Antiplasmodial and Cytotoxic Cytochalasins from an Endophytic Fungus, Nemania sp. UM10M, Isolated from a Diseased Torreya taxifolia Leaf
Abstract
:1. Introduction
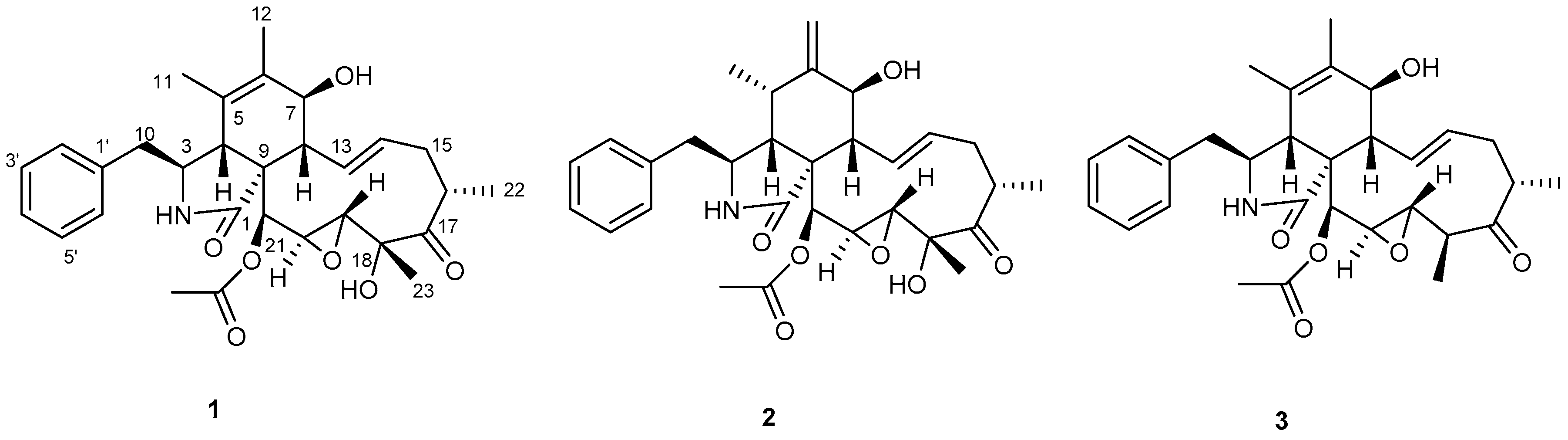
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal Material
3.3. Fermentation, Extraction, and Purification
3.4. Biological Assays
3.4.1. In Vitro Antiplasmodial Assay
3.4.2. In Vitro Phytotoxicity Assay
3.4.3. In Vitro Cytotoxicity Assay
3.4.4. In Vivo Antimalarial Assay
3.5. DNA Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization (WHO). World Malaria Report; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- Lim, L.; McFadden, G.I. The evolution, metabolism, and functions of the apicoplast. Philos. Trans. R. Soc. B 2010, 365, 749–763. [Google Scholar] [CrossRef] [PubMed]
- Ralph, S.A.; D’Ombrain, M.C.; McFadden, G.I. The apicoplast as an antimalarial drug target. Drug Resist. Updates 2001, 4, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Ralph, S.A.; van Dooren, G.G.; Waller, R.F.; Crawford, M.J.; Fraunholz, M.J.; Foth, B.J.; Tonkin, C.J.; Roos, D.S.; McFadden, G.I. Tropical infectious diseases: Metabolic maps and functions of the Plasmodium falciparum apicoplast. Nat. Rev. Microbiol. 2004, 2, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Waller, R.F.; McFadden, G.I. The apicoplast: A review of the derived plastid of apicomplexan parasites. Curr. Issues Mol. Biol. 2005, 7, 57–80. [Google Scholar] [PubMed]
- Dhal, E.L.; Rosenthal, P.J. Multiple antibiotics extert delayed effects against the Plasmodium falciparum apicoplast. Antimicrob. Agents Chemother. 2007, 51, 3485–3490. [Google Scholar] [CrossRef] [PubMed]
- Botté, C.Y.; Dubar, F.; McFadden, G.I.; Maréchal, E.; Biot, C. Plasmodium falciparum Apicoplast Drugs: Targets or Off-Targets? Chem. Rev. 2012, 112, 1269–1283. [Google Scholar] [CrossRef] [PubMed]
- Franck, E.D.; Duke, S. Natural compounds as next-generation herbicides. Plant Physiol. 2014, 166, 1090–1105. [Google Scholar] [CrossRef]
- Roberts, F.; Roberts, C.W.; Johnson, J.J.; Kyle, D.E.; Krell, T.; Coggins, J.R.; Coombs, G.H.; Milhous, W.K.; Tzipori, S.; Ferguson, D.J.; et al. Evidence for the shikimate pathway in apicomplexan parasites. Nature 1998, 393, 801–805. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. Non-mevalonate isoprenoid biosynthesis enzymes, genes, and inhibitors. Biochem. Soc. Trans. 2000, 28, 785–789. [Google Scholar] [CrossRef]
- Bajsa, J.; Singh, K.; Nanayakkara, D.; Duke, S.O.; Rimando, A.M.; Evidente, A.; Tekwani, B.L. A Survey of synthetic and natural phytotoxic compounds and phytoalexins as potential antimalarial compounds. Biol. Pharm. Bull. 2007, 30, 1740–1744. [Google Scholar] [CrossRef]
- Herath, H.M.T.B.; Herath, W.H.M.W.; Carvalho, P.; Khan, S.I.; Tekwani, B.L.; Duke, S.O.; Tomaso-Peterson, M.; Nanayakkara, N.P.D. Biologically active tetranorditerpenoids from the fungus Sclerotinia homoeocarpa causal agent of dollar spot in turfgrass. J. Nat. Prod. 2009, 72, 2091–2097. [Google Scholar] [CrossRef] [PubMed]
- Kumarihamy, M.; Fronczek, F.R.; Ferreira, D.; Jacob, M.; Khan, S.I.; Nanayakkara, N.P.D. Bioactive 1,4-dihydroxy-5-phenyl-2-pyridinone alkaloids from Septoria pistaciarum. J. Nat. Prod. 2010, 73, 1250–1253. [Google Scholar] [CrossRef] [PubMed]
- Kumarihamy, M.; Khan, S.I.; Jacob, M.; Tekwani, B.L.; Duke, S.O.; Ferreira, D.; Nanayakkara, N.P.D. Antiprotozoal and antimicrobial compounds from Septoria pistaciarum. J. Nat. Prod. 2012, 75, 883–889. [Google Scholar] [CrossRef] [PubMed]
- Edwards, R.L.; Jonglaekha, N.; Anandini, K.; Maitland, D.J.; Mekkamol, S.; Nugent, L.K.; Phosri, C.; Rodtong, S.; Ruchichachorn, N.; Sangvichien, E.; et al. The Xylariaceae as phytopathogens. Recent Res. Dev. Plant Sci. 2003, 1, 1–19. [Google Scholar]
- Isaka, M.; Jaturapat, A.; Kladwang, W.; Punya, J.; Lertwerawat, Y.; Tanticharoen, M.; Thebtaranonth, Y. Antiplasmodial compounds from the wood-decayed fungus Xylaria sp. BCC 1067. Planta Med. 2000, 66, 473–475. [Google Scholar] [CrossRef] [PubMed]
- Dieckmann-Schuppert, A.; Franklin, R.M. Compounds binding to cytoskeletal proteins are active against Plasmodium falciparum in vitro. Cell Biol. Int. Rep. 1989, 13, 411–418. [Google Scholar] [CrossRef]
- Dobrowolski, J.M.; Sibley, L.D. Toxoplasma invasion of mammalian cells is powered by the actin cytoskeleton of the parasite. Cell 1996, 84, 933–939. [Google Scholar] [CrossRef]
- Zhang, Q.; Xiao, J.; Sun, Q.-Q.; Qin, J.-C.; Pescitelli, G.; Gao, J.-M. Characterization of cytochalasins from the endophytic sp. and their biological functions. J. Agric. Food Chem. 2014, 62, 10962–10969. [Google Scholar] [CrossRef]
- Hussain, H.; Kliche-Spory, C.; Al-Harrasi, A.; Al-Rawahi, A.; Abbas, G.; Green, I.R.; Schulz, B.; Krohn, K.; Shah, A. Antimicrobial constituents from three endophytic fungi. Asian Pac. J. Trop. Med. 2014, 7, S224–S227. [Google Scholar] [CrossRef]
- Berestetskiy, A.; Dmitriev, A.; Mitina, G.; Lisker, I.; Andolfi, A.; Evidente, A. Nonenolides and cytochalasins with phytotoxic activity against Cirsium arvense and Sonchus arvensis: A structure-activity relationships study. Phytochemistry 2008, 69, 953–960. [Google Scholar] [CrossRef]
- Scherlach, K.; Boettger, D.; Remme, N.; Hertweck, C. The chemistry and biology of cytochalasans. Nat. Prod. Rep. 2010, 27, 869–886. [Google Scholar] [CrossRef] [PubMed]
- Trendowski, M. Using cytochalasins to improve current chemotherapeutic approaches. Anti-Cancer Agents Med. Chem. (Former. Curr. Med. Chem.-Anti-Cancer Agents) 2015, 15, 327–335. [Google Scholar] [CrossRef]
- Abate, D.; Abraham, W.-R.; Meyer, H. Cytochalasins and phytotoxins from the fungus Xylaria obovata. Phytochemistry 1997, 44, 1443–1448. [Google Scholar] [CrossRef]
- Espada, A.; Rivera-Sagredo, A.; de la Fuente, J.M.; Hueso-Rodríguez, J.A.; Elson, S.W. New cytochalasins from the fungus Xylaria hypoxylon. Tetrahedron 1997, 53, 6485–6492. [Google Scholar] [CrossRef]
- Song, Y.X.; Wang, J.; Li, S.W.; Cheng, B.; Li, L.; Chen, B.; Liu, L.; Lin, Y.C.; Gu, Y.C. Metabolites of the mangrove fungus Xylaria sp. BL321 from the South China Sea. Planta Med. 2012, 78, 172–176. [Google Scholar] [CrossRef]
- Okoyea, F.B.C.; Nworuc, C.S.; Debbaba, A.; Esimone, C.O.; Proksch, P. Two new cytochalasins from an endophytic fungus, KL-1.1 isolated from Psidium guajava leaves. Phytochem. Lett. 2015, 14, 51–55. [Google Scholar] [CrossRef]
- Chen, Z.; Chen, Y.; Huang, H.; Yang, H.; Zhang, W.; Sun, Y.; Wen, J. Cytochalasin P1, a new cytochalasin from the marine-derived fungus Xylaria sp. SOF11. Z. Naturforsch. C 2017, 72, 129–132. [Google Scholar] [CrossRef]
- Jikai, L.; Zejun, D.; Zhihui, D.; Xianghua, W.; Peiqui, L. Neoengleromycin, a novel compound from Engleromyces goetzii. Helv. Chim. Acta 2002, 85, 1439–1442. [Google Scholar] [CrossRef]
- Shi, L.-M.; Zhan, Z.-J. Structural revision of 19, 20-epoxycytochalasin D and its cytotoxic activity. J. Chem. Res. 2007, 3, 144–145. [Google Scholar] [CrossRef]
- Drewry, L.D.; Sibley, L.D. Toxoplasma actin is required for efficient host cell invasion. mBio 2015, 6, e00557-15. [Google Scholar] [CrossRef]
- Baum, J.; Papenfuss, A.T.; Baum, B.; Speed, T.P.; Cowman, A.F. Regulation of apicomplexan actin-based motility. Nat. Rev. Microbiol. 2006, 4, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Lemgruber, L.; Tay, C.L.; Baum, J.; Meissner, M. Multiple essential functions of Plasmodium falciparum actin-1 during malaria blood-stage development. BMC Biol. 2017, 15, 70. [Google Scholar] [CrossRef] [PubMed]
- Bousquet, P.F.; Paulsen, L.A.; Fondy, C.; Lipski, K.M.; Loucy, K.J.; Fondy, T.P. Effects of cytochalasin B in culture and in vivo on murine Madison 109 lung carcinoma and on B16 melanoma. Cancer Res. 1990, 50, 1431–1439. [Google Scholar] [PubMed]
- Trendowski, M.; Mitchell, J.M.; Corsette, C.M.; Acquafondata, C.; Fondy, T.P. Chemotherapy in vivo against M109 murine lung carcinoma with cytochalasin B by localized, systemic, and liposomal administration. Investig. New Drugs 2015, 33, 280–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trendowski, M.; Mitchell, J.M.; Corsette, C.M.; Acquafondata, C.; Fondy, T.P. Chemotherapy with cytochalasin congeners in vitro and in vivo against murine models. Investig. New Drugs 2015, 33, 290–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bharate, S.B.; Khan, S.I.; Yunus, N.A.M.; Chauthe, S.K.; Jacob, M.R.; Tekwani, B.L.; Khan, I.A.; Singh, I.P. Antiprotozoal and antimicrobial activities of O-alkylated and formylated acylphloroglucinols. Bioorg. Med. Chem. 2007, 15, 87–96. [Google Scholar] [CrossRef]
- Dayan, F.E.; Romagni, J.G.; Duke, S.O. Investigating the mode of action of natural phytotoxins. J. Chem. Ecol. 2000, 26, 2079–2094. [Google Scholar] [CrossRef]
- Mustafa, J.; Khan, S.I.; Ma, G.; Walker, L.; Khan, I.A. Synthesis and anticancer activities of fatty acid analogs of podophyllotoxin. Lipids 2004, 39, 167–172. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., Whit, T.J., Eds.; Academic Press, Inc.: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Altschul, S.F.; Gish, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Nicholas, K.B.; Nicholas, H.B.; Deerfield, D.W., II. A Tool for Editing and Annotating Multiple Sequence Alignments, Version 2.7. Distributed by the Authors. 1997. Available online: http://www.psc.edu/biomed/genedoc (accessed on 15 October 2006).
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |


| Carbon | 1 | 3 | ||
|---|---|---|---|---|
| δCb | δHa (J in Hz) | δCb | δHa (J in Hz) | |
| 1 | 175.0 | 175.1 | ||
| 3 | 61.0 | 3.35, t (7.3) | 61.1 | 3.32, m |
| 4 | 49.9 | 2.50, brs | 49.9 | 2.43, brs |
| 5 | 126.4 | 128.5 | ||
| 6 | 131.8 | 131.7 | ||
| 7 | 68.1 | 3.76, brd (9.9) | 68.1 | 3.77, d (9.7) |
| 8 | 48.7 | 2.25, dd (10.1,10.0) | 49.0 | 2.27, dd, (10.1, 10.1) |
| 9 | 52.0 | 51.8 | ||
| 10 | 44.1 | 3.03, d (7.4) | 44.4 | 3.04, bd, (7.5) |
| 11 | 16.7 | 1.19, s | 14.1 | 1.26, s |
| 12 | 13.8 | 1.62, s | 13.9 | 1.65, s |
| 13 | 131.4 | 5.99, dd (15.5, 10.2) | 131.0 | 6.24, dd (15.5, 10.4) |
| 14 | 132.9 | 5.68, ddd (15.6, 9.7, 5.8) | 133.7 | 5.61, m |
| 15 | 37.5 | 2.63, dd (22.7, 11.9), 2.14, m c | 37.6 | 2.53, dd (25.3, 12.2), 2.14 m c |
| 16 | 41.7 | 3.27, m c | 42.9 | 2.98, d (1.8) |
| 17 | 215.3 | 216.9 | ||
| 18 | 76.3 | 52.1 | 2.18, m c | |
| 19 | 59.9 | 3.25, brs c | 58.6 | 3.01, m c |
| 20 | 53.2 | 3.38, brs | 57.6 | 3.38, d (1.7) |
| 21 | 72.0 | 5.75, s | 72.3 | 5.69, s |
| 22 | 18.9 | 1.21, d (7.6) | 18.8 | 1.12, d (6.7) |
| 23 | 21.5 | 1.54, s | 17.1 | 1.32, d (6.9) |
| 24 | 170.4 | 170.5 | ||
| 25 | 20.5 | 2.18, s | 20.7 | 2.15, s |
| 1′ | 137.3 | 136.9 | ||
| 2′,6′ | 129.2 | 7.28, m | 129.4 | 7.26, m |
| 3′,5′ | 128.6 | 7.33, m | 128.7 | 7.33, m |
| 4′ | 126.8 | 7.25, m | 126.9 | 7.25, m |
| Compound | Chloroquine-Sensitive (D6)-Strain | Chloroquine-Resistant (W2)-Strain | Cytotoxicity to Vero Cells | ||
|---|---|---|---|---|---|
| IC50 µM (ng/mL) | S. I. | IC50 µM (ng/mL) | S. I. | IC50 µM | |
| 19,20-Epoxycytochalasin C (1) | 0.07 (37) | >128.6 | 0.05 (28) | >170 | NC |
| 19,20-Epoxycytochalasin D (2) | 0.04 (22) | >216.3 | 0.04 (20) | >238 | NC |
| 18-Deoxy-19,20-epoxy-cytochalasin C (3) | 0.56 (280) | >17 | 0.19 (100) | >47.6 | NC |
| Chloroquine a | 0.03 (16) | >297.5 | 0.31 (160) | >29.8 | NC |
| Artemisinin a | 0.02 (5.6) | >850 | 0.01 (3.0) | >1586.6 | NC |
| Compound | Lettuce | Bentgrass |
|---|---|---|
| 19,20-Epoxycytochalasin C (1) | 3 | 2 |
| 19,20-Epoxycytochalasin D (2) | 3 | 3 |
| 18-Deoxy-19,20-epoxycytochalasin C (3) | 3 | 4 |
| Compound | SK-MEL | KB | BT-549 | SK-OV-3 | LLC-PK11 |
|---|---|---|---|---|---|
| 19,20-Epoxycytochalasin C (1) | 8.02 | NC | NC | NC | NC |
| 19,20-Epoxycytochalasin D (2) | NC | NC | 7.84 | NC | 8.4 |
| 18-Deoxy-19,20-epoxycytochalasin C (3) | NC | NC | 6.89 | NC | NC |
| Doxorubicin a | 1.29 | 2.12 | 1.83 | 1.47 | 1.28 |
| Treatment (PO) | Dose (mg/kg × # days Post Infection) | % Parasitemia Suppression b | Survival c | Day of Death | MST d | Cure f | |
|---|---|---|---|---|---|---|---|
| Day 5 | Day 7 | ||||||
| Vehicle | ×3 | - | - | 0/5 | 14/14/13/13/5 | 11.8 | 0/5 |
| Chloroquine a | 100 × 3 | 100 | 100 | 5/5 | 28/28/28/28/28 | 28 | 2/5 |
| 1 | 100 × 3 | 33.9 | 71.4 | 0/5 | 17/5/3/1/0 | 5.2 | 0/2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumarihamy, M.; Ferreira, D.; Croom, E.M., Jr.; Sahu, R.; Tekwani, B.L.; Duke, S.O.; Khan, S.; Techen, N.; Nanayakkara, N.P.D. Antiplasmodial and Cytotoxic Cytochalasins from an Endophytic Fungus, Nemania sp. UM10M, Isolated from a Diseased Torreya taxifolia Leaf. Molecules 2019, 24, 777. https://doi.org/10.3390/molecules24040777
Kumarihamy M, Ferreira D, Croom EM Jr., Sahu R, Tekwani BL, Duke SO, Khan S, Techen N, Nanayakkara NPD. Antiplasmodial and Cytotoxic Cytochalasins from an Endophytic Fungus, Nemania sp. UM10M, Isolated from a Diseased Torreya taxifolia Leaf. Molecules. 2019; 24(4):777. https://doi.org/10.3390/molecules24040777
Chicago/Turabian StyleKumarihamy, Mallika, Daneel Ferreira, Edward M. Croom, Jr., Rajnish Sahu, Babu L. Tekwani, Stephen O. Duke, Shabana Khan, Natascha Techen, and N. P. Dhammika Nanayakkara. 2019. "Antiplasmodial and Cytotoxic Cytochalasins from an Endophytic Fungus, Nemania sp. UM10M, Isolated from a Diseased Torreya taxifolia Leaf" Molecules 24, no. 4: 777. https://doi.org/10.3390/molecules24040777
APA StyleKumarihamy, M., Ferreira, D., Croom, E. M., Jr., Sahu, R., Tekwani, B. L., Duke, S. O., Khan, S., Techen, N., & Nanayakkara, N. P. D. (2019). Antiplasmodial and Cytotoxic Cytochalasins from an Endophytic Fungus, Nemania sp. UM10M, Isolated from a Diseased Torreya taxifolia Leaf. Molecules, 24(4), 777. https://doi.org/10.3390/molecules24040777








