Serum Metabolomics Identifies Altered Bioenergetics, Signaling Cascades in Parallel with Exposome Markers in Crohn’s Disease
Abstract
:1. Introduction
2. Results
2.1. Global Metabolomic Profile of the Crohn’s Disease
2.2. Integrated Pathway Analysis
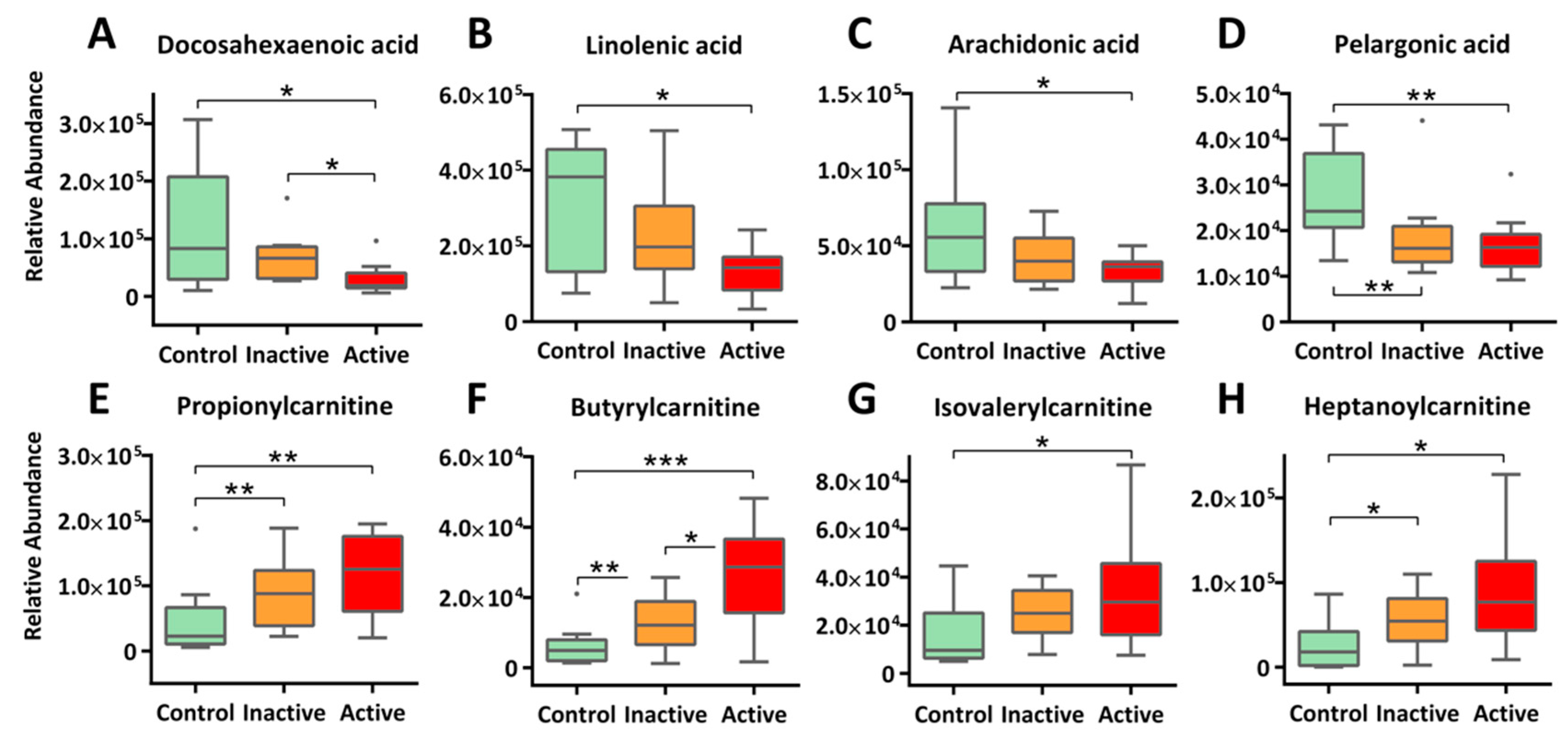
2.3. Enhanced β-Oxidation of Fatty Acids and Altered Lipid Cascades
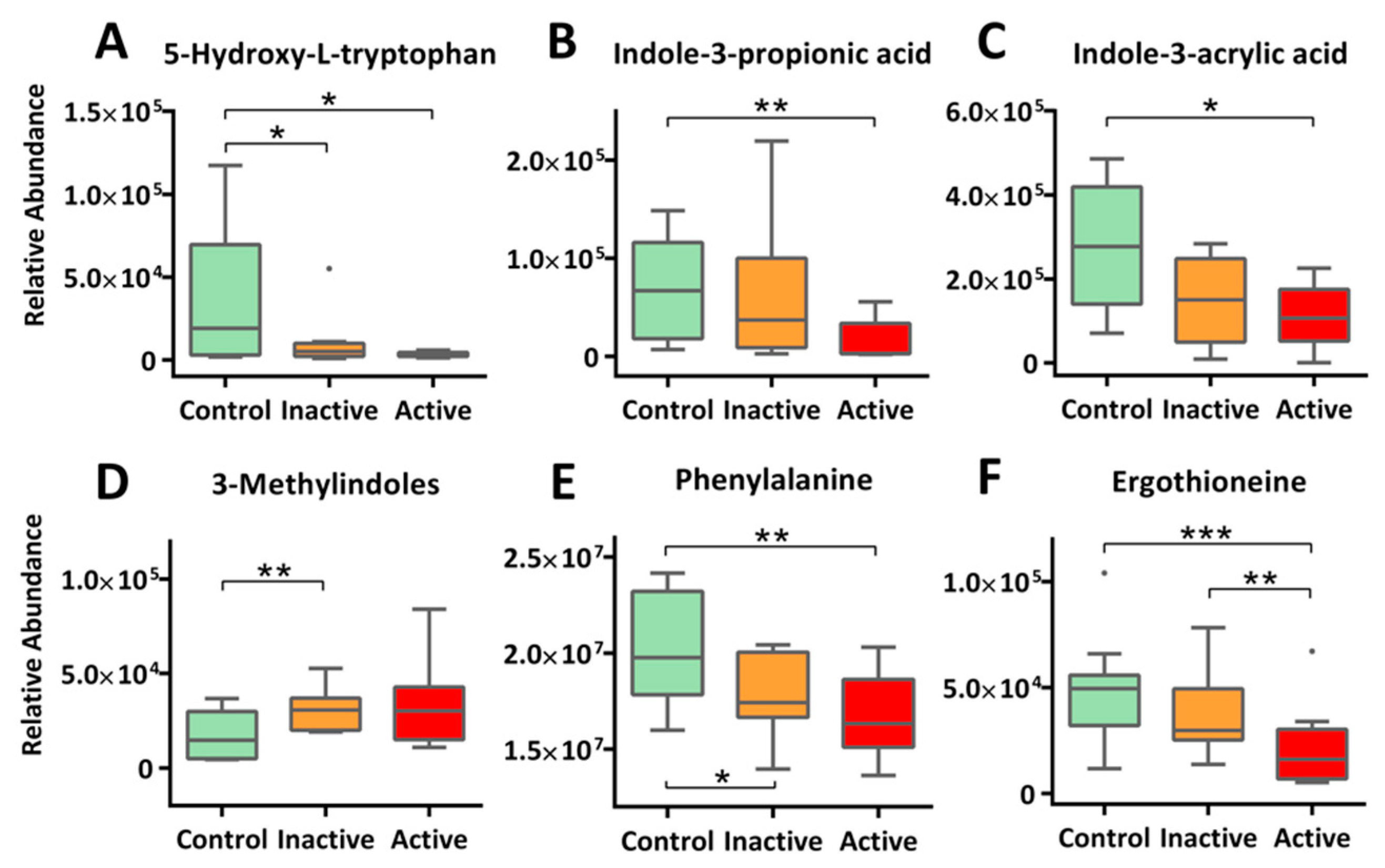
2.4. Altered Amino Acid Metabolism
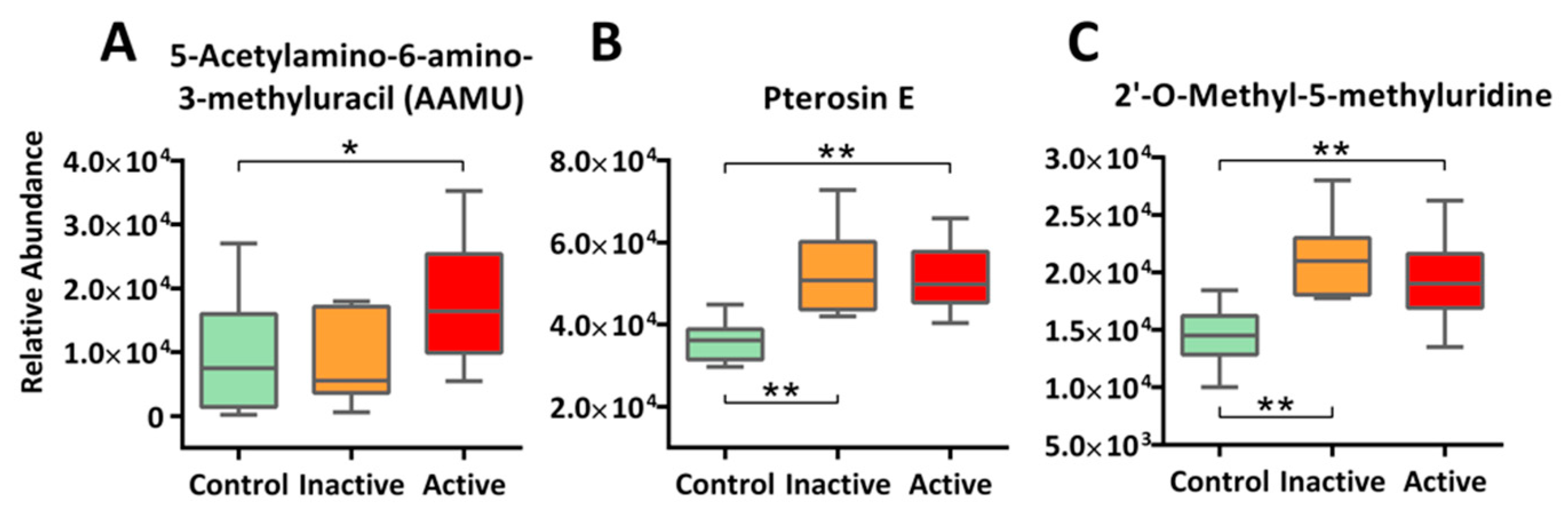
2.5. Xenobiotics and Novel Metabolites: A Call for Exposomics
3. Discussion
4. Materials and Methods
4.1. Characteristics of Participants
4.2. Sample Preparation
4.3. Metabolomic Profiling by LC-MS
4.4. Data Processing and Statistical Analysis
4.5. Compound Identification
4.6. Integrated Pathway Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Torres, J.; Mehandru, S.; Colombel, J.-F.; Peyrin-Biroulet, L. Crohn’s disease. Lancet 2017, 389, 1741–1755. [Google Scholar] [CrossRef]
- Ungaro, R.; Mehandru, S.; Allen, P.B.; Peyrin-Biroulet, L.; Colombel, J.-F. Ulcerative colitis. Lancet 2017, 389, 1756–1770. [Google Scholar] [CrossRef]
- Ng, S.C.; Shi, H.Y.; Hamidi, N.; Underwood, F.E.; Tang, W.; Benchimol, E.I.; Panaccione, R.; Ghosh, S.; Wu, J.C.Y.; Chan, F.K.L.; et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: A systematic review of population-based studies. Lancet 2017, 390, 2769–2778. [Google Scholar] [CrossRef]
- Huang, H.; Vangay, P.; McKinlay, C.E.; Knights, D. Multi-omics analysis of inflammatory bowel disease. Immunol. Lett. 2014, 162 Pt A, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.H.; Achkar, J.P. Gene-environment interactions in inflammatory bowel disease pathogenesis. Curr. Opin. Gastroenterol. 2015, 31, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Cader, M.Z.; Kaser, A. Recent advances in inflammatory bowel disease: Mucosal immune cells in intestinal inflammation. Gut 2013, 62, 1653–1664. [Google Scholar] [CrossRef] [PubMed]
- Abegunde, A.T.; Muhammad, B.H.; Bhatti, O.; Ali, T. Environmental risk factors for inflammatory bowel diseases: Evidence based literature review. World J. Gastroenterol. 2016, 22, 6296–6317. [Google Scholar] [CrossRef]
- Loddo, I.; Romano, C. Inflammatory Bowel Disease: Genetics, Epigenetics, and Pathogenesis. Front. Immunol. 2015, 6, 551. [Google Scholar] [CrossRef]
- Nagalingam, N.A.; Lynch, S.V. Role of the microbiota in inflammatory bowel diseases. Inflamm. Bowel Dis. 2012, 18, 968–984. [Google Scholar] [CrossRef]
- Hold, G.L.; Smith, M.; Grange, C.; Watt, E.R.; El-Omar, E.M.; Mukhopadhya, I. Role of the gut microbiota in inflammatory bowel disease pathogenesis: What have we learnt in the past 10 years? World J. Gastroenterol. 2014, 20, 1192–1210. [Google Scholar] [CrossRef]
- Wlodarska, M.; Kostic, A.D.; Xavier, R.J. An integrative view of microbiome-host interactions in inflammatory bowel diseases. Cell Host Microbe 2015, 17, 577–591. [Google Scholar] [CrossRef] [PubMed]
- Mahid, S.S.; Minor, K.S.; Soto, R.E.; Hornung, C.A.; Galandiuk, S. Smoking and inflammatory bowel disease: A meta-analysis. Mayo Clin. Proc. 2006, 81, 1462–1471. [Google Scholar] [CrossRef] [PubMed]
- Soubieres, A.A.; Poullis, A. Emerging Biomarkers for the Diagnosis and Monitoring of Inflammatory Bowel Diseases. Inflamm. Bowel Dis. 2016, 22, 2016–2022. [Google Scholar] [CrossRef] [Green Version]
- Madsen, R.; Lundstedt, T.; Trygg, J. Chemometrics in metabolomic—A review in human disease diagnosis. Anal. Chim. Acta 2010, 659, 23–33. [Google Scholar] [CrossRef]
- Williams, H.R.; Willsmore, J.D.; Cox, I.J.; Walker, D.G.; Cobbold, J.F.; Taylor-Robinson, S.D.; Orchard, T.R. Serum metabolic profiling in inflammatory bowel disease. Dig. Dis. Sci. 2012, 57, 2157–2165. [Google Scholar] [CrossRef] [PubMed]
- Murdoch, T.B.; Fu, H.; MacFariane, S.; Sydora, B.C.; Fedorak, R.N.; Slupsky, C.M. Urinary metabolic profiles of inflammatory bowel disease in interleukin-10 gene-deficient mice. Anal. Chem. 2008, 80, 5524–5531. [Google Scholar] [CrossRef] [PubMed]
- Bezabeh, T.; Somorjai, R.L.; Smith, I.C. MR metabolomics of fecal extracts: Applications in the study of bowel diseases. Magn. Reson. Chem. 2009, 47 (Suppl. 1), S54–S61. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Raftery, D. Comparing and combining NMR spectroscopy and mass spectrometry in metabolomics. Anal. Bioanal. Chem. 2007, 387, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Kolho, K.L.; Pessia, A.; Jaakkola, T.; de Vos, W.M.; Velagapudi, V. Faecal and Serum Metabolomics in Paediatric Inflammatory Bowel Disease. J. Crohns Colitis 2017, 11, 321–334. [Google Scholar] [CrossRef]
- Scoville, E.A.; Allaman, M.M.; Brown, C.T.; Motley, A.K.; Horst, S.N.; Williams, C.S.; Koyama, T.; Zhao, Z.; Adams, D.W.; Beaulieu, D.B.; et al. Alterations in Lipid, Amino Acid, and Energy Metabolism Distinguish Crohn’s Disease from Ulcerative Colitis and Control Subjects by Serum Metabolomic Profiling. Metabolomics 2018, 14, 17. [Google Scholar] [CrossRef]
- Lehninger, A.L.; Nelson, D.L.; Cox, M.M. Lehninger Principles of Biochemistry; W.H. Freeman: New York, NY, USA, 2013. [Google Scholar]
- Yano, J.M.; Yu, K.; Donaldson, G.P.; Shastri, G.G.; Ann, P.; Ma, L.; Nagler, C.R.; Ismagilov, R.F.; Mazmanian, S.K.; Hsiao, E.Y. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 2015, 161, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Xu, K.; Liu, H.; Liu, G.; Bai, M.; Peng, C.; Li, T.; Yin, Y. Impact of the Gut Microbiota on Intestinal Immunity Mediated by Tryptophan Metabolism. Front. Cell. Infect. Microbiol. 2018, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Graff, L.A.; Walker, J.R.; Bernstein, C.N. Depression and anxiety in inflammatory bowel disease: A review of comorbidity and management. Inflamm. Bowel Dis. 2009, 15, 1105–1118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arsenescu, R.; Arsenescu, V.; Zhong, J.; Nasser, M.; Melinte, R.; Dingle, R.W.; Swanson, H.; de Villiers, W.J. Role of the xenobiotic receptor in inflammatory bowel disease. Inflamm. Bowel Dis. 2011, 17, 1149–1162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monteleone, I.; MacDonald, T.T.; Pallone, F.; Monteleone, G. The aryl hydrocarbon receptor in inflammatory bowel disease: Linking the environment to disease pathogenesis. Curr. Opin. Gastroenterol. 2012, 28, 310–313. [Google Scholar] [CrossRef] [PubMed]
- Lanis, J.M.; Alexeev, E.E.; Curtis, V.F.; Kitzenberg, D.A.; Kao, D.J.; Battista, K.D.; Gerich, M.E.; Glover, L.E.; Kominsky, D.J.; Colgan, S.P. Tryptophan metabolite activation of the aryl hydrocarbon receptor regulates IL-10 receptor expression on intestinal epithelia. Mucosal Immunol. 2017, 10, 1133–1144. [Google Scholar] [CrossRef] [Green Version]
- He, F.; Wu, C.; Li, P.; Li, N.; Zhang, D.; Zhu, Q.; Ren, W.; Peng, Y. Functions and Signaling Pathways of Amino Acids in Intestinal Inflammation. Biomed Res. Int. 2018, 2018, 9171905. [Google Scholar] [CrossRef]
- Nagarjun, S.; Dhadde, S.B.; Veerapur, V.P.; Thippeswamy, B.S.; Chandakavathe, B.N. Ameliorative effect of chromium-d-phenylalanine complex on indomethacin-induced inflammatory bowel disease in rats. Biomed. Pharmacother. 2017, 89, 1061–1066. [Google Scholar] [CrossRef]
- Huff, C.D.; Witherspoon, D.J.; Zhang, Y.; Gatenbee, C.; Denson, L.A.; Kugathasan, S.; Hakonarson, H.; Whiting, A.; Davis, C.T.; Wu, W.; et al. Crohn’s disease and genetic hitchhiking at IBD5. Mol. Biol. Evol. 2012, 29, 101–111. [Google Scholar] [CrossRef]
- Serrano Leon, A.; Amir Shaghaghi, M.; Yurkova, N.; Bernstein, C.N.; El-Gabalawy, H.; Eck, P. Single-nucleotide polymorphisms in SLC22A23 are associated with ulcerative colitis in a Canadian white cohort. Am. J. Clin. Nutr. 2014, 100, 289–294. [Google Scholar] [CrossRef] [Green Version]
- Paul, B.D.; Snyder, S.H. The unusual amino acid L-ergothioneine is a physiologic cytoprotectant. Cell Death Differ. 2010, 17, 1134–1140. [Google Scholar] [CrossRef] [PubMed]
- Weimann, A.; Sabroe, M.; Poulsen, H.E. Measurement of caffeine and five of the major metabolites in urine by high-performance liquid chromatography/tandem mass spectrometry. J. Mass Spectrom. 2005, 40, 307–316. [Google Scholar] [CrossRef]
- Wild, C.P. Complementing the Genome with an “Exposome”: The Outstanding Challenge of Environmental Exposure Measurement in Molecular Epidemiology. Cancer Epidemiol. Biomark. Prev. 2005, 14, 1847–1850. [Google Scholar] [CrossRef] [PubMed]
- Rappaport Stephen, M.; Barupal Dinesh, K.; Wishart, D.; Vineis, P.; Scalbert, A. The Blood Exposome and Its Role in Discovering Causes of Disease. Environ. Health Persp. 2014, 122, 769–774. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A.; Takai, D. The Role of DNA Methylation in Mammalian Epigenetics. Science 2001, 293, 1068–1070. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, S.; Watanabe, T.; Murao, K.; Ishikura, H.; Yamaizumi, Z.; Nishimura, S.; Miyazawa, T. Molecular mechanism of codon recognition by tRNA species with modified uridine in the first position of the anticodon. Proc. Natl. Acad. Sci. USA 1985, 82, 4905–4909. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Knutson, C.G.; Wishnok, J.S.; Fox, J.G.; Tannenbaum, S.R. Serum metabolomics in a Helicobacter hepaticus mouse model of inflammatory bowel disease reveal important changes in the microbiome, serum peptides, and intermediary metabolism. J. Proteome Res. 2012, 11, 4916–4926. [Google Scholar] [CrossRef]
- Nishiumi, S.; Izumi, Y.; Yoshida, M. Alterations in Docosahexaenoic Acid-Related Lipid Cascades in Inflammatory Bowel Disease Model Mice. Digestive Dis. Sci. 2018, 63, 1485–1496. [Google Scholar] [CrossRef]
- Cabre, E.; Manosa, M.; Gassull, M.A. Omega-3 fatty acids and inflammatory bowel diseases—A systematic review. Br. J. Nutr. 2012, 107 (Suppl. 2), S240–S252. [Google Scholar] [CrossRef]
- Tabbaa, M.; Golubic, M.; Roizen, M.; Bernstein, A. Docosahexaenoic Acid, Inflammation, and Bacterial Dysbiosis in Relation to Periodontal Disease, Inflammatory Bowel Disease, and the Metabolic Syndrome. Nutrients 2013, 5, 3299–3310. [Google Scholar] [CrossRef] [Green Version]
- Maceyka, M.; Harikumar, K.B.; Milstien, S.; Spiegel, S. Sphingosine-1-phosphate signaling and its role in disease. Trends Cell Biol. 2012, 22, 50–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suh, J.H.; Degagné, É.; Gleghorn, E.E.; Setty, M.; Rodriguez, A.; Park, K.T.; Verstraete, S.G.; Heyman, M.B.; Patel, A.S.; Irek, M.; et al. Sphingosine-1-Phosphate Signaling and Metabolism Gene Signature in Pediatric Inflammatory Bowel Disease: A Matched-case Control Pilot Study. Inflamm. Bowel Dis. 2018, 24, 1321–1334. [Google Scholar] [CrossRef] [PubMed]
- Round, J.L.; Mazmanian, S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009, 9, 313. [Google Scholar] [CrossRef] [PubMed]
- Clemente Jose, C.; Ursell Luke, K.; Parfrey Laura, W.; Knight, R. The Impact of the Gut Microbiota on Human Health: An Integrative View. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vijay-Kumar, M.; Aitken, J.D.; Carvalho, F.A.; Cullender, T.C.; Mwangi, S.; Srinivasan, S.; Sitaraman, S.V.; Knight, R.; Ley, R.E.; Gewirtz, A.T. Metabolic Syndrome and Altered Gut Microbiota in Mice Lacking Toll-Like Receptor 5. Science 2010, 328, 228–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamada, N.; Seo, S.-U.; Chen, G.Y.; Núñez, G. Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 2013, 13, 321. [Google Scholar] [PubMed]
- Louis, P.; Hold, G.L.; Flint, H.J. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2014, 12, 661. [Google Scholar] [CrossRef] [PubMed]
- Wikoff, W.R.; Anfora, A.T.; Liu, J.; Schultz, P.G.; Lesley, S.A.; Peters, E.C.; Siuzdak, G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. USA 2009, 106, 3698–3703. [Google Scholar] [CrossRef] [Green Version]
- Venkatesh, M.; Mukherjee, S.; Wang, H.; Li, H.; Sun, K.; Benechet, A.P.; Qiu, Z.; Maher, L.; Redinbo, M.R.; Phillips, R.S.; et al. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity 2014, 41, 296–310. [Google Scholar] [CrossRef]
- Hasnain, S.Z.; Tauro, S.; Das, I.; Tong, H.; Chen, A.C.H.; Jeffery, P.L.; McDonald, V.; Florin, T.H.; McGuckin, M.A. IL-10 Promotes Production of Intestinal Mucus by Suppressing Protein Misfolding and Endoplasmic Reticulum Stress in Goblet Cells. Gastroenterology 2013, 144, 357–368. [Google Scholar] [CrossRef]
- Wlodarska, M.; Luo, C.; Kolde, R.; d’Hennezel, E.; Annand, J.W.; Heim, C.E.; Krastel, P.; Schmitt, E.K.; Omar, A.S.; Creasey, E.A.; et al. Indoleacrylic Acid Produced by Commensal Peptostreptococcus Species Suppresses Inflammation. Cell Host Microbe 2017, 22, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, M.T.; Carlson, J.R. Microbial metabolites of tryptophan in the intestinal tract with special reference to skatole. Am. J. Clin. Nutr. 1979, 32, 173–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whitehead, T.R.; Price, N.P.; Drake, H.L.; Cotta, M.A. Catabolic Pathway for the Production of Skatole and Indoleacetic Acid by the Acetogen Clostridium drakei, Clostridium scatologenes, and Swine Manure. Appl. Environ. Microbiol. 2008, 74, 1950–1953. [Google Scholar] [CrossRef] [PubMed]
- Cook, K.L.; Rothrock, J.M.J.; Loughrin, J.H.; Doerner, K.C. Characterization of skatole-producing microbial populations in enriched swine lagoon slurry. FEMS Microbiol. Ecol. 2007, 60, 329–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birdsall, T.C. 5-Hydroxytryptophan: A clinically-effective serotonin precursor. Altern. Med. Rev. J. Clin. Therap. 1998, 3, 271–280. [Google Scholar]
- Rogers, G.B.; Keating, D.J.; Young, R.L.; Wong, M.L.; Licinio, J.; Wesselingh, S. From gut dysbiosis to altered brain function and mental illness: Mechanisms and pathways. Mol. Psychiatry 2016, 21, 738. [Google Scholar] [CrossRef] [PubMed]
- Hinz, M.; Stein, A.; Uncini, T. 5-HTP efficacy and contraindications. Neuropsychiatr. Dis. Treat. 2012, 8, 323–328. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, T.; Masuo, Y.; Takahashi, S.; Nakamichi, N.; Kato, Y. Organic cation transporter Octn1-mediated uptake of food-derived antioxidant ergothioneine into infiltrating macrophages during intestinal inflammation in mice. Drug Metab. Pharmacokinet. 2015, 30, 231–239. [Google Scholar] [CrossRef]
- Kato, Y.; Kubo, Y.; Iwata, D.; Kato, S.; Sudo, T.; Sugiura, T.; Kagaya, T.; Wakayama, T.; Hirayama, A.; Sugimoto, M.; et al. Gene knockout and metabolome analysis of carnitine/organic cation transporter OCTN1. Pharm. Res. 2010, 27, 832–840. [Google Scholar] [CrossRef]
- Brown, A.C.; Rampertab, S.D.; Mullin, G.E. Exisiting dietary guidelines for Crohn’s disease and ulcerative colitis. Expert Rev. Gastroenterol. Hepatol. 2011, 5, 411–425. [Google Scholar] [CrossRef]
- Magee, E.A.; Edmond, L.M.; Tasker, S.M.; Kong, S.C.; Curno, R.; Cummings, J.H. Associations between diet and disease activity in ulcerative colitis patients using a novel method of data analysis. Nutr. J. 2005, 4, 7. [Google Scholar] [CrossRef]
- Burisch, J.; Pedersen, N.; Cukovic-Cavka, S.; Turk, N.; Kaimakliotis, I.; Duricova, D.; Bortlik, M.; Shonova, O.; Vind, I.; Avnstrom, S.; et al. Environmental factors in a population-based inception cohort of inflammatory bowel disease patients in Europ—An ECCO-EpiCom study. J. Crohns Colitis 2014, 8, 607–616. [Google Scholar] [CrossRef] [PubMed]
- Barthel, C.; Wiegand, S.; Scharl, S.; Scharl, M.; Frei, P.; Vavricka, S.R.; Fried, M.; Sulz, M.C.; Wiegand, N.; Rogler, G.; et al. Patients’ perceptions on the impact of coffee consumption in inflammatory bowel disease: Friend or foe?—A patient survey. Nutr. J. 2015, 14, 78. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.A.; Low, D.; Kamba, A.; Llado, V.; Mizoguchi, E. Oral caffeine administration ameliorates acute colitis by suppressing chitinase 3-like 1 expression in intestinal epithelial cells. J. Gastroenterol. 2014, 49, 1206–1216. [Google Scholar] [CrossRef] [PubMed]
- Michielan, A.; D’Inca, R. Intestinal Permeability in Inflammatory Bowel Disease: Pathogenesis, Clinical Evaluation, and Therapy of Leaky Gut. Mediat. Inflamm. 2015, 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing Mass Spectrometry Data for Metabolite Profiling Using Nonlinear Peak Alignment, Matching, and Identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef]
- Tsugawa, H.; Kind, T.; Nakabayashi, R.; Yukihira, D.; Tanaka, W.; Cajka, T.; Saito, K.; Fiehn, O.; Arita, M. Hydrogen Rearrangement Rules: Computational MS/MS Fragmentation and Structure Elucidation Using MS-FINDER Software. Anal. Chem. 2016, 88, 7946–7958. [Google Scholar] [CrossRef]
- Vaniya, A.; Samra, S.N.; Palazoglu, M.; Tsugawa, H.; Fiehn, O. Using MS-FINDER for identifying 19 natural products in the CASMI 2016 contest. Phytochem. Lett. 2017, 21, 306–312. [Google Scholar] [CrossRef]
- Xia, J.; Sinelnikov, I.V.; Han, B.; Wishart, D.S. MetaboAnalyst 3.0—Making metabolomics more meaningful. Nucleic Acids Res. 2015, 43, W251–W257. [Google Scholar] [CrossRef]
- Barupal, D.K.; Haldiya, P.K.; Wohlgemuth, G.; Kind, T.; Kothari, S.L.; Pinkerton, K.E.; Fiehn, O. MetaMapp: Mapping and visualizing metabolomic data by integrating information from biochemical pathways and chemical and mass spectral similarity. BMC Bioinform. 2012, 13, 99. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lai, Y.; Xue, J.; Liu, C.-W.; Gao, B.; Chi, L.; Tu, P.; Lu, K.; Ru, H. Serum Metabolomics Identifies Altered Bioenergetics, Signaling Cascades in Parallel with Exposome Markers in Crohn’s Disease. Molecules 2019, 24, 449. https://doi.org/10.3390/molecules24030449
Lai Y, Xue J, Liu C-W, Gao B, Chi L, Tu P, Lu K, Ru H. Serum Metabolomics Identifies Altered Bioenergetics, Signaling Cascades in Parallel with Exposome Markers in Crohn’s Disease. Molecules. 2019; 24(3):449. https://doi.org/10.3390/molecules24030449
Chicago/Turabian StyleLai, Yunjia, Jingchuan Xue, Chih-Wei Liu, Bei Gao, Liang Chi, Pengcheng Tu, Kun Lu, and Hongyu Ru. 2019. "Serum Metabolomics Identifies Altered Bioenergetics, Signaling Cascades in Parallel with Exposome Markers in Crohn’s Disease" Molecules 24, no. 3: 449. https://doi.org/10.3390/molecules24030449
APA StyleLai, Y., Xue, J., Liu, C.-W., Gao, B., Chi, L., Tu, P., Lu, K., & Ru, H. (2019). Serum Metabolomics Identifies Altered Bioenergetics, Signaling Cascades in Parallel with Exposome Markers in Crohn’s Disease. Molecules, 24(3), 449. https://doi.org/10.3390/molecules24030449







