Woutersen, S. Evidence for intramolecular antiparallel β-sheet structure in alpha-synuclein fibrils from a combination of two-dimensional infrared spectroscopy and atomic force microscopy. Sci. Rep. 2017, 7, 41051. [Google Scholar] [CrossRef] [PubMed]
- Maiti, N.C.; Apetri, M.M.; Zagorski, M.G.; Carey, P.R.; Anderson, V.E. Raman spectroscopic characterization of secondary structure in natively unfolded proteins: α-synuclein. J. Am. Chem. Soc. 2004, 126, 2399–2408. [Google Scholar] [CrossRef] [PubMed]
- Apetri, M.M.; Maiti, N.C.; Zagorski, M.G.; Carey, P.R.; Anderson, V.E. Secondary structure of α-synuclein oligomers: Characterization by raman and atomic force microscopy. J. Mol. Biol. 2006, 355, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Flynn, J.D.; McGlinchey, R.P.; Walker, R.L.; Lee, J.C. Structural features of α-synuclein amyloid fibrils revealed by Raman spectroscopy. J. Biol. Chem. 2018, 293, 767–776. [Google Scholar] [CrossRef]
- Schweitzer-Stenner, R. Advances in vibrational spectroscopy as a sensitive probe of peptide and protein structure—A critical review. Vib. Spec. 2006, 42, 98–117. [Google Scholar] [CrossRef]
- Asher, S.A.; Ianoul, A.; Mix, G.; Boyden, M.N.; Karnoup, A.; Diem, M.; Schweitzer-Stenner, R. Dihedral psi angle dependence of the amide III vibration: A uniquely sensitive UV resonance Raman secondary structural probe. J. Am. Chem. Soc. 2001, 123, 11775–11781. [Google Scholar] [CrossRef]
- Ianoul, A.; Boyden, M.N.; Asher, S.A. Dependence of the peptide amide III vibration on the phi dihedral angle. J. Am. Chem. Soc. 2001, 123, 7433–7434. [Google Scholar] [CrossRef] [PubMed]
- Papanikolopoulou, K.; Mills-Henry, I.; Thol, S.L.; Wang, Y.; Gross, A.A.; Kirschner, D.A.; Decatur, S.M.; King, J. Formation of amyloid fibrils in vitro by human γD-crystallin and its isolated domains. Mol. Vis. 2008, 14, 81–89. [Google Scholar] [PubMed]
- Gustiananda, M.; Haris, P.I.; Milburn, P.J.; Gready, J.E. Copper-induced conformational change in a marsupial prion protein repeat peptide probed using FTIR spectroscopy. FEBS Lett. 2002, 512, 38–42. [Google Scholar] [CrossRef]
- Sokolowski, F.; Naumann, D. FTIR study on thermal denaturation and aggregation of recombinant hamster prion protein SHaPrP90–232. Vib. Spec. 2005, 38, 39–44. [Google Scholar] [CrossRef]
- Heck, B.S.; Doll, F.; Hauser, K. Length-dependent conformational transitions of polyglutamine repeats as molecular origin of fibril initiation. Biophys. Chem. 2014, 185, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Dzwolak, W.; Muraki, T.; Kato, M.; Taniguchi, Y. Chain-length dependence of α-helix to β-sheet transition in polylysine: Model of protein aggregation studied by temperature-tuned FTIR spectroscopy. Biopolymers 2004, 73, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Markiewicz, B.N.; Oyola, R.; Du, D.; Gai, F. Aggregation Gatekeeper and Controlled Assembly of Trpzip β-Hairpins. Biochemistry 2014, 53, 1146–1154. [Google Scholar] [CrossRef]
- Zhang, T.O.; Alperstein, A.M.; Zanni, M.T. Amyloid β-Sheet Secondary Structure Identified in UV-Induced Cataracts of Porcine Lenses using 2D IR Spectroscopy. J. Mol. Biol. 2017, 429, 1705–1721. [Google Scholar] [CrossRef]
- Fang, C.; Hochstrasser, R.M. Two-Dimensional Infrared Spectra of the 13C 18O Isotopomers of Alanine Residues in an α-Helix. J. Phys. Chem. B 2005, 109, 18652–18663. [Google Scholar] [CrossRef]
- Arkin, I.T. Isotope-edited IR spectroscopy for the study of membrane proteins. Curr. Opin. Chem. Biol. 2006, 10, 394–401. [Google Scholar] [CrossRef]
- Middleton, C.T.; Woys, A.M.; Mukherjee, S.S.; Zanni, M.T. Residue-specific structural kinetics of proteins through the union of isotope labeling, mid-IR pulse shaping, and coherent 2D IR spectroscopy. Methods 2010, 52, 12–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tadesse, L.; Nazarbaghi, R.; Walters, L. Isotopically Enhanced Infrared Spectroscopy: A Novel Method for Examining Secondary Structure at Specific Sites in Conformationally Heterogeneous Peptides. J. Am. Chem. Soc. 1991, 113, 7036–7037. [Google Scholar] [CrossRef]
- Decatur, S.M.; Antonic, J. Isotope-Edited Infrared Spectroscopy of Helical Peptides. J. Am. Chem. Soc. 1999, 121, 11914–11915. [Google Scholar] [CrossRef]
- Decatur, S.M. Elucidation of residue-level structure and dynamics of polypeptides via isotope-edited infrared spectroscopy. Acc. Chem. Res. 2006, 39, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Middleton, C.T.; Singh, S.; Reddy, A.S.; Woys, A.M.; Strasfeld, D.B.; Marek, P.; Raleigh, D.P.; de Pablo, J.J.; Zanni, M.T.; et al. 2DIR spectroscopy of human amylin fibrils reflects stable β-sheet structure. J. Am. Chem. Soc. 2011, 133, 16062–16071. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.Y.; Getahun, Z.; Wang, T.; DeGrado, W.F.; Gai, F. Time-resolved infrared study of the helix-coil transition using 13C-labeled helical peptides. J. Am. Chem. Soc. 2001, 123, 12111–12112. [Google Scholar] [CrossRef] [PubMed]
- Backus, E.H.; Bloem, R.; Pfister, R.; Moretto, A.; Crisma, M.; Toniolo, C.; Hamm, P. Dynamical transition in a small helical peptide and its implication for vibrational energy transport. J. Phys. Chem. B 2009, 113, 13405–13409. [Google Scholar] [CrossRef]
- Backus, E.H.; Bloem, R.; Donaldson, P.M.; Ihalainen, J.A.; Pfister, R.; Paoli, B.; Caflisch, A.; Hamm, P. 2D-IR study of a photoswitchable isotope-labeled α-helix. J. Phys. Chem. B 2010, 114, 3735–3740. [Google Scholar] [CrossRef]
- Remorino, A.; Korendovych, I.V.; Wu, Y.; DeGrado, W.F.; Hochstrasser, R.M. Residue-specific vibrational echoes yield 3D structures of a transmembrane helix dimer. Science 2011, 332, 1206–1209. [Google Scholar] [CrossRef]
- Ghosh, A.; Qiu, J.; DeGrado, W.F.; Hochstrasser, R.M. Tidal surge in the M2 proton channel, sensed by 2D IR spectroscopy. Proc. Natl. Acad. Sci. USA 2011, 108, 6115–6120. [Google Scholar] [CrossRef]
- Petty, S.A.; Decatur, S.M. Experimental evidence for the reorganization of β-strands within aggregates of the Aβ(16–22) peptide. J. Am. Chem. Soc. 2005, 127, 13488–13489. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, G.; Polavarapu, P.L. Isotope-assisted vibrational circular dichroism investigations of amyloid β peptide fragment, Aβ(16–22). J. Struct. Biol. 2011, 176, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Liu, L.; Axelsen, P.H.; Hochstrasser, R.M. Two-dimensional infrared spectra of isotopically diluted amyloid fibrils from Aβ40. Proc. Natl. Acad. Sci. USA 2008, 105, 7720–7725. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, W.; Sgourakis, N.G.; Li, Z.; Garcia, A.E.; Mukamel, S. Discriminating early stage Aβ42 monomer structures using chirality-induced 2D IR spectroscopy in a simulation study. Proc. Natl. Acad. Sci. USA 2010, 107, 15687–15692. [Google Scholar] [CrossRef] [PubMed]
- El Khoury, Y.; Dorlet, P.; Faller, P.; Hellwig, P. New insights into the coordination of Cu (II) by the amyloid-β 16 peptide from Fourier transform IR spectroscopy and isotopic labeling. J. Phys. Chem. B 2011, 115, 14812–14821. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, K.; Odaka, A.; Suzuki, N.; Ihara, Y. GM1 ganglioside-bound amyloid β-protein (Aβ): A possible form of preamyloid in Alzheimer’s disease. Nat. Med. 1995, 1, 1062–1066. [Google Scholar] [CrossRef] [PubMed]
- Okada, Y.; Okubo, K.; Ikeda, K.; Yano, Y.; Hoshino, M.; Hayashi, Y.; Kiso, Y.; Itoh-Watanabe, H.; Naito, A.; Matsuzaki, K. Toxic Amyloid Tape: A Novel Mixed Antiparallel/Parallel β-Sheet Structure Formed by Amyloid β-Protein on GM1 Clusters. ACS Chem. Neurosci. 2018. [Google Scholar] [CrossRef]
- Fukunaga, S.; Ueno, H.; Yamaguchi, T.; Yano, Y.; Hoshino, M.; Matsuzaki, K. GM1 cluster mediates formation of toxic Aβ fibrils by providing hydrophobic environments. Biochemistry 2012, 51, 8125–8131. [Google Scholar] [CrossRef]
- Kim, Y.S.; Liu, L.; Axelsen, P.H.; Hochstrasser, R.M. 2D IR provides evidence for mobile water molecules in β-amyloid fibrils. Proc. Natl. Acad. Sci. USA 2009, 106, 17751–17756. [Google Scholar] [CrossRef]
- Ma, J.; Komatsu, H.; Kim, Y.S.; Liu, L.; Hochstrasser, R.M.; Axelsen, P.H. Intrinsic structural heterogeneity and long-term maturation of amyloid β peptide fibrils. ACS Chem. Neurosci. 2013, 4, 1236–1243. [Google Scholar] [CrossRef]
- Falvo, C.; Zhuang, W.; Kim, Y.S.; Axelsen, P.H.; Hochstrasser, R.M.; Mukamel, S. Frequency distribution of the amide-I vibration sorted by residues in amyloid fibrils revealed by 2D-IR measurements and simulations. J. Phys. Chem. B 2012, 116, 3322–3330. [Google Scholar] [CrossRef]
- Shim, S.H.; Gupta, R.; Ling, Y.L.; Strasfeld, D.B.; Raleigh, D.P.; Zanni, M.T. Two-dimensional IR spectroscopy and isotope labeling defines the pathway of amyloid formation with residue-specific resolution. Proc. Natl. Acad. Sci. USA 2009, 106, 6614–6619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woys, A.M.; Almeida, A.M.; Wang, L.; Chiu, C.C.; McGovern, M.; de Pablo, J.J.; Skinner, J.L.; Gellman, S.H.; Zanni, M.T. Parallel β-sheet vibrational couplings revealed by 2D IR spectroscopy of an isotopically labeled macrocycle: Quantitative benchmark for the interpretation of amyloid and protein infrared spectra. J. Am. Chem. Soc. 2012, 134, 19118–19128. [Google Scholar] [CrossRef] [PubMed]
- Dunkelberger, E.B.; Buchanan, L.E.; Marek, P.; Cao, P.; Raleigh, D.P.; Zanni, M.T. Deamidation accelerates amyloid formation and alters amylin fiber structure. J. Am. Chem. Soc. 2012, 134, 12658–12667. [Google Scholar] [CrossRef]
- Buchanan, L.E.; Dunkelberger, E.B.; Tran, H.Q.; Cheng, P.N.; Chiu, C.C.; Cao, P.; Raleigh, D.P.; de Pablo, J.J.; Nowick, J.S.; Zanni, M.T. Mechanism of IAPP amyloid fibril formation involves an intermediate with a transient β-sheet. Proc. Natl. Acad. Sci. USA 2013, 110, 19285–19290. [Google Scholar] [CrossRef] [PubMed]
- Serrano, A.L.; Lomont, J.P.; Tu, L.H.; Raleigh, D.P.; Zanni, M.T. A Free Energy Barrier Caused by the Refolding of an Oligomeric Intermediate Controls the Lag Time of Amyloid Formation by hIAPP. J. Am. Chem. Soc. 2017, 139, 16748–16758. [Google Scholar] [CrossRef] [Green Version]
- Maj, M.; Lomont, J.P.; Rich, K.L.; Alperstein, A.M.; Zanni, M.T. Site-specific detection of protein secondary structure using 2D IR dihedral indexing: A proposed assembly mechanism of oligomeric hIAPP. Chem. Sci. 2018, 9, 463–474. [Google Scholar] [CrossRef]
- Middleton, C.T.; Marek, P.; Cao, P.; Chiu, C.C.; Singh, S.; Woys, A.M.; de Pablo, J.J.; Raleigh, D.P.; Zanni, M.T. Two-dimensional infrared spectroscopy reveals the complex behaviour of an amyloid fibril inhibitor. Nat. Chem. 2012, 4, 355–360. [Google Scholar] [CrossRef] [Green Version]
- Cao, P.; Meng, F.; Abedini, A.; Raleigh, D.P. The ability of rodent islet amyloid polypeptide to inhibit amyloid formation by human islet amyloid polypeptide has important implications for the mechanism of amyloid formation and the design of inhibitors. Biochemistry 2010, 49, 872–881. [Google Scholar] [CrossRef]
- Moran, S.D.; Woys, A.M.; Buchanan, L.E.; Bixby, E.; Decatur, S.M.; Zanni, M.T. Two-dimensional IR spectroscopy and segmental 13C labeling reveals the domain structure of human γD-crystallin amyloid fibrils. Proc. Natl. Acad. Sci. USA 2012, 109, 3329–3334. [Google Scholar] [CrossRef]
- Moran, S.D.; Decatur, S.M.; Zanni, M.T. Structural and sequence analysis of the human γD-crystallin amyloid fibril core using 2D IR spectroscopy, segmental 13C labeling, and mass spectrometry. J. Am. Chem. Soc. 2012, 134, 18410–18416. [Google Scholar] [CrossRef] [PubMed]
- Lam, A.R.; Moran, S.D.; Preketes, N.K.; Zhang, T.O.; Zanni, M.T.; Mukamel, S. Study of the γD-crystallin protein using two-dimensional infrared (2D IR) spectroscopy: Experiment and simulation. J. Phys. Chem. B 2013, 117, 15436–15443. [Google Scholar] [CrossRef] [PubMed]
- Moran, S.D.; Zhang, T.O.; Zanni, M.T. An alternative structural isoform in amyloid-like aggregates formed from thermally denatured human γD-crystallin. Protein Sci. 2014, 23, 321–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, R.A.; Barber-Armstrong, W.; Decatur, S.M. The organization and assembly of a β-sheet formed by a prion peptide in solution: An isotope-edited FTIR study. J. Am. Chem. Soc. 2003, 125, 13674–13675. [Google Scholar] [CrossRef] [PubMed]
- Petty, S.A.; Adalsteinsson, T.; Decatur, S.M. Correlations among morphology, β-sheet stability, and molecular structure in prion peptide aggregates. Biochemistry 2005, 44, 4720–4726. [Google Scholar] [CrossRef] [PubMed]
- Petty, S.A.; Decatur, S.M. Intersheet rearrangement of polypeptides during nucleation of β-sheet aggregates. Proc. Natl. Acad. Sci. USA 2005, 102, 14272–14277. [Google Scholar] [CrossRef] [PubMed]
- Chi, H.; Welch, W.R.; Kubelka, J.; Keiderling, T.A. Insight into the packing pattern of β2 fibrils: A model study of glutamic acid rich oligomers with 13C isotopic edited vibrational spectroscopy. Biomacromolecules 2013, 14, 3880–3891. [Google Scholar] [CrossRef]
- Buchanan, L.E.; Carr, J.K.; Fluitt, A.M.; Hoganson, A.J.; Moran, S.D.; de Pablo, J.J.; Skinner, J.L.; Zanni, M.T. Structural motif of polyglutamine amyloid fibrils discerned with mixed-isotope infrared spectroscopy. Proc. Natl. Acad. Sci. USA 2014, 111, 5796–5801. [Google Scholar] [CrossRef] [Green Version]
- Paul, C.; Wang, J.; Wimley, W.C.; Hochstrasser, R.M.; Axelsen, P.H. Vibrational coupling, isotopic editing, and β-sheet structure in a membrane-bound polypeptide. J. Am. Chem. Soc. 2004, 126, 5843–5850. [Google Scholar] [CrossRef]
- Londergan, C.H.; Wang, J.; Axelsen, P.H.; Hochstrasser, R.M. Two-dimensional infrared spectroscopy displays signatures of structural ordering in peptide aggregates. Biophys. J. 2006, 90, 4672–4685. [Google Scholar] [CrossRef]
- Barth, A. The infrared absorption of amino acid side chains. Prog. Biophys. Mol. Biol. 2000, 74, 141–173. [Google Scholar] [CrossRef]
- Bloem, R.; Koziol, K.; Waldauer, S.A.; Buchli, B.; Walser, R.; Samatanga, B.; Jelesarov, I.; Hamm, P. Ligand binding studied by 2D IR spectroscopy using the azidohomoalanine label. J. Phys. Chem. B 2012, 116, 13705–13712. [Google Scholar] [CrossRef] [PubMed]
- Getahun, Z.; Huang, C.Y.; Wang, T.; De Leon, B.; DeGrado, W.F.; Gai, F. Using nitrile-derivatized amino acids as infrared probes of local environment. J. Am. Chem. Soc. 2003, 125, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Waegele, M.M.; Culik, R.M.; Gai, F. Site-Specific Spectroscopic Reporters of the Local Electric Field, Hydration, Structure, and Dynamics of Biomolecules. J. Phys. Chem. Lett. 2011, 2, 2598–2609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindquist, B.A.; Furse, K.E.; Corcelli, S.A. Nitrile groups as vibrational probes of biomolecular structure and dynamics: An overview. Phys. Chem. Chem. Phys. 2009, 11, 8119–8132. [Google Scholar] [CrossRef] [PubMed]
- Fafarman, A.T.; Webb, L.J.; Chuang, J.I.; Boxer, S.G. Site-specific conversion of cysteine thiols into thiocyanate creates an IR probe for electric fields in proteins. J. Am. Chem. Soc. 2006, 128, 13356–13357. [Google Scholar] [CrossRef] [PubMed]
- Van Wilderen, L.J.; Kern-Michler, D.; Müller-Werkmeister, H.M.; Bredenbeck, J. Vibrational dynamics and solvatochromism of the label SCN in various solvents and hemoglobin by time dependent IR and 2D-IR spectroscopy. Phys. Chem. Chem. Phys. 2014, 16, 19643–19653. [Google Scholar] [CrossRef] [Green Version]
- Miller, C.S.; Ploetz, E.A.; Cremeens, M.E.; Corcelli, S.A. Carbon-deuterium vibrational probes of peptide conformation: Alanine dipeptide and glycine dipeptide. J. Chem. Phys. 2009, 130, 125103. [Google Scholar] [CrossRef]
- Yu, W.; Dawson, P.E.; Zimmermann, J.; Romesberg, F.E. Carbon-deuterium bonds as probes of protein thermal unfolding. J. Phys. Chem. B 2012, 116, 6397–6403. [Google Scholar] [CrossRef]
- Hoffman, K.W.; Romei, M.G.; Londergan, C.H. A new Raman spectroscopic probe of both the protonation state and noncovalent interactions of histidine residues. J. Phys. Chem. A 2013, 117, 5987–5996. [Google Scholar] [CrossRef]
- Bazewicz, C.G.; Liskov, M.T.; Hines, K.J.; Brewer, S.H. Sensitive, site-specific, and stable vibrational probe of local protein environments: 4-azidomethyl-l-phenylalanine. J. Phys. Chem. B 2013, 117, 8987–8993. [Google Scholar] [CrossRef] [PubMed]
- Lipkin, J.S.; Song, R.; Fenlon, E.E.; Brewer, S.H. Modulating Accidental Fermi Resonance: What a Difference a Neutron Makes. J. Phys. Chem. Lett. 2011, 2011, 1672–1676. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Viloca, M.; Nam, K.; Alhambra, C.; Gao, J. Solvent and protein effects on the vibrational frequency shift and energy relaxation of the azide ligand in carbonic anhydrase. J. Phys. Chem. B 2004, 108, 13501–13512. [Google Scholar] [CrossRef]
- Ye, S.; Huber, T.; Vogel, R.; Sakmar, T.P. FTIR analysis of GPCR activation using azido probes. Nat. Chem. Biol. 2009, 5, 397–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tucker, M.J.; Gai, X.S.; Fenlon, E.E.; Brewer, S.H.; Hochstrasser, R.M. 2D IR photon echo of azido-probes for biomolecular dynamics. Phys. Chem. Chem. Phys. 2011, 13, 2237–2241. [Google Scholar] [CrossRef] [PubMed]
- Thielges, M.C.; Axup, J.Y.; Wong, D.; Lee, H.S.; Chung, J.K.; Schultz, P.G.; Fayer, M.D. Two-dimensional IR spectroscopy of protein dynamics using two vibrational labels: A site-specific genetically encoded unnatural amino acid and an active site ligand. J. Phys. Chem. B 2011, 115, 11294–11304. [Google Scholar] [CrossRef]
- Wolfshorndl, M.P.; Baskin, R.; Dhawan, I.; Londergan, C.H. Covalently bound azido groups are very specific water sensors, even in hydrogen-bonding environments. J. Phys. Chem. B 2012, 116, 1172–1179. [Google Scholar] [CrossRef]
- Oh, K.I.; Lee, J.H.; Joo, C.; Han, H.; Cho, M. β-azidoalanine as an IR probe: Application to amyloid Aβ(16–22) aggregation. J. Phys. Chem. B 2008, 112, 10352–10357. [Google Scholar] [CrossRef]
- Velarde, L.; Wang, H.F. Capturing inhomogeneous broadening of the -CN stretch vibration in a Langmuir monolayer with high-resolution spectra and ultrafast vibrational dynamics in sum-frequency generation vibrational spectroscopy (SFG-VS). J. Chem. Phys. 2013, 139, 084204. [Google Scholar] [CrossRef]
- Waegele, M.M.; Tucker, M.J.; Gai, F. 5-Cyanotryptophan as an Infrared Probe of Local Hydration Status of Proteins. Chem. Phys. Lett. 2009, 478, 249–253. [Google Scholar] [CrossRef]
- Tucker, M.J.; Kim, Y.S.; Hochstrasser, R.M. 2D IR photon echo study of the anharmonic coupling in the OCN region of phenyl cyanate. Chem. Phys. Lett. 2009, 470, 80–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weeks, C.L.; Jo, H.; Kier, B.; DeGrado, W.F.; Spiro, T.G. Cysteine-linked aromatic nitriles as UV resonance Raman probes of protein structure. J. Raman Specrosc. 2012, 43, 1244–1249. [Google Scholar] [CrossRef]
- Tucker, M.J.; Oyola, R.; Gai, F. Conformational distribution of a 14-residue peptide in solution: A fluorescence resonance energy transfer study. J. Phys. Chem. B 2005, 109, 4788–4795. [Google Scholar] [CrossRef] [PubMed]
- Tucker, M.J.; Oyola, R.; Gai, F. A novel fluorescent probe for protein binding and folding studies: P-cyano-phenylalanine. Biopolymers 2006, 83, 571–576. [Google Scholar] [CrossRef] [PubMed]
- Aprilakis, K.N.; Taskent, H.; Raleigh, D.P. Use of the novel fluorescent amino acid p-cyanophenylalanine offers a direct probe of hydrophobic core formation during the folding of the N-terminal domain of the ribosomal protein L9 and provides evidence for two-state folding. Biochemistry 2007, 46, 12308–12313. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Yin, H.; Qiu, J.; Tucker, M.J.; DeGrado, W.F.; Gai, F. Using two fluorescent probes to dissect the binding, insertion, and dimerization kinetics of a model membrane peptide. J. Am. Chem. Soc. 2009, 131, 3816–3817. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, J.; Thielges, M.C.; Seo, Y.J.; Dawson, P.E.; Romesberg, F.E. Cyano groups as probes of protein microenvironments and dynamics. Angew. Chem. Int. Ed. 2011, 50, 8333–8337. [Google Scholar] [CrossRef]
- Chung, J.K.; Thielges, M.C.; Fayer, M.D. Conformational dynamics and stability of HP35 studied with 2D IR vibrational echoes. J. Am. Chem. Soc. 2012, 134, 12118–12124. [Google Scholar] [CrossRef]
- Suydam, I.T.; Snow, C.D.; Pande, V.S.; Boxer, S.G. Electric fields at the active site of an enzyme: Direct comparison of experiment with theory. Science 2006, 313, 200–204. [Google Scholar] [CrossRef]
- Fafarman, A.T.; Boxer, S.G. Nitrile bonds as infrared probes of electrostatics in ribonuclease S. J. Phys. Chem. B 2010, 114, 13536–13544. [Google Scholar] [CrossRef]
- Lindquist, B.A.; Corcelli, S.A. Nitrile Groups as Vibrational Probes: Calculations of the CN Infrared Absorption Line Shape of Acetonitrile in Water and Tetrahydrofuran. J. Phys. Chem. B 2008, 112, 6301–6303. [Google Scholar] [CrossRef] [PubMed]
- Weeks, C.L.; Polishchuk, A.; Getahun, Z.; Degrado, W.F.; Spiro, T.G. Investigation of an unnatural amino acid for use as a resonance Raman probe: Detection limits, solvent and temperature dependence of the vCN band of 4-cyanophenylalanine. J. Raman Spectrosc. 2008, 39, 1606–1613. [Google Scholar] [CrossRef] [PubMed]
- Schultz, K.C.; Supekova, L.; Ryu, Y.; Xie, J.; Perera, R.; Schultz, P.G. A genetically encoded infrared probe. J. Am. Chem. Soc. 2006, 128, 13984–13985. [Google Scholar] [CrossRef] [PubMed]
- Miyake-Stoner, S.J.; Miller, A.M.; Hammill, J.T.; Peeler, J.C.; Hess, K.R.; Mehl, R.A.; Brewer, S.H. Probing protein folding using site-specifically encoded unnatural amino acids as FRET donors with tryptophan. Biochemistry 2009, 48, 5953–5962. [Google Scholar] [CrossRef] [PubMed]
- Tucker, M.J.; Tang, J.; Gai, F. Probing the kinetics of membrane-mediated helix folding. J. Phys. Chem. B 2006, 110, 8105–8109. [Google Scholar] [CrossRef]
- Glasscock, J.M.; Zhu, Y.; Chowdhury, P.; Tang, J.; Gai, F. Using an amino acid fluorescence resonance energy transfer pair to probe protein unfolding: Application to the villin headpiece subdomain and the LysM domain. Biochemistry 2008, 47, 11070–11076. [Google Scholar] [CrossRef]
- Taskent-Sezgin, H.; Chung, J.; Patsalo, V.; Miyake-Stoner, S.J.; Miller, A.M.; Brewer, S.H.; Mehl, R.A.; Green, D.F.; Raleigh, D.P.; Carrico, I. Interpretation of p-cyanophenylalanine fluorescence in proteins in terms of solvent exposure and contribution of side-chain quenchers: A combined fluorescence, IR and molecular dynamics study. Biochemistry 2009, 48, 9040–9046. [Google Scholar] [CrossRef]
- Taskent-Sezgin, H.; Marek, P.; Thomas, R.; Goldberg, D.; Chung, J.; Carrico, I.; Raleigh, D.P. Modulation of p-cyanophenylalanine fluorescence by amino acid side chains and rational design of fluorescence probes of α-helix formation. Biochemistry 2010, 49, 6290–6295. [Google Scholar] [CrossRef]
- Marek, P.; Mukherjee, S.; Zanni, M.T.; Raleigh, D.P. Residue-specific, real-time characterization of lag-phase species and fibril growth during amyloid formation: A combined fluorescence and IR study of p-cyanophenylalanine analogs of islet amyloid polypeptide. J. Mol. Biol. 2010, 400, 878–888. [Google Scholar] [CrossRef]
- Inouye, H.; Gleason, K.A.; Zhang, D.; Decatur, S.M.; Kirschner, D.A. Differential effects of Phe19 and Phe20 on fibril formation by amyloidogenic peptide Aβ16–22 (Ac-KLVFFAE-NH2). Proteins 2010, 78, 2306–2321. [Google Scholar] [CrossRef]
- Liu, H.; Lantz, R.; Cosme, P.; Rivera, N.; Andino, C.; Gonzalez, W.G.; Terentis, A.C.; Wojcikiewicz, E.P.; Oyola, R.; Miksovska, J.; et al. Site-specific dynamics of amyloid formation and fibrillar configuration of Aβ(1–23) using an unnatural amino acid. Chem. Commun. 2015, 51, 7000–7003. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Morris, C.; Lantz, R.; Kent, T.W.; Elbassal, E.A.; Wojcikiewicz, E.P.; Du, D. Residue-Specific Dynamics and Local Environmental Changes in Aβ40 Oligomer and Fibril Formation. Angew. Chem. Int. Ed. 2018, 57, 8017–8021. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Cho, M. Vibrational solvatochromism and electrochromism of infrared probe molecules containing C≡O, C≡N, C=O, or C-F vibrational chromophore. J. Chem. Phys. 2011, 134, 154513. [Google Scholar] [CrossRef] [PubMed]
- Pazos, I.M.; Ghosh, A.; Tucker, M.J.; Gai, F. Ester carbonyl vibration as a sensitive probe of protein local electric field. Angew. Chem. Int. Ed. 2014, 53, 6080–6084. [Google Scholar] [CrossRef] [PubMed]
- Pazos, I.M.; Ma, J.; Mukherjee, D.; Gai, F. Ultrafast Hydrogen-Bonding Dynamics in Amyloid Fibrils. J. Phys. Chem. B 2018, 122, 11023–11029. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, I.A.; Gai, F. Simple method to introduce an ester infrared probe into proteins. Prot. Sci. 2017, 26, 375–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]

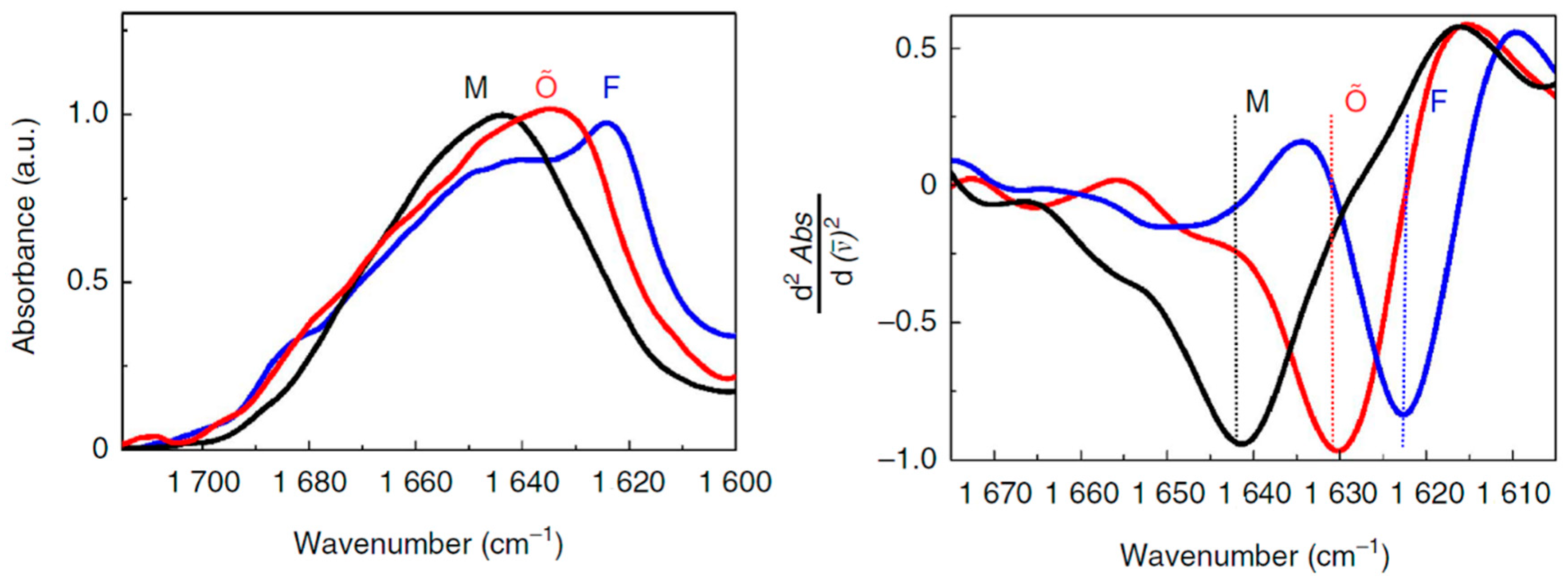


| Secondary Structure | Band Position in H2O/cm−1 | Band Position in D2O/cm−1 | ||
|---|---|---|---|---|
| Average | Extremes | Average | Extremes | |
| α-helix | 1654 | 1648–1657 | 1652 | 1642–1660 |
| β-sheet | 1633 | 1623–1641 | 1630 | 1615–1638 |
| β-sheet | 1684 | 1674–1695 | 1679 | 1672–1694 |
| Turns | 1672 | 1662–1686 | 1671 | 1653–1691 |
| Disordered | 1654 | 1642–1657 | 1645 | 1639–1654 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Lantz, R.; Du, D. Vibrational Approach to the Dynamics and Structure of Protein Amyloids. Molecules 2019, 24, 186. https://doi.org/10.3390/molecules24010186
Li H, Lantz R, Du D. Vibrational Approach to the Dynamics and Structure of Protein Amyloids. Molecules. 2019; 24(1):186. https://doi.org/10.3390/molecules24010186
Chicago/Turabian StyleLi, Haoqian, Richard Lantz, and Deguo Du. 2019. "Vibrational Approach to the Dynamics and Structure of Protein Amyloids" Molecules 24, no. 1: 186. https://doi.org/10.3390/molecules24010186
APA StyleLi, H., Lantz, R., & Du, D. (2019). Vibrational Approach to the Dynamics and Structure of Protein Amyloids. Molecules, 24(1), 186. https://doi.org/10.3390/molecules24010186