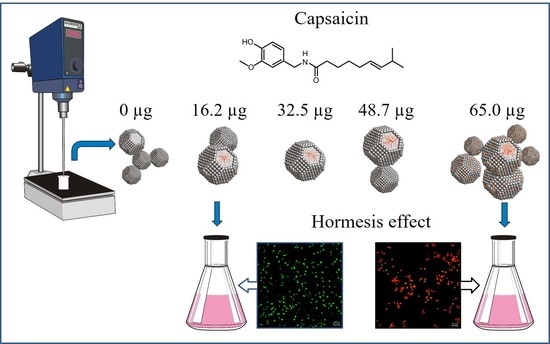
Generation of BSA-capsaicin Nanoparticles and Their Hormesis Effect on the Rhodotorula mucilaginosa Yeast
Abstract
:1. Introduction
2. Results and Discussion
2.1. Yield and Efficiency of the BSA-Capsaicin Nanoparticles
2.2. Structural Changes of BSA-Capsaicin Nanoparticles Characterized by FTIR
2.3. Morphology of Nanoparticles
2.4. Effective Diameter (Ed), Aspect Ratio (Ar) and Shape Factor (Sf) of the Nanoparticles
2.5. ζ-Potential, Polydispersity Index (PDI) and Hydrodynamic Diameter of Aggregates
2.6. Isolation and Identification of R. mucilaginosa
2.7. Hormesis Effect of NPs BSA-Capsaicin in R. mucilaginosa
2.8. Cellular Uptake and Release of Capsaicin
3. Materials and Methods
3.1. Reagents and Chemicals
3.2. Preparation of BSA-Capsaicin Nanoparticles
3.3. Determination of ζ-Potential, Polydispersity Index (PDI) and Hydrodynamic Diameter of Aggregates
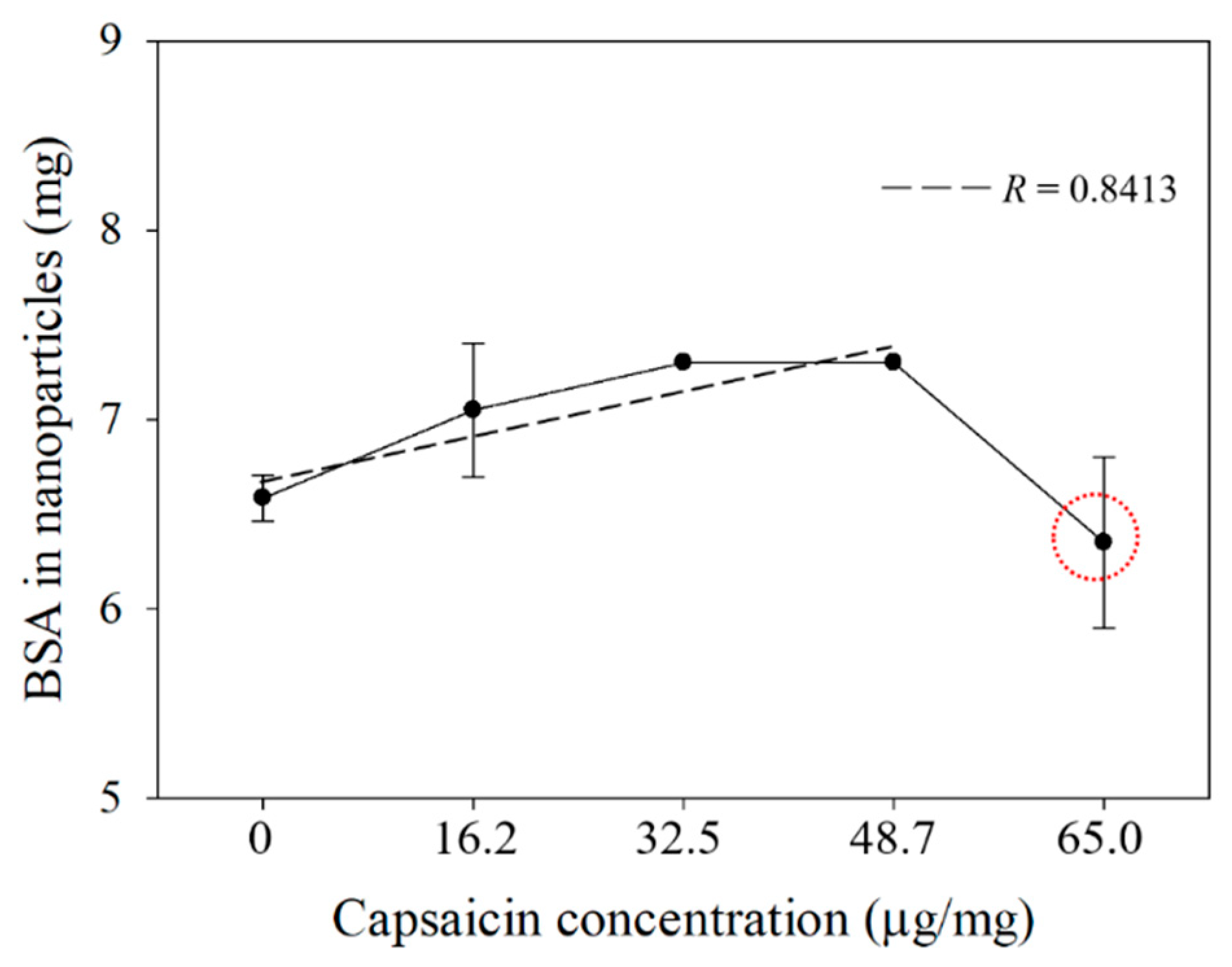
3.4. Quantification of BSA Transformed in Nanoparticles and Encapsulated Capsaicin
3.5. Fourier Transform Infrared Spectroscopy (FTIR) of BSA-Capsaicin Nanoparticles
3.6. Transmission Electron Microscopy (TEM) and Morphometric Analysis of Nanoparticles
3.7. Yeast Sampling and Genetic Identification of R. mucilaginosa
3.8. Nanoparticle Staining with Rhodamine B and Uptake by R. mucilaginosa
3.9. Determination of Viability of R. mucilaginosa
3.10. BSA-Capsaicin Nanoparticles Uptake by R. mucilaginosa and Intracellular Distribution
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Perucka, I.; Oleszek, W. Extraction and determination of capsaicinoids in fruit of hot pepper Capsicum annuum L. by spectrophotometry and high-performance liquid chromatography. Food Chem. 2000, 71, 287–291. [Google Scholar] [CrossRef]
- Kraikruan, W.; Sangchote, S.; Sukprakarn, S. Effect of capsaicin on germination of Colletotrichum capsici conidia. Nat. Sci. 2008, 42, 417–422. [Google Scholar]
- Antonious, F.G.; Lobel, L.; Kochhar, T.; Berke, T.; Jarret, R.L. Antioxidants in Capsicum chinense: Variation among countries of origin. J. Environ. Sci. Health B 2009, 44, 621–626. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, K. Biological activities of red pepper (Capsicum annuum) and its pungent principle capsaicin: A review. Crit. Rev. Food Sci. Nutr. 2016, 56, 1488–1500. [Google Scholar] [CrossRef] [PubMed]
- Kopec, E.S.; DeBellis, J.R.; Irwin, S.R. Chemical analysis of freshly prepared and stored capsaicin solutions: Implications for tussigenic challenges. Pulm. Pharmacol. Ther. 2002, 15, 529–534. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, S.; Xu, S.; Xue, Y.; Lou, J. Structure and properties analysis of microcapsulated capsaicin prepared by diphase emulsion method. Adv. Mat. Res. 2011, 239–242, 3182–3185. [Google Scholar] [CrossRef]
- Wang, J.; Dong, X.; Chena, S.; Lou, J. Microencapsulation of capsaicin by solvent evaporation method and thermal stability study of microcapsules. Colloid J. 2013, 75, 26–33. [Google Scholar] [CrossRef]
- Sánchez-Segura, L.; García-Armenta, E.; de Jesús Perea-Flores, M.; Téllez-Medina, D.I.; Carpio-Pedroza, J.C.; Hernández-Sánchez, H.; Alamilla-Beltrán, L.; Gutiérrez-López, G.F. Food Nanoscience and Nanotechnology, 1st ed.; Springer Science+Business Media: New York, NY, USA, 2015; pp. 187–203. [Google Scholar]
- Sánchez-Segura, L.; Ochoa-Alejo, N.; Carriles, N.; Zavala-García, E.L. Development of bovine serum albumin-capsaicin nanoparticles for biotechnological applications. Appl. Nanosci. 2018, 8, 1877–1886. [Google Scholar] [CrossRef]
- Langer, K.; Balthasar, S.; Vogel, V.; Dinauer, N.; von Briesen, H.; Schubert, D. Optimization of the preparation process for human serum albumin (HSA) nanoparticles. Int. J. Pharm. 2003, 257, 169–180. [Google Scholar] [CrossRef]
- Weber, C.; Coester, C.; Kreuter, J.; Langer, K. Desolvation process and surface characterisation of protein nanoparticles. Int. J. Pharm. 2000, 194, 91–102. [Google Scholar] [CrossRef]
- Wirth, F.; Goldani, Z.L. Epidemiology of Rhodotorula: An emerging pathogen. Interdiscip. Perspect. Infecs. Dis. 2012, 1–7. [Google Scholar]
- Falces-Romero, I.; Cendejas-Bueno, E.; Romero-Gómez, M.P.; García-Rodríguez, J. Isolation of Rhodotorula mucilaginosa from blood cultures in a tertiary care hospital. Mycoses 2018, 61, 35–39. [Google Scholar] [CrossRef]
- Villar, J.M.; Delgado, J.D.J.; Velasco, C.G. Fungemia due to Rhodotorula mucilaginosa in an immunocompetent, critically ill patient. J. Infect. Chemother. 2012, 18, 581–583. [Google Scholar] [CrossRef]
- Kurita, S.; Kitagawa, E.; Kim, H.C.; Momose, Y.M.; Iwahashi, H. Studies on the antimicrobial mechanisms of capsaicin using yeast DNA microarray. Biosci. Biotechnol. Biochem. 2002, 66, 532–536. [Google Scholar] [CrossRef]
- Baler, K.; Martin, O.A.; Carignano, M.A.; Ameer, G.A.; Vila, J.A.; Szleifer, I. Electrostatic unfolding and interactions of albumin driven by pH changes: A molecular dynamics study. J. Phys. Chem. B 2014, 118, 921–930. [Google Scholar] [CrossRef]
- De Freitas, G.B.L.; De Almeida, D.J.; Carraro, E.; Kerppers, I.I.; Martins, G.A.G.; Mainardes, R.M.; Khalil, N.M.; Messias-Reason, I.J.T. Formulation, characterization, and in vitro/in vivo studies of capsaicin-loaded albumin nanoparticles. Mater. Sci. Eng. C 2018, 93, 70–79. [Google Scholar] [CrossRef]
- Luebbert, C.C.; Clarke, T.M.; Pointet, R.; Frahm, G.E.; Tam, S.; Lorbetskie, B.; Sauvé, S.; Johnston, W.J.M. Nanoparticle size and production efficiency are affected by the presence of fatty acids during albumin nanoparticle fabrication. PLoS ONE 2017, 12, e0189814. [Google Scholar] [CrossRef]
- Bhalekar, M.; Upadhaya, P.; Madgulkar, A. Formulation and characterization of solid lipid nanoparticles for an anti-retroviral drug darunavir. Appl. Nanosci. 2017, 7, 47–57. [Google Scholar] [CrossRef] [Green Version]
- Jincheng, W.; Xiaoyu, Z.; Siahao, C. Preparation and properties of nanoencapsulated capsaicin by complex coacervation method. Chem. Eng. Commun. 2010, 197, 919–933. [Google Scholar] [CrossRef]
- Peng, W.; Jiang, X.Y.; Zhu, Y.; Omari-Siaw, E.; Deng, W.W.; Yu, J.N.; Xu, X.M.; Zhang, W.M. Oral delivery of capsaicin using MPEG-PCL nanoparticles. Acta Pharmacol. Sin. 2015, 36, 139–148. [Google Scholar] [CrossRef]
- Leela, J.S.P.P.; Hemamalini, R.; Muthu, S.; Al-Saadi, A.A. Spectroscopic investigation (FTIR spectrum), NBO, HOMO–LUMO energies, NLO and thermodynamic properties of 8-Methyl-N-vanillyl-6-nonenamideby DFT methods. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2015, 146, 177–186. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, L.; Devi, P.; Shridhar, D.M.; Naik, C.G. Use of Fourier Transform Infrared (FTIR) spectroscopy to study cadmium-induced changes in Padina tetrastromatica (Hauck). Anal. Chem. Insights. 2008, 3, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Escogido, M.D.L.; Gonzalez-Mondragon, E.G.; Vazquez-Tzompantzi, E. Chemical and pharmacological aspects of capsaicin. Molecules 2011, 16, 1253–1270. [Google Scholar] [CrossRef] [PubMed]
- Bronze-Uhle, E.S.; Costa, B.C.; Ximenes, V.F.; Lisboa-Filho, P.N. Synthetic nanoparticles of bovine serum albumin with entrapped salicylic acid. Nanotechnol. Sci. Appl. 2017, 10, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Anand, B.G.; Dubey, K.; Shekhawat, D.S.; Kar, K. Capsaicin-coated silver nanoparticles inhibit amyloid fibril formation of serum albumin. Biochemistry 2016, 55, 3345–3348. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, L.; Prenna, G.; Okolicsanyi, L.; Gautier, A. Electron staining with uranyl acetate. Possible role of free amino groups. J. Histochem. Cytochem. 1971, 19, 161–188. [Google Scholar] [CrossRef] [PubMed]
- Maghsoudi, A.; Shojaosadati, S.A.; Farahani, E.V. 5-Fluorouracil-loaded BSA nanoparticles: Formulation optimization and in vitro release study. AAPS. Pharm. Sci. Tech. 2008, 9, 1092–1096. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, X.; Zu, Y.; Han, X.; Ge, Y.; Wang, W.; Yu, X. A novel active targeting preparation, vinorelbine tartrate (VLBT) encapsulated by folate-conjugated bovine serum albumin (BSA) nanoparticles: Preparation, characterization and in vitro release study. Materials 2012, 5, 2403–2422. [Google Scholar] [CrossRef]
- Choi, A.J.; Kim, C.J.; Cho, Y.J.; Hwang, J.K.; Kim, C.T. Characterization of capsaicin-loaded nanoemulsions stabilized with alginate and chitosan by self-assembly. Food Bioprocess Tech. 2011, 4, 1119–1126. [Google Scholar] [CrossRef]
- Ikeda, S.; Morris, V.J. Fine-stranded and particulate aggregates of heat-denatured whey protein visualized by atomic force microscopy. Biomacromolecules 2002, 3, 382–389. [Google Scholar] [CrossRef]
- Li, R.; Wu, Z.; Wangb, Y.; Ding, L.; Wang, Y. Role of pH-induced structural change in protein aggregation in foam fractionation of bovine serum albumin. Biotechnol. Rep. (Amst) 2016, 9, 46–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mattson, M.P. Hormesis defined. Ageing. Res. Rev. 2008, 7, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Kato-Noguchi, H.; Tanaka, Y. Effects of capsaicin on plant growth. Biol. Plantarum. 2003, 47, 157–159. [Google Scholar] [CrossRef]
- Young, S.L.; Sarda, X.; Rosenberg, M. Microencapsulating properties of whey proteins. 2. Combination of whey protein with carbohydrates. J. Dairy Sci. 1993, 76, 2878–2885. [Google Scholar] [CrossRef]
- Jahanban-Esfahlan, A.; Dastmalchi, S.; Davaran, S. A simple improved desolvation method for the rapid preparation of albumin nanoparticles. Int. J. Biol. Macromol. 2016, 91, 703–709. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.H.; Nazari, M.; Raval, G.; Khan, Z.; Patel, H.; Heerklotz, H. Utilizing zeta potential measurements to study the effective charge, membrane partitioning, and membrane permeation of the lipopeptide surfactin. Biochim. Biophys. Acta. 2014, 1838, 2306–2312. [Google Scholar] [CrossRef] [Green Version]
- Sganzerla, M.; Coutinho, P.J.; Tavares de Melo, M.A.; Godoy, T.H. Fast method for capsaicinoids analysis from Capsicum chinense fruits. Food Rest. Int. 2014, 64, 718–725. [Google Scholar] [CrossRef]
- Syverud, K.; Chinga, G.; Per Olav, J.; Ingebjorg, L.; Knut, W. Analysis of lint particles from full-scale printing trials. Appita J. 2007, 60, 286–290. [Google Scholar]
- Parakhonskiy, B.; Zyuzin, V.M.; Yashchenok, A.; Carregal-Romero, S.; Rejman, J.; Möhwald, H.; Parak, J.W.; Skirtach, G.A. The influence of the size and aspect ratio of anisotropic, porous CaCO3 particles on their uptake by cells. J. Nanobiotechnology 2015, 53, 1–13. [Google Scholar] [CrossRef]
- Bouwman, M.A.; Bosma, C.J.; Vonk, P.; Wesselingh, A.J.; Frijlink, W.H. Which shape factor(s) best describe granules? Powder Technol. 2004, 146, 66–72. [Google Scholar] [CrossRef]
- Hoffman, C.S.; Winston, F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 1987, 57, 267–272. [Google Scholar] [CrossRef]
- Toju, H.; Tanabe, A.S.; Yamamoto, S.; Sato, H. High-Coverage ITS primers for the DNA-based identification of Ascomycetes and Basidiomycetes in environmental samples. PLoS ONE 2012, 7, e40863. [Google Scholar] [CrossRef] [PubMed]
- Manzo-Valencia, M.K.; Valdés-Santiago, L.; Sánchez-Segura, L.; Guzmán-de-Peña, D.L. Naphthalene acetic acid potassium salt (NAA-K+) affects conidial germination, sporulation, mycelial growth, cell surface morphology, and viability of Fusarium oxysporum f. sp. radici-lycopersici and F. oxysporum f. sp. cubense in vitro. J. Agric. Food. Chem. 2016, 64, 8315–8323. [Google Scholar]
- Yang, Q.; Li, Y.; Apaliya, M.T.; Zheng, X.; Serwah, B.N.A.; Zhang, X.; Zhang, H. The response of Rhodotorula mucilaginosa to patulin based on lysine crotonylation. Front. Microbiol. 2018, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |










| Capsaicin Concentration (µg/mg) | BSA Nanoparticles Yield (%) a | Encapsulated Efficiency (%) b |
|---|---|---|
| 0 | 72.4 ± 1.3 | 0.0 ± 0.0 |
| 16.2 | 77.5 ± 3.8 | 60.9 ± 8.3 |
| 32.5 | 80.3 ± 0.0 | 70.8 ± 2.9 |
| 48.7 | 80.3 ± 0.0 | 72.2 ± 0.9 |
| 65.0 | 69.8 ± 4.9 | 55.1 ± 10.4 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Arreguin, A.; Carriles, R.; Ochoa-Alejo, N.; López, M.G.; Sánchez-Segura, L. Generation of BSA-capsaicin Nanoparticles and Their Hormesis Effect on the Rhodotorula mucilaginosa Yeast. Molecules 2019, 24, 2800. https://doi.org/10.3390/molecules24152800
Sánchez-Arreguin A, Carriles R, Ochoa-Alejo N, López MG, Sánchez-Segura L. Generation of BSA-capsaicin Nanoparticles and Their Hormesis Effect on the Rhodotorula mucilaginosa Yeast. Molecules. 2019; 24(15):2800. https://doi.org/10.3390/molecules24152800
Chicago/Turabian StyleSánchez-Arreguin, Alejandro, Ramón Carriles, Neftalí Ochoa-Alejo, Mercedes G. López, and Lino Sánchez-Segura. 2019. "Generation of BSA-capsaicin Nanoparticles and Their Hormesis Effect on the Rhodotorula mucilaginosa Yeast" Molecules 24, no. 15: 2800. https://doi.org/10.3390/molecules24152800