HPLC-ESI-MSn Identification and NMR Characterization of Glucosyloxybenzyl 2R-Benzylmalate Deriva-Tives from Arundina Graminifolia and Their Anti-Liver Fibrotic Effects In Vitro
Abstract
:1. Introduction
2. Results and Discussion
2.1. Structure Elucidation of New Compounds
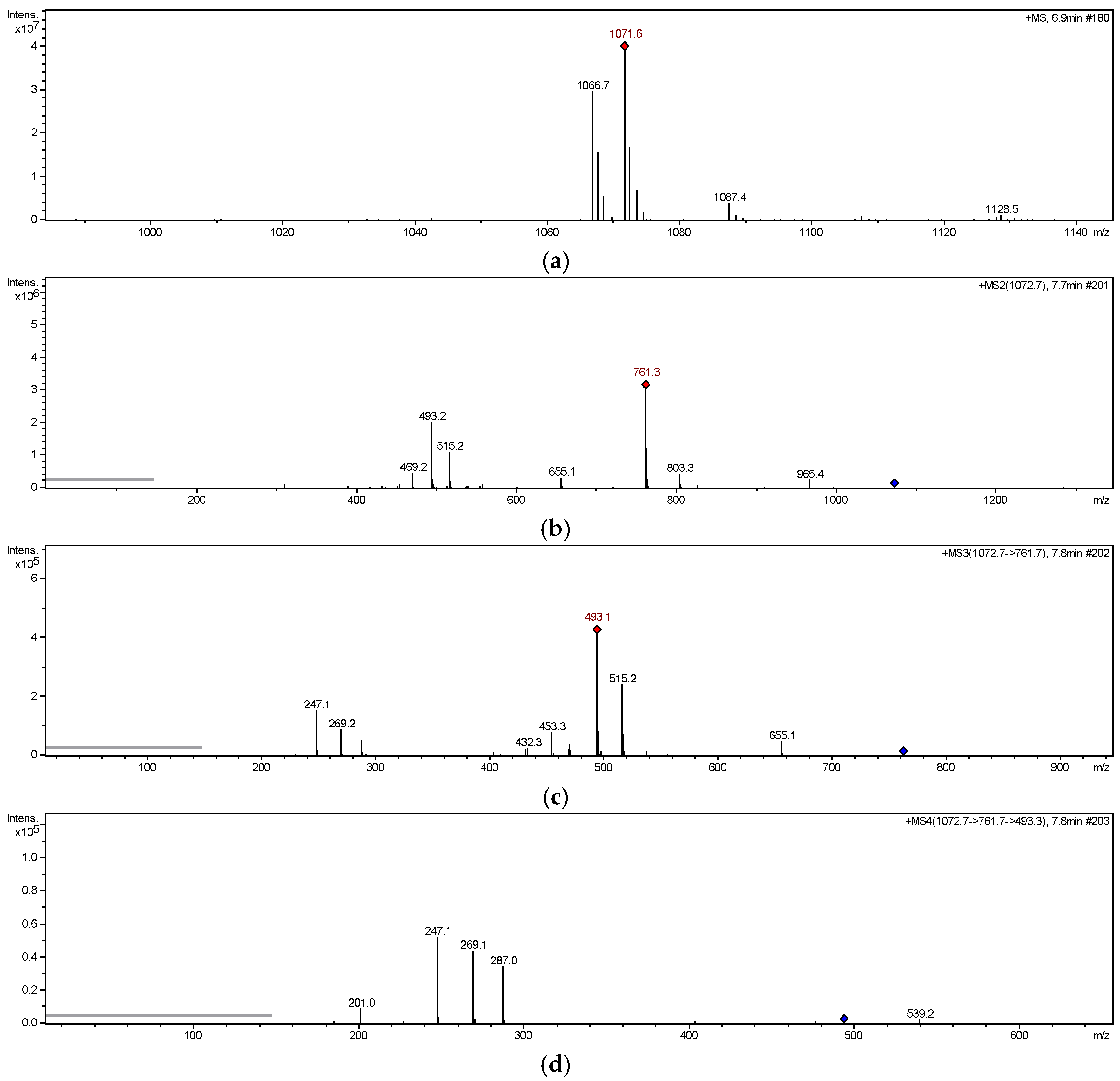
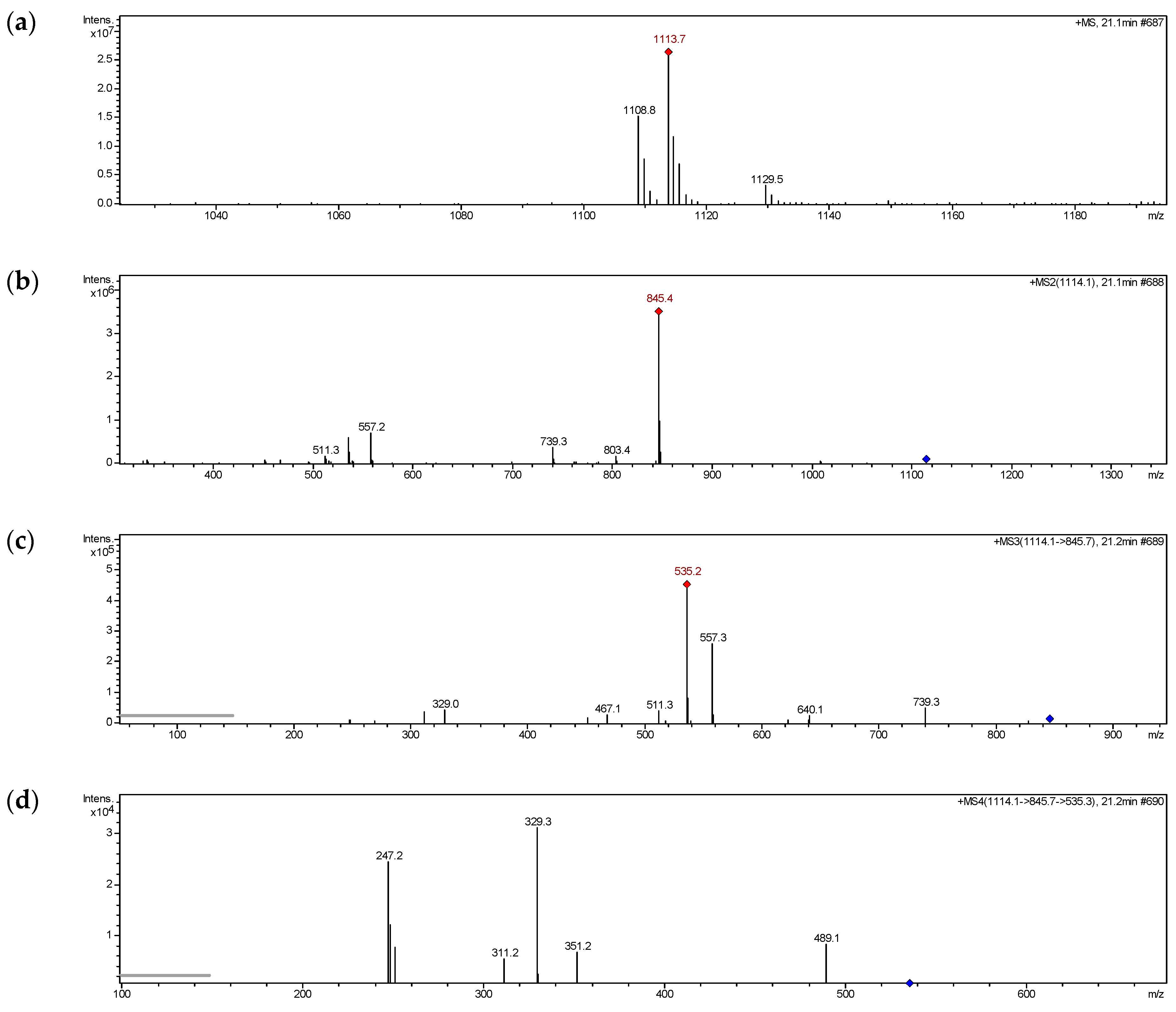
2.2. MS Fragmentation Pattern
2.2.1. MS Fragmentation Pathway of Glucosyloxybenzyl 2R-Benzylmalate Derivatives Isolated
2.2.2. Structural Prediction of Glucosyloxybenzyl 2R-Benzylmalates Unreported
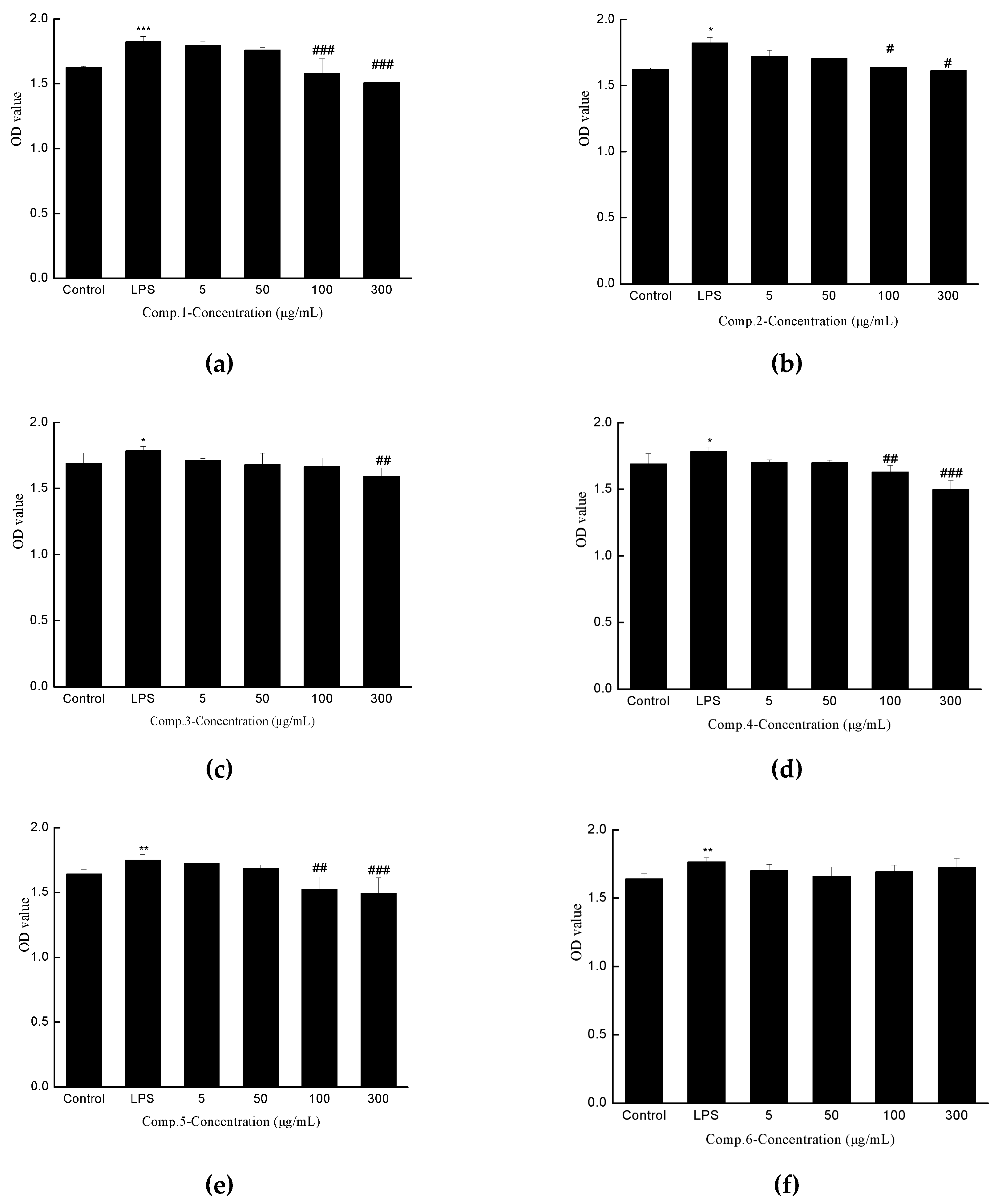
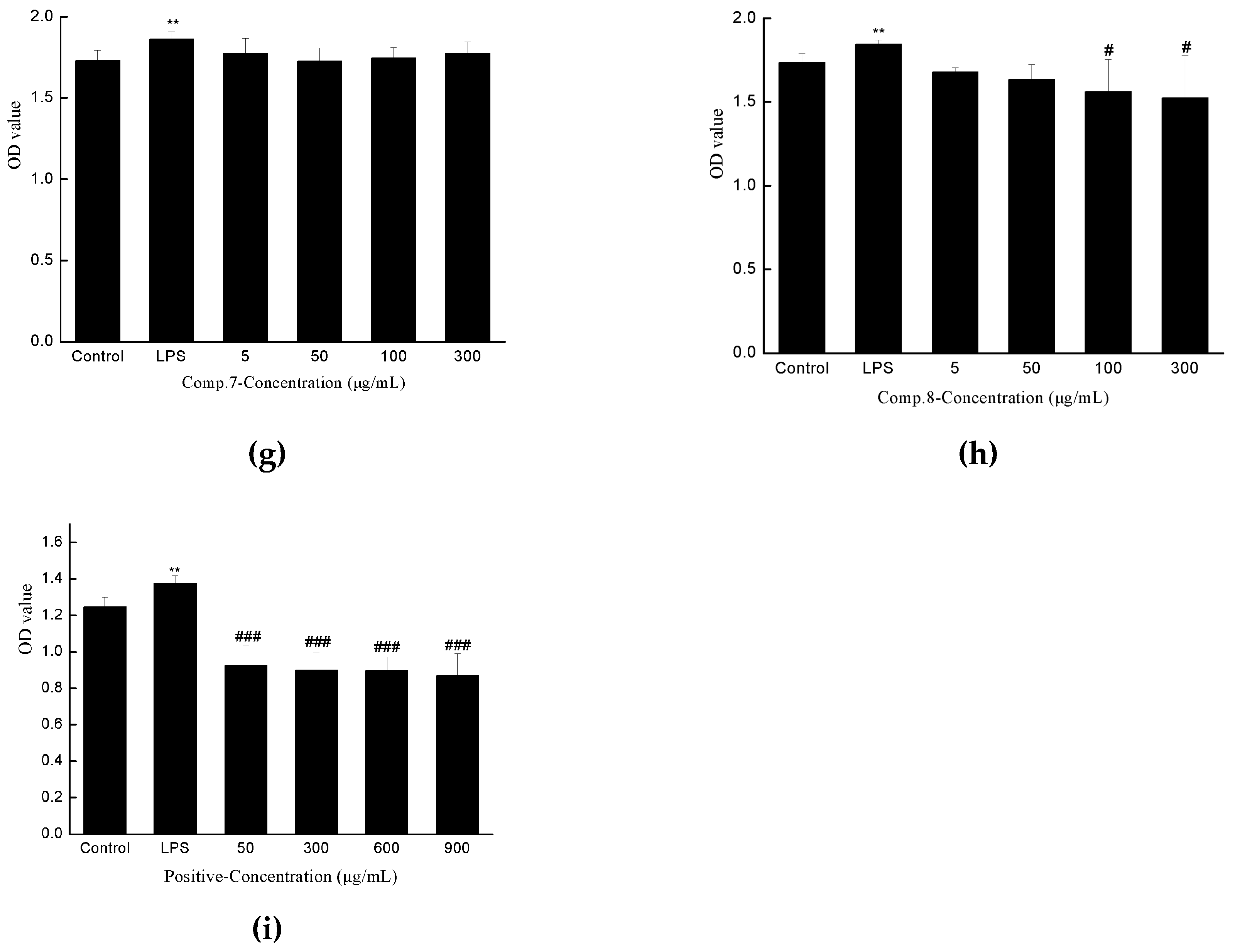
2.3. Anti-hepatic Fibrosis Activity
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Plant Material
3.3. Extraction and Isolation
3.4. Cell Proliferation Inhibition Assay
3.4.1. Chemical and Reagents
3.4.2. In vitro Evaluation of Anti-Liver Fibrotic Activity
4. Conclusions
5. Patents
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Jiang Su. New Medical College. Dictionary of Chinese Traditional Medicine; Shanghai Science and Technology Press: Shanghai, China, 1986; pp. 455–456. [Google Scholar]
- Li, Y.K.; Zhou, B.; Ye, Y.Q.; Du, G.; Niu, D.Y.; Meng, C.Y.; Gao, X.M.; Hu, Q.F. Two new diphenylethylenes from Arundina graminifolia and their cytotoxicity. Bull. Korean Chem. Soc. 2013, 34, 3257–3260. [Google Scholar] [CrossRef]
- Hu, Q.F.; Zhou, B.; Ye, Y.Q.; Jiang, Z.Y.; Huang, X.Z.; Li, Y.K.; Du, G.; Yang, G.Y.; Gao, X.M. Cytotoxic deoxybenzoins and diphenylethylenes from Arundina graminifolia. J. Nat. Product. 2013, 76, 1854–1859. [Google Scholar] [CrossRef] [PubMed]
- Meng, C.Y.; Niu, D.Y.; Li, Y.K.; Zhou, B.; Ye, Y.Q.; Du, G.; Hu, Q.F.; Gao, X.M. A new cytotoxic stilbenoid from Arundina graminifolia. Asian J. Chem. 2014, 26, 2411–2413. [Google Scholar] [CrossRef]
- Hu, Q.F.; Zhou, B.; Huang, J.M.; Gao, X.M.; Shu, L.D.; Yang, G.Y.; Che, C.T. Antiviral phenolic compounds from Arundina gramnifolia. J. Nat. Prod. 2013, 76, 292–296. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.R.; Xu, S.T.; Wei, J.; Shi, H.L.; Hu, Q.F. Phenolic compounds from Arundina graminifolia and their anti-tobacco mosaic virus activities. Asian J. Chem. 2013, 25, 2747–2749. [Google Scholar] [CrossRef]
- Li, L.; Xu, W.X.; Liu, C.B.; Zhang, C.M.; Zhao, W.; Shang, S.Z.; Deng, L.; Guo, Y.D. A new antiviral phenolic compound from Arundina gramnifolia. Asian J. Chem. 2015, 27, 3525–3526. [Google Scholar] [CrossRef]
- Li, Y.K.; Yang, L.Y.; Shu, L.D.; Shen, Y.Q.; Hu, Q.F.; Xia, Z.Y. Flavonoid compounds from Arundina graminifolia. Asian J. Chem. 2013, 25, 4922–4924. [Google Scholar] [CrossRef]
- Shu, L.D.; Shen, Y.Q.; Yang, L.Y.; Gao, X.M.; Hu, Q.F. Flavonoids derivatives from Arundina graminifolia and their cytotoxicity. Asian J. Chem. 2013, 25, 8358–8360. [Google Scholar] [CrossRef]
- Niu, D.Y.; Han, J.M.; Kong, W.S.; Cui, Z.W.; Hu, Q.F.; Gao, X.M. Antiviral fluorenone derivatives from Arundina gramnifolia. Asian J. Chem. 2013, 25, 9514–9516. [Google Scholar] [CrossRef]
- Zhu, H.; Song, Q.S. Chemical components of Arundina graminifolia. Nat. Prod. Res. Dev. 2008, 20, 5–7. [Google Scholar]
- Meng, W.W.; Nan, G.; Lin, F.K.; Zhao, H.W.; Deng, Y.L.; Dai, R.J.; Chen, Y.; Huang, Y.M.; Zhao, Y.H. Effect of Baogan Yihao on Liver Fibrosis in Rats Induced by CCl4 and High-Fat Feeding. Trans. Beijing Inst. Technol. 2014, 34, 767–770. [Google Scholar]
- He, P.P.; Song, C.C.; Meng, W.W.; Deng, Y.L.; Dai, R.J.; Lin, F.K.; Chen, Y.; Zhao, Y.H.; Huang, Y.M. Protective Effect of Baogan Yihao on Acute and Chronic Hepatic Injury. Chin. J. Exp. Tradit. Med. Formul. 2012, 18, 195–198. [Google Scholar]
- Dai, R.J.; Su, J.; Wang, F.; Lin, F.K.; Lv, F.; Chen, Y.; Meng, W.W.; Deng, Y.L. The Protective Effects of BGYH on Mice Acute Liver Injury Induced by D-Galn and LPS. Trans. Beijing Inst. Technol. 2015, 35 (Suppl. 1), 67–71. [Google Scholar]
- Liu, X.J.; Su, J.; Shi, Y.; Guo, Y.; Suheryani, I.; Zhao, S.C.; Deng, Y.L.; Meng, W.W.; Chen, Y.; Sun, L.L.; Dai, R.J. Herbal Formula, Baogan Yihao (BGYH), Prevented Dimethyl-nitrosamine (DMN)-Induced Liver Injury in Rats. Drug Dev. Res. 2017, 78, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Auberon, F.; Olatunji, O.J.; Krisa, S.; Herbette, G.; Antheaume, C.; Bonte, F.; Merillon, J.M.; Lobstein, A. Arundinosides AG, new glucosyloxybenzyl 2R-benzylmalate derivatives from the aerial parts of Arundina graminifolia. Fitoterapia 2018, 125, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Morikawa, T.; Xie, H.; Matsuda, H.; Yoshikawa, M. Glucosyloxybenzyl 2-Isobutylmalates from the Tubers of Gymnadenia conopsea. J. Nat. Prod. 2006, 69, 881–886. [Google Scholar] [CrossRef] [PubMed]
- Tatsis, C.E.; Boeren, S.; Exarchou, V.; Troganis, A.N.; Vervoort, J.; Gerothanassis, I.P. Identification of the major constituents of Hypericum perforatum by LC/SPE/NMR and/or LC/MS. Phytochemistry 2007, 68, 383–393. [Google Scholar] [CrossRef]
- Song, J.; Dai, R.J.; Deng, Y.L.; Lv, F. Rapid structure prediction by HPLC-ESI-MSn of twenty-five polyoxypregnane tetraglycosides from Dregea sinensis with NMR confirmation of eight structures. Phytochemistry 2018, 147, 147–157. [Google Scholar] [CrossRef]
- Cai, M.; Zhou, Y.; Gesang, S.L.; Bianba, C.; Ding, L.S. Chemical fingerprint analysis of rhizomes of Gymnadenia conopsea by HPLC–DAD–MSn. J. Chromatogr. B 2006, 844, 301–307. [Google Scholar] [CrossRef] [Green Version]
- Kao, Y.H.; Chen, P.H.; Wu, T.Y.; Lin, Y.C.; Tsai, M.S.; Lee, P.H.; Tai, T.S.; Chang, H.R.; Sun, C.K. Lipopolysaccharides induce Smad2 phosphorylation through PI3K/Akt and MAPK cascades in HSC-T6 hepatic stellate cells. Life Sci. 2017, 184, 37–46. [Google Scholar] [CrossRef]
- Kang, K.B.; Jin, B.J.; Kim, J.W.; Kim, H.W.; Sung, S.H. Ceanothane- and lupane-type triterpene esters from the roots of Hovenia dulcis, and their antiproliferative activity on HSC-T6 cells. Phytochemistry 2017, 142, 60. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |
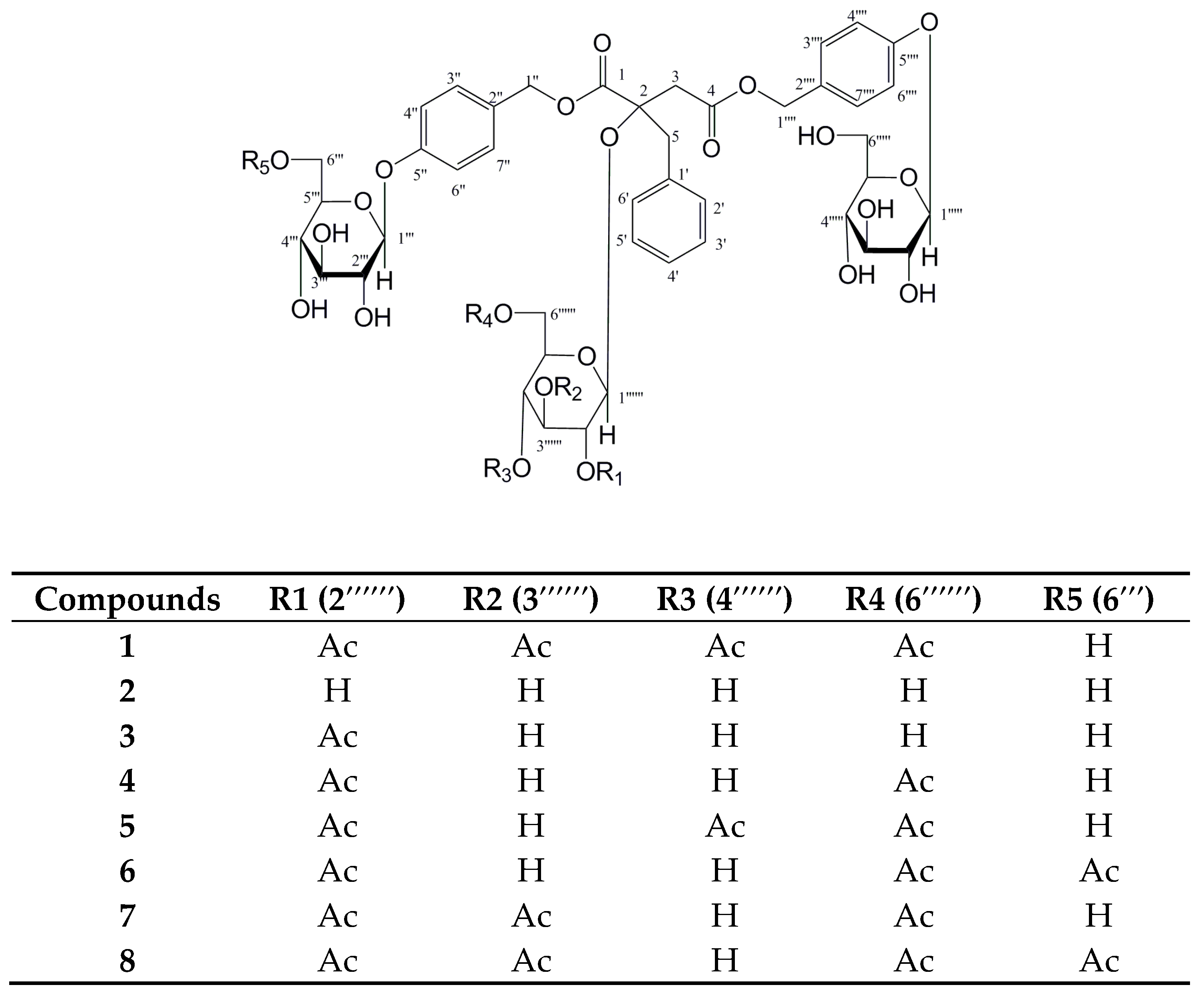
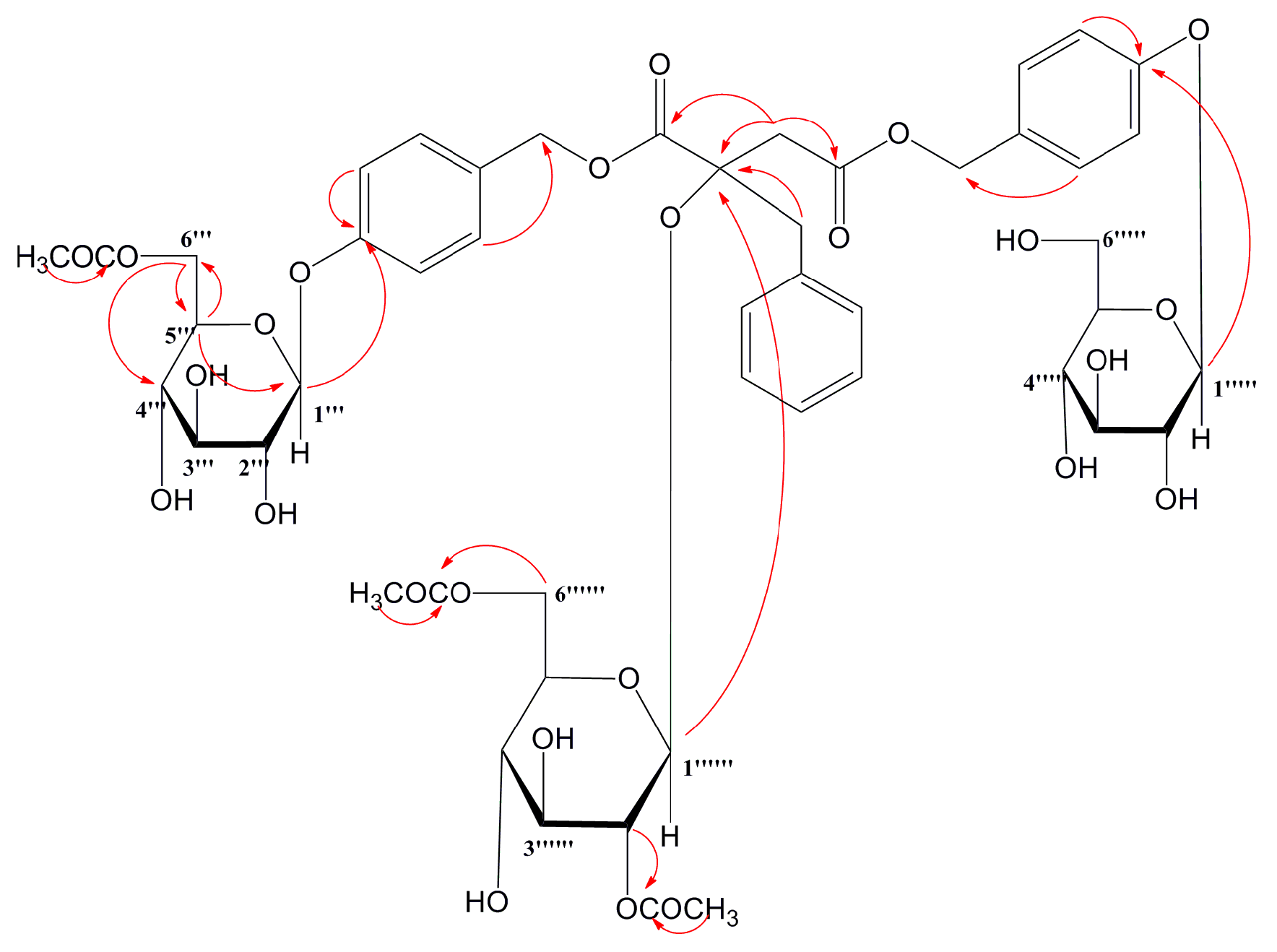

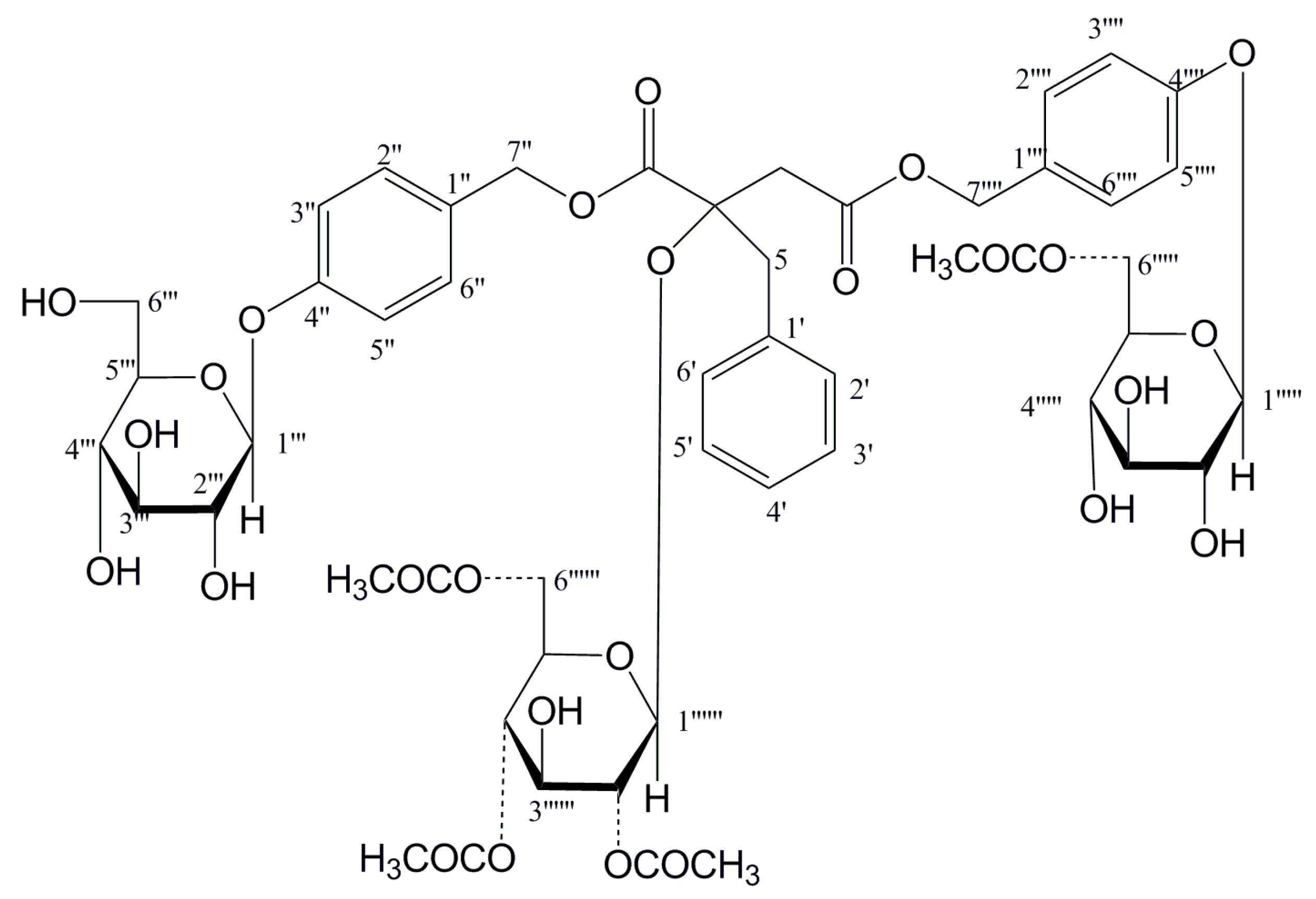
| Position | 2 | 5 | 6 | 8 |
|---|---|---|---|---|
| 3 | 2.96 (d, 17.7); 2.82 (d, 17.7) | 2.96 (d, 15); 2.90 (d, 15) | 2.96 (d, 17.8); 2.90 (d, 17.8) | 2.96 (d, 17.8); 2.92 (d, 17.8) |
| 5 | 3.17 (m); 3.06 (m) | 3.10 (d, 14); 3.02 (d, 14) | 3.10(d, 14); 3.02 (d, 14) | 3.10 (d, 14); 3.02 (d, 14) |
| 2′, 6′ | 7.19 (m) | 7.18 (m) | 7.18 (m) | 7.18 (m) |
| 3′, 5′ | 7.16 (m) | 7.02 (m) | 7.01 (d, 8.7) | 7.01 (d, 8.7) |
| 4′ | 7.19 (m) | 7.18 (m) | 7.17 (m) | 7.17 (m) |
| 1″ | 4.99 (d, 7.9); 4.90 (d, 7.9) | 5.00 (d, 12); 4.92 (d, 12) | 5.00 (d, 12); 4.93 (d,12) | 5.00(d,12); 4.93 (d,12) |
| 3″, 7″ | 7.27 (d, 8.4) | 7.27 (d, 8.3) | 7.27 (d, 8.7) | 7.27 (d, 8.7) |
| 4″, 6″ | 7.01 (d, 8.5) | 7.02 (m) | 7.01 (d, 8.7) | 7.01 (d, 8.7) |
| Glc-1′′′ | 4.87 (d, 7.5) | 4.91 (d, 7.9) | 4.92 (d, 7.5) | 4.92 (d, 7.7) |
| Glc-2′′′ | 3.27 (m) | 3.26 (m) | 3.27 (m) | 3.27 (m) |
| Glc-3′′′ | 3.24 (m) | 3.23 (m) | 3.22 (m) | 3.22 (m) |
| Glc-4′′′ | 3.16 (m) | 3.16 (m) | 3.18 (m) | 3.18 (m) |
| Glc-5′′′ | 3.27 (m) | 3.26 (m) | 3.27 (m) | 3.27 (m) |
| Glc-6′′′ | 3.68 (m); 3.47 (m) | 3.68 (m); 3.46 (m) | 4.27 (m); 4.08 (m) | 4.27 (m); 4.08 (m) |
| Glc-6′′′-COCH3 | - | - | 1.99 (s) | 1.99 (s) |
| 1′′′′ | 5.01 (d, 12); 4.90 (d, 12) | 5.04 (d, 12); 4.91 (d, 12) | 5.00 (d, 12); 4.99 (d, 12) | 5.00 (d,12); 4.99 (d,12) |
| 3′′′′, 7′′′′ | 7.25 (d, 8.4) | 7.23 (d, 8.3) | 7.23 (d, 8.7) | 7.23 (d, 8.7) |
| 4′′′′, 6′′′′ | 7.02 (d, 8.5) | 7.02 (m) | 7.02 (d, 8.7) | 7.02 (d, 8.7) |
| Glc-1′′′′′ | 4.87 (d, 7.5) | 4.86 (d, 7.7) | 4.92 (d, 7.5) | 4.87 (d, 7.5) |
| Glc-2′′′′′ | 3.27 (m) | 3.16 (m) | 3.30 (m) | 3.30 (m) |
| Glc-3′′′′′ | 3.24 (m) | 3.23 (m) | 3.22 (m) | 3.22 (m) |
| Glc-4′′′′′ | 3.16 (m) | 3.16 (m) | 3.18 (m) | 3.18 (m) |
| Glc-5′′′′′ | 3.27 (m) | 3.26 (m) | 3.27 (m) | 3.27 (m) |
| Glc-6′′′′′ | 3.69 (m); 3.47 (m) | 3.68 (m); 3.46 (m) | 3.68 (m); 3.46 (m) | 3.68 (m); 3.46 (m) |
| Glc-1′′′′′′ | 4.66 (d, 7.8) | 4.94 (d, 8.0) | 4.86 (d, 7.6) | 5.05 (d, 8.0) |
| Glc-2′′′′′′ | 3.03 (m) | 4.68 (m) | 4.56 (m) | 4.69 (m) |
| Glc-3′′′′′′ | 3.39 (m) | 3.46 (m) | 3.68 (m) | 4.93 (m) |
| Glc-4′′′′′′ | 4.36 (m) | 4.64 (m) | 3.46 (m) | 3.46 (m) |
| Glc-5′′′′′′ | 3.52 (m) | 3.45 (m) | 3.61 (m) | 3.61 (m) |
| Glc-6′′′′′′ | 3.68 (m); 3.38 (m) | 4.05 (m); 3.76 (m) | 4.08 (m); 4.05 (m) | 4.08 (m); 4.05 (m) |
| Glc-2′′′′′′-COCH3 | - | 1.70 (s) | 1.72 (s) | 1.65 (s) |
| Glc-3′′′′′′-COCH3 | - | - | - | 1.92 (s) |
| Glc-4′′′′′′-COCH3 | - | 2.01 (s) | - | |
| Glc-6′′′′′′-COCH3 | - | 1.92 (s) | 1.92 (s) | 1.99 (s) |
| Position | 2 | 5 | 6 | 8 |
|---|---|---|---|---|
| 1 | 170.9 | 170.1 | 170.5 | 170.1 |
| 2 | 79.8 | 80.7 | 81.0 | 81.3 |
| 3 | 40.4 | 40.9 | 41.0 | 41.0 |
| 4 | 169.8 | 169.5 | 170.1 | 170.1 |
| 5 | 42.0 | 43.6 | 43.8 | 43.7 |
| 1′ | 135.5 | 135.0 | 135.5 | 135.4 |
| 2′, 6′ | 127.9 | 127.9 | 128.3 | 128.3 |
| 3′, 5′ | 130.6 | 130.5 | 130.9 | 130.9 |
| 4′ | 126.7 | 126.7 | 127.1 | 127.1 |
| 1″ | 66.3 | 66.2 | 66.6 | 66.6 |
| 2″ | 128.9 | 128.8 | 129.3 | 129.2 |
| 3″, 7″ | 129.9 | 129.9 | 130.3 | 130.3 |
| 4″, 6″ | 116.2 | 116.2 | 116.7 | 116.7 |
| 5″ | 157.4 | 157.4 | 157.9 | 157.9 |
| Glc-1′′′ | 100.3 | 100.3 | 100.5 | 100.5 |
| Glc-2′′′ | 76.8 | 76.6 | 77.1 | 77.1 |
| Glc-3′′′ | 73.2 | 73.2 | 73.5 | 73.6 |
| Glc-4′′′ | 69.6 | 69.7 | 70.1 | 70.2 |
| Glc-5′′′ | 76.6 | 76.6 | 76.8 | 76.8 |
| Glc-6′′′ | 60.7 | 60.7 | 63.9 | 63.8 |
| Glc-6′′′-COCH3 | - | - | 170.7; 21.2 | 170.7; 21.1 |
| 1′′′′ | 65.6 | 65.7 | 66.2 | 66.2 |
| 2′′′′ | 128.8 | 128.6 | 129.3 | 129.3 |
| 3′′′′, 7′′′′ | 129.9 | 129.9 | 130.4 | 130.4 |
| 4′′′′, 6′′′′ | 116.1 | 116.2 | 116.5 | 116.5 |
| 5′′′′ | 157.4 | 157.4 | 157.6 | 157.6 |
| Glc-1′′′′′ | 100.3 | 100.4 | 100.8 | 100.8 |
| Glc-2′′′′′ | 76.8 | 77.0 | 77.5 | 77.5 |
| Glc-3′′′′′ | 73.2 | 73.2 | 73.6 | 73.4 |
| Glc-4′′′′′ | 69.6 | 70.3 | 70.1 | 70.1 |
| Glc-5′′′′′ | 76.6 | 76.6 | 76.8 | 76.8 |
| Glc-6′′′′′ | 60.7 | 60.7 | 61.1 | 61.1 |
| Glc-1′′′′′′ | 98.3 | 96.7 | 97.0 | 96.7 |
| Glc-2′′′′′′ | 77.0 | 70.9 | 73.8 | 71.2 |
| Glc-3′′′′′′ | 73.7 | 70.9 | 74.0 | 75.1 |
| Glc-4′′′′′′ | 69.7 | 72.8 | 73.7 | 67.9 |
| Glc-5′′′′′′ | 76.7 | 70.6 | 74.1 | 74.1 |
| Glc-6′′′′′′ | 61.2 | 61.8 | 63.3 | 62.9 |
| Glc-2′′′′′′-COCH3 | - | 169.7; 20.5 | 169.8; 21.0 | 169.6; 20.7 |
| Glc-3′′′′′′-COCH3 | - | - | - | 170.3; 21.0 |
| Glc-4′′′′′′-COCH3 | - | 169.3; 20.8 | - | - |
| Glc-6′′′′′′-COCH3 | - | 170; 20.7 | 170.7; 21.1 | 170.1;21.2 |
| Compounds | Molecular Formula | HR-ESI-MS [M + NH4]+ | ESI-MS1: [M + Na]+ | ESI-MSn |
|---|---|---|---|---|
| 1 | C51H62O26 | 1108.3861 | 1113 | 845, 577, 515, 497, 371, 353, 311, 251, 247 |
| 2 | C43H54O22 | 940.3453 | 945 | 677, 515, 409, 247 |
| 3 | C45H56O23 | 982.3554 | 987 | 719, 515, 451, 247 |
| 4 | C47H58O24 | 1024.3665 | 1029 | 761, 515, 493, 287, 269, 247, 227 |
| 5 | C49H60O25 | 1066.3766 | 1071 | 803, 535, 329, 311, 269, 247 |
| 6 | C49H60O25 | 1066.3764 | 1071 | 761, 515,493, 287, 269, 247 |
| 7 | C49H60O25 | 1066.3766 | 1071 | 803, 535, 515, 329, 311, 247, 209 |
| 8 | C51H62O26 | 1108.3869 | 1113 | 803, 535, 515, 329, 311, 247, 209 |

| Peaks | RT | MS1 [M + Na]+ | MSn | G1 | G2 | G3 |
|---|---|---|---|---|---|---|
| A1 | 3.9 min | 1006 | 1029, 761, 515, 493, 287, 269, 247 | 2Ac | ||
| A2 | 6.4 min | 1048 | 1071, 803, 535, 515, 329, 311 | 3Ac | ||
| A3 | 7.3 min | 1048 | 1072, 761, 515, 493, 287, 269, 247 | Ac | 2Ac | |
| A4 | 12.7 min | 1090 | 1113, 845, 535, 329, 311 | Ac | 3Ac | |
| A5 | 13.9 min | 1090 | 1113, 803, 535, 329, 311, 247 | Ac | 3Ac | |
| A6 | 14.0 min | 1052 | 1071, 677, 515, 247 | Cin | ||
| A7 | 22.2 min | 1094 | 1117, 849, 645, 247 | Cin | Ac | |
| A8 | 21.1 min | 1090 | 1113, 845, 535, 329, 247 | Ac | 3Ac | |
| A9 | 28.0 min | 1094 | 1117, 719, 451, 247 | Cin | Ac | |
| B1 | 3.7 min | 964 | 987, 719, 515, 247 | Ac | ||
| B2 | 4.3 min | 964 | 987, 719, 515, 247 | Ac | ||
| B3 | 7.0 min | 760 | 783, 515, 247 | OH | ||
| B4 | 9.0 min | 1006 | 1029, 761, 557, 451, 247 | Ac | Ac | |
| B5 | 11.0 min | 1006 | 1029, 719, 515, 451, 247 | Ac | Ac | |
| B6 | 12.6 min | 1006 | 1029, 761, 515, 493, 287, 269, 247 | 2Ac | ||
| C1 | 6.4 min | 1048 | 1071, 761, 557, 451, 247 | Ac | Ac | Ac |
| C2 | 8.9 min | 1048 | 1071, 761, 515, 493, 287, 269, 247 | Ac | 2Ac | |
| C3 | 11.9 min | 1048 | 1071, 803, 535, 329, 311, 269, 247 | 3Ac | ||
| D1 | 16.0 min | 1252 | 1275, 845, 577, 371, 353, 311 | 4Ac | ||
| D2 | 8.5 min | 1052 | 1075, 677, 515, 247 | Cin | ||
| D3 | 7.0 min | 1052 | 1075, 677, 515, 247 | Cin | ||
| D4 | 15.6 min | 1094 | 1117, 719, 515, 451, 247 | Cin | Ac | |
| D5 | 18.6 min | 1094 | 1117, 719, 515, 451, 247 | Cin | Ac | |
| D6 | 19.3 min | 1094 | 1117, 719, 515, 451, 247 | Cin | Ac |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Q.; Sun, F.; Deng, Y.; Dai, R.; Lv, F. HPLC-ESI-MSn Identification and NMR Characterization of Glucosyloxybenzyl 2R-Benzylmalate Deriva-Tives from Arundina Graminifolia and Their Anti-Liver Fibrotic Effects In Vitro. Molecules 2019, 24, 525. https://doi.org/10.3390/molecules24030525
Liu Q, Sun F, Deng Y, Dai R, Lv F. HPLC-ESI-MSn Identification and NMR Characterization of Glucosyloxybenzyl 2R-Benzylmalate Deriva-Tives from Arundina Graminifolia and Their Anti-Liver Fibrotic Effects In Vitro. Molecules. 2019; 24(3):525. https://doi.org/10.3390/molecules24030525
Chicago/Turabian StyleLiu, Qingqing, Feiyi Sun, Yulin Deng, Rongji Dai, and Fang Lv. 2019. "HPLC-ESI-MSn Identification and NMR Characterization of Glucosyloxybenzyl 2R-Benzylmalate Deriva-Tives from Arundina Graminifolia and Their Anti-Liver Fibrotic Effects In Vitro" Molecules 24, no. 3: 525. https://doi.org/10.3390/molecules24030525




