Statistical Methods for Rapid Quantification of Proteins, Lipids, and Carbohydrates in Nordic Microalgal Species Using ATR–FTIR Spectroscopy
Abstract
1. Introduction
2. Results and Discussion
2.1. Algal Biomass Composition Based on Classical Extractions and ATR–FTIR Spectral Analysis
2.2. Statistical Methods (ULRA, OPLS, and MCR–ALS) Can Facilitate the Prediction of Protein-, Lipid- and Carbohydrate-Content in Microalgae
2.2.1. Method 1: ULRA, based on FTIR spectral band intensities
2.2.2. Method 2: OPLS Based on the Fingerprint Region of FTIR Spectra
2.2.3. Method 3: MCR–ALS Based on the Fingerprint Region of FTIR Spectra
3. Materials and Methods
3.1. Algal Cultivation and Sampling Preparation
3.2. Chemical Extraction of Proteins, Lipids, and Carbohydrates
3.3. ATR–FTIR Spectroscopy and Spectral Data Processing
3.4. Modeling and Statistical Analysis
3.4.1. ULRA
3.4.2. OPLS
3.4.3. MCR–ALS
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A
| Cultivation Time | Cv 13-1 | Ca RW10 | So UTEX | Ds RUC-2 | Cs 3-4 | Ds 2-6 | Ss B2-2 |
|---|---|---|---|---|---|---|---|
| (day) | Nitrogen (mM) | ||||||
| 1 | 5.4 ± 0.09 | 5.3 ± 0.11 | 5.1 ± 0.17 | 5.2 ± 0.12 | 5.0 ± 0.17 | 5.2 ± 0.2 | 4.7 ± 0.08 |
| 2 | 5.1 ± 0.01 | 4.9 ± 0.27 | 4.8 ± 0.54 | 4.9 ± 0.25 | 4.9 ± 0.12 | 5.2 ± 0.21 | 4.5 ± 0.11 |
| 3 | 4.6 ± 0.16 | 4.7 ± 0.12 | 3.8 ± 0.79 | 4.2 ± 0.50 | 4.5 ± 0.16 | 5.2 ± 0.13 | 4.4 ± 0.03 |
| 4 | 4.3 ± 0.33 | 4.6 ± 0.10 | 2.6 ± 0.83 | 2.6 ± 0.57 | 3.4 ± 0.19 | 5.2 ± 0.21 | 4.1 ± 0.02 |
| 5 | 3.2 ± 0.10 | 3.8 ± 0.22 | 1.4 ± 0.44 | 0.0 | 0.8 ± 0.32 | 4.9 ± 0.20 | 3.7 ± 0.11 |
| 6 | 2.3 ± 0.51 | 2.1 ± 0.24 | 0.0 | - | 0.0 | 4.3 ± 0.04 | 3.6 ± 0.06 |
| 7 | 0.0 | 1.1 ± 0.21 | - | - | - | 3.0 ± 0.05 | 2.7 ± 0.04 |
| 8 | - | 0.0 | - | - | - | 0.0 | 2.3 ± 0.25 |
| 9 | 1.5 ± 0.30 | ||||||
| 10 | 0.0 | ||||||
| Sample | Proteins (% DW) | Lipids (% DW) | Carbohydrates (% DW) |
|---|---|---|---|
| Cv 13-1 R1 D0 | 21.6 ± 0.5 | 1.5 ± 0.2 | 13.8 ± 0.1 |
| Cv 13-1 R1 D2 | 18.6 ± 1.6 | 3.7 ± 1.0 | 38.9 ± 1.2 |
| Cv 13-1 R1 D4 | 18.7 ± 0.6 | 7.6 ± 0.4 | 37.0 ± 2.1 |
| Cv 13-1 R1 D6 | 20.3 ± 0.1 | 8.6 ± 2.2 | 34.7 ± 1.1 |
| Cv 13-1 R1 D8 | 17.2 ± 0.5 | 12.6 ± 0.5 | 34.4 ± 0.1 |
| Cv 13-1 R2 D0 | 23.8 ± 0.3 | 1.3 ± 0.3 | 13.3 ± 1.2 |
| Cv 13-1 R2 D2 | 16.6 ± 2.4 | 5.0 ± 0.6 | 40.2 ± 0.4 |
| Cv 13-1 R2 D4 | 18.5 ± 0.2 | 6.8 ± 2.1 | 37.6 ± 1.8 |
| Cv 13-1 R2 D6 | 20.5 ± 0.1 | 7.1 ± 1.2 | 38.3 ± 0.9 |
| Cv 13-1 R2 D8 | 21.2 ± 1.6 | 10.3 ± 1.6 | 35.5 ± 1.3 |
| Ca RW10 R1 D0 | 40.1 ± 2.9 | 0.2 ± 0.2 | 29.9 ± 0.1 |
| Ca RW10 R1 D2 | 27.2 ± 0.4 | 0.6 ± 0.0 | 26.9 ± 0.4 |
| Ca RW10 R1 D4 | 18.2 ±1.7 | 3.7 ± 1.0 | 38.2 ± 1.0 |
| Ca RW10 R1 D6 | 15.9 ± 1.0 | 6.2 ± 0.4 | 53.6 ± 0.2 |
| Ca RW10 R1 D8 | 16.9 ± 0.6 | 9.3 ± 0.6 | 55.3 ± 1.0 |
| Ca RW10 R1 D0 | 47.5 ± 1.3 | 0.0 ± 0.0 | 26.8 ± 2.7 |
| Ca RW10 R2 D2 | 25.2 ± 2.1 | 1.0 ± 0.0 | 28.6 ± 0.7 |
| Ca RW10 R2 D4 | 18.0 ± 0.2 | 4.2 ± 0.1 | 40.3 ± 0.6 |
| Ca RW10 R2 D6 | 18.2 ± 2.60 | 6.3 ± 0.1 | 50.7 ± 3.8 |
| Ca RW10 R2 D8 | 18.7 ± 3.4 | 9.1 ± 0.4 | 52.5 ± 1.0 |
| Ds RUC-2 R1 D0 | 37.7 ± 0.8 | 0.5 ± 0.1 | 28.8 ± 1.0 |
| Ds RUC-2 R1 D2 | 13.0 ± 1.4 | 2.9 ± 0.8 | 32.6 ± 1.2 |
| Ds RUC-2 R1 D4 | 8.7 ± 2.3 | 5.0 ± 0.9 | 37.2 ± 2.1 |
| Ds RUC-2 R1 D6 | 11.7 ± 04 | 7.2 ± 1.1 | 41.9 ± 1.8 |
| Ds RUC-2 R2 D0 | 37.4 ± 3.7 | 0.8 ± 0.0 | 30.0 ± 3.0 |
| Ds RUC-2 R2 D2 | 13.8 ± 0.8 | 3.8 ± 0.9 | 31.9 ± 3.3 |
| Ds RUC-2 R2 D4 | 9.5 ± 2.2 | 4.8 ± 0.1 | 39.2 ± 1.6 |
| Ds RUC-2 R2 D6 | 11.8 ± 2.1 | 5.7 ± 0.4 | 43.8 ± 0.1 |
| So UTEX R1 D0 | 37.3 ± 1.0 | 0.2 ± 0.0 | 20.7 ± 1.4 |
| So UTEX R1 D2 | 33.3 ± 0.5 | 0.9 ± 0.1 | 25.8 ± 1.2 |
| So UTEX R1 D4 | 19.0 ± 1.9 | 2.2 ± 0.2 | 45.4 ± 0.3 |
| So UTEX R1 D6 | 16.5 ± 2.4 | 3.2 ± 1.6 | 42.4 ± 1.3 |
| So UTEX R2 D0 | 36.8 ± 1.5 | 0.3 ± 0.1 | 21.1 ± 2.3 |
| So UTEX R2 D2 | 33.3 ± 2.3 | 1.2 ± 0.3 | 29.0 ± 3.7 |
| So UTEX R2 D4 | 20.7 ± 1.1 | 2.6 ± 0.5 | 46.0 ± 0.7 |
| So UTEX R2 D6 | 16.4 ± 0.7 | 3.6 ± 0.5 | 39.9 ± 2.8 |
| Cs 3-4 R1 D0 | 23.6 ± 0.9 | 0.6 ± 0.1 | 37.3 ± 1.9 |
| Cs 3-4 R1 D2 | 17.5 ± 3.1 | 1.5 ± 0.0 | 39.5 ± 1.0 |
| Cs 3-4 R1 D4 | 16.0 ± 0.5 | 2.9 ± 1.3 | 42.5 ± 0.2 |
| Cs 3-4 R1 D6 | 13.7 ± 0.2 | 3.5 ± 0.1 | 44.7 ± 1.2 |
| Cs 3-4 R2 D0 | 21.3 ± 1.6 | 0.9 ± 0.4 | 37.1 ± 2.2 |
| Cs 3-4 R2 D2 | 15.2 ± 3.2 | 2.0 ± 0.1 | 38.5 ± 0.5 |
| Cs 3-4 R2 D4 | 14.1 ± 0.1 | 3.3 ± 0.5 | 41.6 ± 0.1 |
| Cs 3-4 R2 D6 | 11.4 ± 2.9 | 3.3 ± 0.3 | 46.6 ± 2.5 |
| Ss B2-2 R1 D0 | 37.2 ± 0.9 | 0.4 ± 0.0 | 26.3 ± 2.2 |
| Ss B2-2 R1 D2 | 14.6 ± 2.0 | 1.6 ± 0.1 | 43.0 ± 0.6 |
| Ss B2-2 R1 D4 | 12.6 ± 1.8 | 2.1 ± 0.7 | 47.9 ± 2.2 |
| Ss B2-2 R1 D6 | 14.4 ± 1.8 | 3.4 ± 0.4 | 47.2 ± 2.2 |
| Ss B2-2 R2 D0 | 36.6 ± 1.9 | 0.2 ± 0.1 | 25.7 ± 0.8 |
| Ss B2-2 R2 D2 | 16.8 ± 1.3 | 2.4 ± 0.7 | 46.0 ± 0.8 |
| Ss B2-2 R2 D4 | 12.7 ± 0.4 | 1.9 ± 0.1 | 48.4 ± 1.3 |
| Ss B2-2 R2 D6 | 12.8 ± 0.3 | 3.1 ± 0.0 | 47.7 ± 2.5 |
| Ds 2-6 R1 D0 | 28.1 ± 1.7 | 0.8 ± 0.0 | 31.2 ± 1.1 |
| Ds 2-6 R1 D2 | 19.3 ± 0.9 | 1.9 ± 0.8 | 34.5 ± 3.1 |
| Ds 2-6 R1 D4 | 18.8 ± 1.8 | 3.0 ± 0.8 | 31.7 ± 0.1 |
| Ds 2-6 R2 D0 | 24.1 ± 1.4 | 1.6 ± 0.4 | 29.5 ± 3.4 |
| Ds 2-6 R2 D2 | 19.1 ±. 0.5 | 1.9 ± 0.4 | 30.7 ± 2.3 |
| Ds 2-6 R2 D4 | 21.1 ± 1.4 | 2.1 ± 0.5 | 32.3 ± 1.7 |


References
- Gouveia, L. From Tiny Microalgae to Huge Biorefineries. Oceanogr. Open Access 2014, 02, 71–94. [Google Scholar] [CrossRef]
- Khan, M.I.; Shin, J.H.; Kim, J.D. The promising future of microalgae: Current status, challenges, and optimization of a sustainable and renewable industry for biofuels, feed, and other products. Microb. Cell Factories 2018, 17, 36. [Google Scholar] [CrossRef]
- Sen Roy, S.; Pal, R. Microalgae in Aquaculture: A Review with Special References to Nutritional Value and Fish Dietetics. Proc. Zool. Soc. 2015, 68, 1–8. [Google Scholar] [CrossRef]
- Caporgno, M.P.; Mathys, A. Trends in Microalgae Incorporation into Innovative Food Products With Potential Health Benefits. Front. Nutr. 2018, 5, 58. [Google Scholar] [CrossRef]
- Madeira, M.S.; Cardoso, C.; Lopes, P.A.; Coelho, D.; Afonso, C.; Bandarra, N.M.; Prates, J.A. Microalgae as feed ingredients for livestock production and meat quality: A review. Livest. Sci. 2017, 205, 111–121. [Google Scholar] [CrossRef]
- Garcia-Gonzalez, J.; Sommerfeld, M. Biofertilizer and biostimulant properties of the microalga Acutodesmus dimorphus. J. Appl. Phycol. 2016, 28, 1051–1061. [Google Scholar] [CrossRef]
- Ferreira, A.; Ribeiro, B.; Ferreira, A.F.; Tavares, M.L.A.; Vladic, J.; Vidović, S.; Cvetkovic, D.; Melkonyan, L.; Avetisova, G.; Goginyan, V.; et al. Scenedesmus obliquus microalga-based biorefinery—From brewery effluent to bioactive compounds, biofuels and biofertilizers—Aiming at a circular bioeconomy. Biofuels Bioprod Biorefining 2019. [Google Scholar] [CrossRef]
- Silva, C.E.D.F.; Bertucco, A. Bioethanol from microalgae and cyanobacteria: A review and technological outlook. Process Biochem. 2016, 51, 1833–1842. [Google Scholar] [CrossRef]
- Matos, C.T.; Gouveia, L.; Morais, A.; Reis, A.; Bogel-Łukasik, R. Green metrics evaluation of isoprene production by microalgae and bacteria. Green Chem. 2013, 15, 2854–2864. [Google Scholar] [CrossRef]
- Mathiot, C.; Ponge, P.; Gallard, B.; Sassi, J.-F.; Delrue, F.; Le Moigne, N. Microalgae starch-based bioplastics: Screening of ten strains and plasticization of unfractionated microalgae by extrusion. Carbohydr. Polym. 2019, 208, 142–151. [Google Scholar] [CrossRef]
- Gouveia, L.; Oliveira, A.C.; Congestri, R.; Bruno, L.; Soares, A.T.; Menezes, R.S.; Filho, N.R.A.; Tzovenis, I. Microalgae-Based Biofuels and Bioproducts: From Feedstock Cultivation to End-Products; Woodhead Publishing: Soston, UK, 2017. [Google Scholar] [CrossRef]
- Ryckebosch, E.; Bruneel, C.; Termote-Verhalle, R.; Goiris, K.; Muylaert, K.; Foubert, I. Nutritional evaluation of microalgae oils rich in omega-3 long chain polyunsaturated fatty acids as an alternative for fish oil. Food Chem. 2014, 160, 393–400. [Google Scholar] [CrossRef]
- Vuppaladadiyam, A.K.; Prinsen, P.; Raheem, A.; Luque, R.; Zhao, M. Microalgae cultivation and metabolites production: A comprehensive review. Biofuels Bioprod Biorefining 2018, 12, 304–324. [Google Scholar] [CrossRef]
- Markou, G.; Nerantzis, E. Microalgae for high-value compounds and biofuels production: A review with focus on cultivation under stress conditions. Biotechnol. Adv. 2013, 31, 1532–1542. [Google Scholar] [CrossRef]
- Breuer, G.; Evers, W.A.C.; De Vree, J.H.; Kleinegris, D.M.M.; Martens, D.E.; Wijffels, R.H.; Lamers, P.P. Analysis of Fatty Acid Content and Composition in Microalgae. J. Vis. Exp. 2013, 5, 1–9. [Google Scholar] [CrossRef]
- Sun, X.; Cao, Y.; Xu, H.; Liu, Y.; Sun, J.; Qiao, D.; Cao, Y. Effect of nitrogen-starvation, light intensity and iron on triacylglyceride/carbohydrate production and fatty acid profile of Neochloris oleoabundans HK-129 by a two-stage process. Bioresour. Technol. 2014, 155, 204–212. [Google Scholar] [CrossRef]
- Zhu, L.D.; Li, Z.H.; Hiltunen, E. Strategies for Lipid Production Improvement in Microalgae as a Biodiesel Feedstock. BioMed Res. Int. 2016, 2016, 1–8. [Google Scholar] [CrossRef]
- Johnson, X.; Alric, J. Central Carbon Metabolism and Electron Transport in Chlamydomonas reinhardtii: Metabolic Constraints for Carbon Partitioning between Oil and Starch. Eukaryot. Cell 2013, 12, 776–793. [Google Scholar] [CrossRef]
- Baker, M.J.; Trevisan, J.; Bassan, P.; Bhargava, R.; Butler, H.J.; Dorling, K.M.; Fielden, P.R.; Fogarty, S.W.; Fullwood, N.J.; Heys, K.A.; et al. Using Fourier transform IR spectroscopy to analyze biological materials. Nat. Protoc. 2014, 9, 1771–1791. [Google Scholar] [CrossRef]
- Kosa, G.; Shapaval, V.; Kohler, A.; Zimmermann, B. FTIR spectroscopy as a unified method for simultaneous analysis of intra- and extracellular metabolites in high-throughput screening of microbial bioprocesses. Microb. Cell Factories 2017, 16, 195. [Google Scholar] [CrossRef]
- Schuster, K.; Mertens, F.; Gapes, J. FTIR spectroscopy applied to bacterial cells as a novel method for monitoring complex biotechnological processes. Vib. Spectrosc. 1999, 19, 467–477. [Google Scholar] [CrossRef]
- Sigee, D.C.; Dean, A.; Levado, E.; Tobin, M.J. Fourier-transform infrared spectroscopy of Pediastrum duplex: Characterization of a micro-population isolated from a eutrophic lake. Eur. J. Phycol. 2002, 37, 19–26. [Google Scholar] [CrossRef]
- Stehfest, K.; Toepel, J.; Wilhelm, C. The application of micro-FTIR spectroscopy to analyze nutrient stress-related changes in biomass composition of phytoplankton algae. Plant Physiol. Biochem. 2005, 43, 717–726. [Google Scholar] [CrossRef]
- Giordano, M.; Kansiz, M.; Heraud, P.; Beardall, J.; Wood, B.; McNaughton, D. Fourier Transform Infrared Spectroscopy as a novel tool to investigate changes in intracellular macromolecular pools in the marine microalga Chaetoceros Muellerii (Bacillariophyceae). J. Phycol. 2001, 37, 271–279. [Google Scholar] [CrossRef]
- Driver, T.; Bajhaiya, A.K.; Allwood, J.W.; Goodacre, R.; Pittman, J.K.; Dean, A.P. Metabolic responses of eukaryotic microalgae to environmental stress limit the ability of FT-IR spectroscopy for species identification. Algal Res. 2015, 11, 148–155. [Google Scholar] [CrossRef]
- Meng, Y.; Yao, C.; Xue, S.; Yang, H. Application of Fourier transform infrared (FT-IR) spectroscopy in determination of microalgal compositions. Bioresour. Technol. 2014, 151, 347–354. [Google Scholar] [CrossRef]
- Pistorius, A.M.; DeGrip, W.J.; Egorova-Zachernyuk, T.A.; Egorova-Zachernyuk, T.A. Monitoring of biomass composition from microbiological sources by means of FT-IR spectroscopy. Biotechnol. Bioeng. 2009, 103, 123–129. [Google Scholar] [CrossRef]
- Dean, A.P.; Sigee, D.C.; Estrada, B.; Pittman, J.K. Using FTIR spectroscopy for rapid determination of lipid accumulation in response to nitrogen limitation in freshwater microalgae. Bioresour. Technol. 2010, 101, 4499–4507. [Google Scholar] [CrossRef]
- Wagner, H.; Liu, Z.; Langner, U.; Stehfest, K.; Wilhelm, C. The use of FTIR spectroscopy to assess quantitative changes in the biochemical composition of microalgae. J. Biophotonics 2010, 3, 557–566. [Google Scholar] [CrossRef]
- Mayers, J.J.; Flynn, K.J.; Shields, R.J. Rapid determination of bulk microalgal biochemical composition by Fourier-Transform Infrared spectroscopy. Bioresour. Technol. 2013, 148, 215–220. [Google Scholar] [CrossRef]
- Ferro, L.; Gentili, F.G.; Funk, C. Isolation and characterization of microalgal strains for biomass production and wastewater reclamation in Northern Sweden. Algal Res. 2018, 32, 44–53. [Google Scholar] [CrossRef]
- Mirabella, F.M. Internal Reflection Spectroscopy: Theory and Applications, 1st ed.; CRC Press: New York, NY, USA, 1992. [Google Scholar]
- Allison, G.G. Application of Fourier Transform Mid-Infrared Spectroscopy (FTIR) for Research into Biomass Feed-Stocks. In Fourier Transforms—New Analytical Approaches and FTIR Strategies; InTech: Rijeka, Croatia, 2011. [Google Scholar] [CrossRef][Green Version]
- He, X.; Dai, J.; Wu, Q. Identification of Sporopollenin as the Outer Layer of Cell Wall in Microalga Chlorella protothecoides. Front. Microbiol. 2016, 7, 257. [Google Scholar] [CrossRef]
- Komaristaya, V.P.; Gorbulin, O.S. Sporopollenin in the composition of cell walls of Dunaliella salina Teod. (Chlorophyta) zygotes. Int. J. Algae 2006, 8, 43–52. [Google Scholar] [CrossRef]
- Kim, D.-Y.; Vijayan, D.; Praveenkumar, R.; Han, J.-I.; Lee, K.; Park, J.-Y.; Chang, W.-S.; Lee, J.-S.; Oh, Y.-K. Cell-wall disruption and lipid/astaxanthin extraction from microalgae: Chlorella and Haematococcus. Bioresour. Technol. 2016, 199, 300–310. [Google Scholar] [CrossRef]
- Martínez, J.M.; Gojkovic, Z.; Ferro, L.; Maza, M.; Álvarez, I.; Raso, J.; Funk, C. Use of pulsed electric field permeabilization to extract astaxanthin from the Nordic microalga Haematococcus pluvialis. Bioresour. Technol. 2019, 289, 121694. [Google Scholar] [CrossRef]
- Felten, J.; Hall, H.; Jaumot, J.; Tauler, R.; De Juan, A.; Gorzsás, A. Vibrational spectroscopic image analysis of biological material using multivariate curve resolution–alternating least squares (MCR-ALS). Nat. Protoc. 2015, 10, 217–240. [Google Scholar] [CrossRef]
- Nicolaï, B.M.; Beullens, K.; Bobelyn, E.; Peirs, A.; Saeys, W.; Theron, K.I.; Lammertyn, J. Nondestructive measurement of fruit and vegetable quality by means of NIR spectroscopy: A review. Postharvest Boil. Technol. 2007, 46, 99–118. [Google Scholar] [CrossRef]
- Ringnér, M. What is principal component analysis? Nat. Biotechnol. 2008, 26, 303–304. [Google Scholar] [CrossRef]
- Mariey, L.; Signolle, J.; Amiel, C.; Travert, J. Discrimination, classification, identification of microorganisms using FTIR spectroscopy and chemometrics. Vib. Spectrosc. 2001, 26, 151–159. [Google Scholar] [CrossRef]
- Wold, S.; Sjöström, M.; Eriksson, L. PLS-regression: A basic tool of chemometrics. Chemom. Intell. Lab. Syst. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Jaumot, J.; Gargallo, R.; De Juan, A.; Tauler, R. A graphical user-friendly interface for MCR-ALS: A new tool for multivariate curve resolution in MATLAB. Chemom. Intell. Lab. Syst. 2005, 76, 101–110. [Google Scholar] [CrossRef]
- De Juan, A.; Jaumot, J.; Tauler, R. Multivariate Curve Resolution (MCR). Solving the mixture analysis problem. Anal. Methods 2014, 6, 4964–4976. [Google Scholar] [CrossRef]
- Gojkovic, Z.; Lindberg, R.H.; Tysklind, M.; Funk, C. Northern green algae have the capacity to remove active pharmaceutical ingredients. Ecotoxicol. Environ. Saf. 2019, 170, 644–656. [Google Scholar] [CrossRef]
- Bischoff, H.W.; Bold, H.C. Some Soil Slgae from Enchanted Rock and Related Algal Species; University of Texas Publishing: Austin, TX, USA, 1963; pp. 1–95. [Google Scholar]
- Garcia-Robledo, E.; Corzo, A.; Papaspyrou, S.; Rodríguez, A.C. A fast and direct spectrophotometric method for the sequential determination of nitrate and nitrite at low concentrations in small volumes. Mar. Chem. 2014, 162, 30–36. [Google Scholar] [CrossRef]
- Slocombe, S.P.; Ross, M.; Thomas, N.; McNeill, S.; Stanley, M.S. A rapid and general method for measurement of protein in micro-algal biomass. Bioresour. Technol. 2013, 129, 51–57. [Google Scholar] [CrossRef]
- Dubois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric Method for Determination of Sugars and Related Substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Duygu, D. (Yalcin) Fourier transform infrared (FTIR) spectroscopy for identification of Chlorella vulgaris Beijerinck 1890 and Scenedesmus obliquus (Turpin) Kützing 1833. Afr. J. Biotechnol. 2012, 11, 3817–3824. [Google Scholar] [CrossRef]
- R Core Team, R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. 2014. Available online: http://www.r-project.org/ (accessed on 10 August 2019).
Sample Availability: Samples of the compounds are available from the authors. |


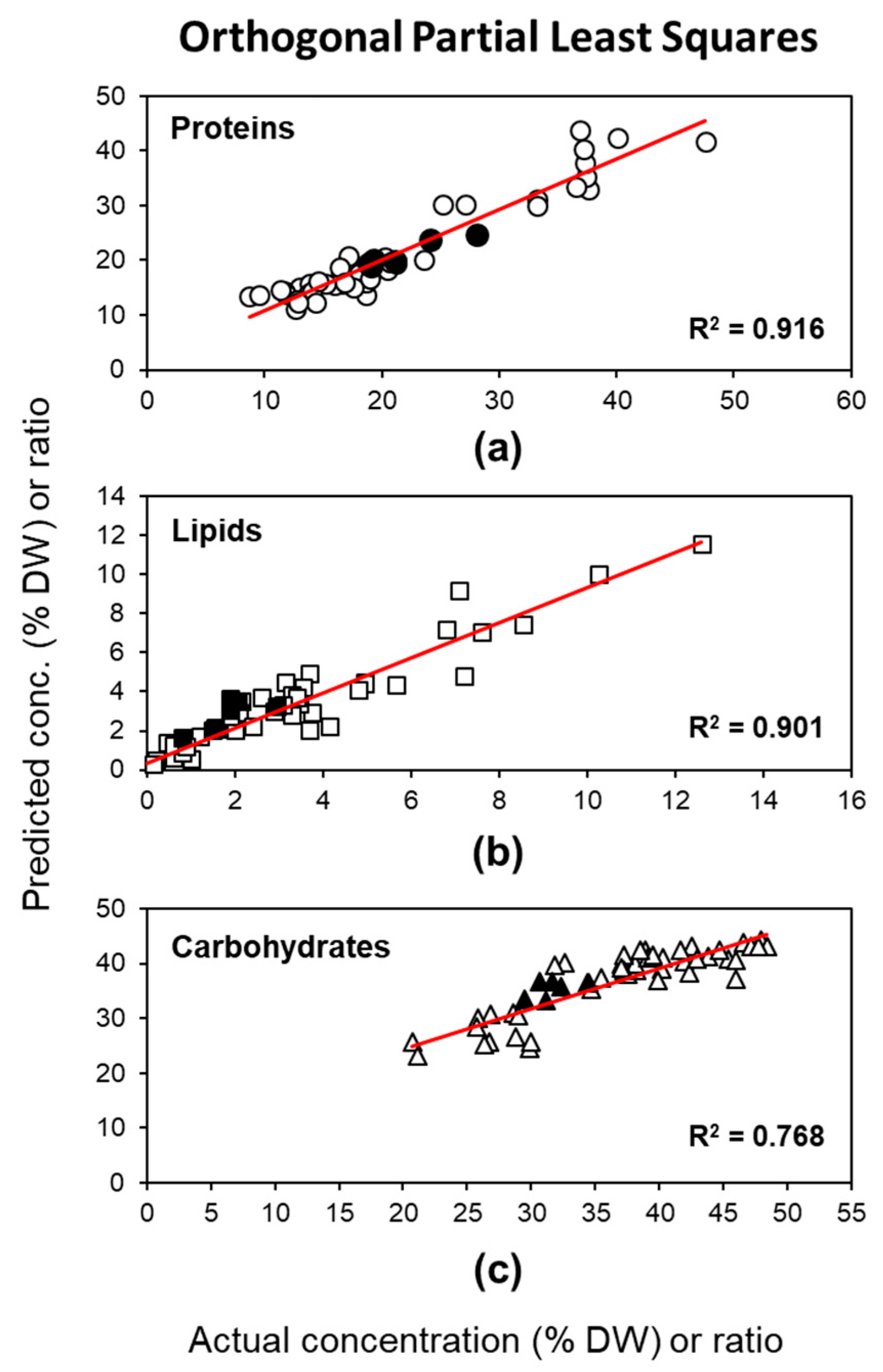

| Univariate Linear Regression Analysis (ULRA) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Y Variable | N | Intercept | slope | R2 | RMSEC | Q2 a | RMSECV a | RPD a | RMSEP d |
| Proteins | 52 | 3.793 | 145.371 | 0.876 | 3.211 | 0.859 | 3.417 | 2.693 | 3.186 |
| Lipids | 50 | −0.027 | 149.447 | 0.816 | 1.189 | 0.798 | 1.246 | 2.248 | 0.726 |
| Carbohydrates | 46 | 0.426 | 66.945 | 0.647 | 4.413 | 0.611 | 4.634 | 1.621 | 9.33 |
| Carbohydrates/Proteins | 46 | 0.027 | 0.375 | 0.844 | 0.428 | 0.833 | 0.442 | 2.476 | 0.684 |
| Orthogonal Partial Least Squares (OPLS) | |||||||||
| Y Variable | N | Components b | R2X(cum) | R2Y(cum) | RMSEC | Q2(cum) c | RMSECV c | RPD c | RMSEP d |
| (model) | 46 | 2 + 2 + 1 | 0.984 | 0.861 | 0.837 | ||||
| Proteins | 46 | 0.916 | 2.944 | 0.898 | 3.073 | 2.994 | 1.48 | ||
| Lipids | 46 | 0.901 | 0.932 | 0.877 | 0.979 | 2.859 | 1.135 | ||
| Carbohydrates | 46 | 0.768 | 3.797 | 0.735 | 3.826 | 1.964 | 4.081 | ||
| Multivariate Curve Resolution Alternating Least Squares (MCR–ALS) | |||||||||
| Y Variable | N | lof PCA (%) | lof exp (%) | R2 | RMSEC | RMSECV a | |||
| (model) | 52 | 0.798 | 5.854 | 0.997 | |||||
| Proteins | 52 | 0.851 | 3.521 | 3.699 | |||||
| Lipids | 50 | 0.768 | 1.335 | 1.392 | |||||
| Carbohydrates | 46 | 0.632 | 4.508 | 4.73 | |||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferro, L.; Gojkovic, Z.; Gorzsás, A.; Funk, C. Statistical Methods for Rapid Quantification of Proteins, Lipids, and Carbohydrates in Nordic Microalgal Species Using ATR–FTIR Spectroscopy. Molecules 2019, 24, 3237. https://doi.org/10.3390/molecules24183237
Ferro L, Gojkovic Z, Gorzsás A, Funk C. Statistical Methods for Rapid Quantification of Proteins, Lipids, and Carbohydrates in Nordic Microalgal Species Using ATR–FTIR Spectroscopy. Molecules. 2019; 24(18):3237. https://doi.org/10.3390/molecules24183237
Chicago/Turabian StyleFerro, Lorenza, Zivan Gojkovic, András Gorzsás, and Christiane Funk. 2019. "Statistical Methods for Rapid Quantification of Proteins, Lipids, and Carbohydrates in Nordic Microalgal Species Using ATR–FTIR Spectroscopy" Molecules 24, no. 18: 3237. https://doi.org/10.3390/molecules24183237
APA StyleFerro, L., Gojkovic, Z., Gorzsás, A., & Funk, C. (2019). Statistical Methods for Rapid Quantification of Proteins, Lipids, and Carbohydrates in Nordic Microalgal Species Using ATR–FTIR Spectroscopy. Molecules, 24(18), 3237. https://doi.org/10.3390/molecules24183237








