Epigallocatechin-3-gallate (EGCG) Alters Histone Acetylation and Methylation and Impacts Chromatin Architecture Profile in Human Endothelial Cells
Abstract
:1. Introduction
2. Results
2.1. Effect of Epigallocatechin-3-gallate (EGCG) on Proliferation of Endothelial Immortalized Cell Line and Primary Cells
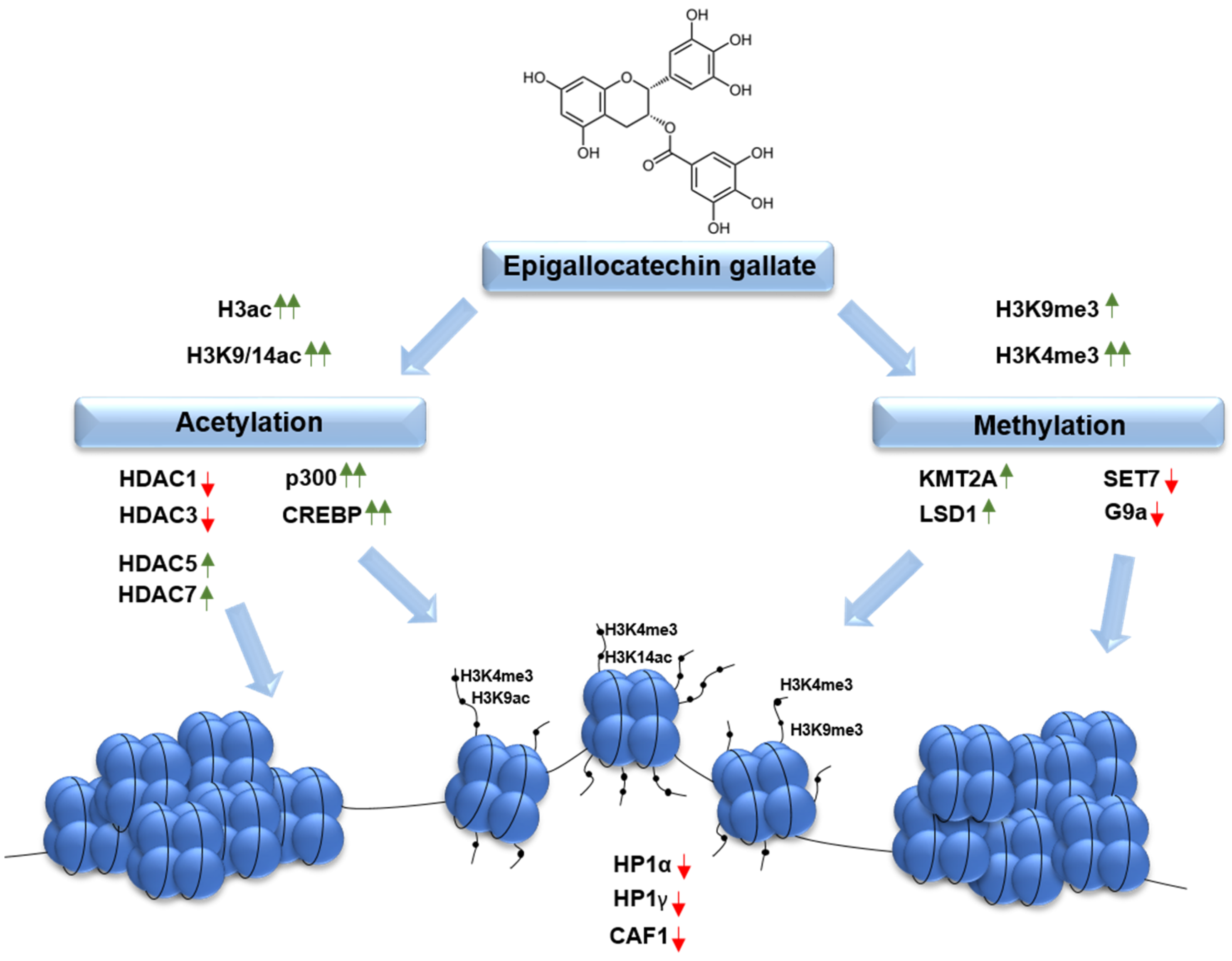
2.2. Epigallocatechin Gallate Alters Acetylation Profile of Histone Core Protein 3 (H3) via Modulation of Expression and Activity of the Major Acetylation Status Drivers
2.3. Effect of Epigallocatechin Gallate on the Methylation Profile of Histone Core Protein 3 (H3) and Gene Expression of the Selected Methylation Status Drivers
2.4. Effect of Long-Term Incubation of Cells with Epigallocatechin Gallate on the Selected Histone Modification Signatures
2.5. Epigallocatechin Gallate Induces Changes in Expression of Chromatin Architecture Determinants: Heterochromatin Protein 1 (HP1) and Chromatin Assembly Factor 1A (CAF1A)
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Cell Proliferation Analysis by Resazurin Reduction Assay
4.3. RNA Isolation, Reverse Transcription and Real-Time PCR
4.4. Western Blotting
4.5. Histone Deacetylase Activity Assay
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Yang, C.S.; Hong, J. Prevention of Chronic Diseases by Tea: Possible Mechanisms and Human Relevance. Annu. Rev. Nutr. 2013, 33, 161–181. [Google Scholar] [CrossRef]
- Meng, J.M.; Cao, S.Y.; Wei, X.L.; Gan, R.Y.; Wang, Y.F.; Cai, S.X.; Xu, X.Y.; Zhang, P.Z.; Li, H.B. Effects and Mechanisms of Tea for the Prevention and Management of Diabetes Mellitus and Diabetic Complications: An Updated Review. Antioxidants 2019, 8, 170. [Google Scholar] [CrossRef] [Green Version]
- Schnekenburger, M.; Dicato, M.; Diederich, M.F. Anticancer potential of naturally occurring immunoepigenetic modulators: A promising avenue? Cancer 2019, 125, 1612–1628. [Google Scholar] [CrossRef] [PubMed]
- Calcagno, D.Q.; Wisnieski, F.; Mota, E.R.D.S.; Sousa, S.B.M.D.; Silva, J.M.C.D.; Leal, M.F.; Gigek, C.O.; Santos, L.C.; Rasmussen, L.T.; Assumpção, P.P.; et al. Role of histone acetylation in gastric cancer: Implications of dietetic compounds and clinical perspectives. Epigenomics 2019, 11, 349–362. [Google Scholar] [CrossRef] [PubMed]
- Donejko, M.; Niczyporuk, M.; Galicka, E.; Przylipiak, A. Anti-cancer properties epigallocatechin-gallate contained in green tea. Postępy Hig. Med. Doświadczalnej 2013, 67, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Bartosikova, L.; Necas, J. Epigallocatechin gallate: A review. Veterinární Med. 2018, 63, 443–467. [Google Scholar] [CrossRef]
- Yang, C.; Wang, H. Cancer Preventive Activities of Tea Catechins. Molecules 2016, 21, 1679. [Google Scholar] [CrossRef]
- Min, K.-J.; Kwon, T.K. Anticancer effects and molecular mechanisms of epigallocatechin-3-gallate. Integr. Med. Res. 2014, 3, 16–24. [Google Scholar] [CrossRef] [Green Version]
- Tachibana, H.; Koga, K.; Fujimura, Y.; Yamada, K. A receptor for green tea polyphenol EGCG. Nat. Struct. Mol. Biol. 2004, 11, 380–381. [Google Scholar] [CrossRef]
- Urusova, D.V.; Shim, J.-H.; Kim, D.J.; Jung, S.K.; Zykova, T.A.; Carper, A.; Bode, A.M.; Dong, Z. Epigallocatechin-gallate Suppresses Tumorigenesis by Directly Targeting Pin1. Cancer Prev. Res. 2011, 4, 1366–1377. [Google Scholar] [CrossRef] [Green Version]
- Tabuchi, M.; Hayakawa, S.; Honda, E.; Ooshima, K.; Itoh, T.; Yoshida, K.; Park, A.-M.; Higashino, H.; Isemura, M.; Munakata, H. Epigallocatechin-3-gallate suppresses transforming growth factor-beta signaling by interacting with the transforming growth factor-beta type II receptor. World J. Exp. Med. 2013, 3, 100. [Google Scholar] [CrossRef]
- Negri, A.; Naponelli, V.; Rizzi, F.; Bettuzzi, S. Molecular Targets of Epigallocatechin—Gallate (EGCG): A Special Focus on Signal Transduction and Cancer. Nutrients 2018, 10, 1936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nandakumar, V.; Vaid, M.; Katiyar, S.K. (-)-Epigallocatechin-3-gallate reactivates silenced tumor suppressor genes, Cip1/p21 and p16INK4a, by reducing DNA methylation and increasing histones acetylation in human skin cancer cells. Carcinogenesis 2011, 32, 537–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, M.Z.; Wang, Y.; Ai, N.; Hou, Z.; Sun, Y.; Lu, H.; Welsh, W.; Yang, C.S. Tea polyphenol (-)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003, 63, 7563–7570. [Google Scholar]
- Singh, B.N.; Shankar, S.; Srivastava, R.K. Green tea catechin, epigallocatechin-3-gallate (EGCG): Mechanisms, perspectives and clinical applications. Biochem. Pharmacol. 2011, 82, 1807–1821. [Google Scholar] [CrossRef] [Green Version]
- Lucock, M.D.; Roach, P.D. The Antifolate Activity of Tea Catechins. Cancer Res. 2005, 65, 8573. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.S.; Quon, M.J.; Kim, J.A. New insights into the mechanisms of polyphenols beyond antioxidant properties; lessons from the green tea polyphenol, epigallocatechin 3-gallate. Redox Biol. 2014, 2, 187–195. [Google Scholar] [CrossRef] [Green Version]
- Choudhury, S.R.; Balasubramanian, S.; Chew, Y.C.; Han, B.; Marquez, V.E.; Eckert, R.L. (-)-Epigallocatechin-3-gallate and DZNep reduce polycomb protein level via a proteasome-dependent mechanism in skin cancer cells. Carcinogenesis 2011, 32, 1525–1532. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Yuan, Y.Y.; Meeran, S.M.; Tollefsbol, T.O. Synergistic epigenetic reactivation of estrogen receptor-α (ERα) by combined green tea polyphenol and histone deacetylase inhibitor in ERα-negative breast cancer cells. Mol. Cancer 2010, 9, 274. [Google Scholar] [CrossRef] [Green Version]
- Groh, I.A.M.; Chen, C.; Lüske, C.; Cartus, A.T.; Esselen, M. Plant Polyphenols and Oxidative Metabolites of the Herbal Alkenylbenzene Methyleugenol Suppress Histone Deacetylase Activity in Human Colon Carcinoma Cells. J. Nutr. Metab. 2013, 2013, 1–10. [Google Scholar] [CrossRef]
- Moseley, V.R.; Morris, J.; Knackstedt, R.W.; Wargovich, M.J. Green tea polyphenol epigallocatechin 3-gallate, contributes to the degradation of DNMT3A and HDAC3 in HCT 116 human colon cancer cells. Anticancer Res. 2013, 33, 5325–5333. [Google Scholar] [PubMed]
- Xu, W.; Wang, F.; Yu, Z.; Xin, F. Epigenetics and Cellular Metabolism. Genet. Epigenetics 2016, 8, S32160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lameirinhas, A.; Miranda-Gonçalves, V.; Henrique, R.; Jerónimo, C. The Complex Interplay between Metabolic Reprogramming and Epigenetic Alterations in Renal Cell Carcinoma. Genes 2019, 10, 264. [Google Scholar] [CrossRef] [Green Version]
- Chisolm, D.A.; Weinmann, A.S. Connections Between Metabolism and Epigenetics in Programming Cellular Differentiation. Annu. Rev. Immunol. 2018, 36, 221–246. [Google Scholar] [CrossRef]
- Lu, C.; Thompson, C.B. Metabolic Regulation of Epigenetics. Cell Metab. 2012, 16, 9–17. [Google Scholar] [CrossRef] [Green Version]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Pirola, L.; Ciesielski, O.; Balcerczyk, A. The Methylation Status of the Epigenome: Its Emerging Role in the Regulation of Tumor Angiogenesis and Tumor Growth, and Potential for Drug Targeting. Cancers 2018, 10, 268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadri-Vakili, G.; Cha, J.H.J. Mechanisms of Disease: Histone modifications in Huntingtons disease. Nat. Clin. Pract. Neurol. 2006, 2, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Wojtala, M.; Pirola, L.; Balcerczyk, A. Modulation of the vascular endothelium functioning by dietary components, the role of epigenetics. BioFactors 2016, 43, 5–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Wang, X.; Han, L.; Zhou, Y.; Sun, S. Green tea polyphenol EGCG reverse cisplatin resistance of A549/DDP cell line through candidate genes demethylation. Biomed. Pharmacother. 2015, 69, 285–290. [Google Scholar] [CrossRef]
- Gu, J.J.; Qiao, K.S.; Sun, P.; Chen, P.; Li, Q. Study of EGCG induced apoptosis in lung cancer cells by inhibiting PI3K/Akt signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 4557–4563. [Google Scholar] [PubMed]
- Wei, R.; Penso, N.E.C.; Hackman, R.M.; Wang, Y.; Mackenzie, G.G. Epigallocatechin-3-Gallate (EGCG) Suppresses Pancreatic Cancer Cell Growth, Invasion, and Migration partly through the Inhibition of Akt Pathway and Epithelial-Mesenchymal Transition: Enhanced Efficacy when Combined with Gemcitabine. Nutrients 2019, 11, 1856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsen, C.A.; Dashwood, R.H. (-)-Epigallocatechin-3-gallate inhibits Met signaling, proliferation, and invasiveness in human colon cancer cells. Arch. Biochem. Biophys. 2010, 501, 52–57. [Google Scholar] [CrossRef] [Green Version]
- Ohga, N.; Hida, K.; Hida, Y.; Muraki, C.; Tsuchiya, K.; Matsuda, K.; Ohiro, Y.; Totsuka, Y.; Shindoh, M. Inhibitory effects of epigallocatechin-3 gallate, a polyphenol in green tea, on tumor-associated endothelial cells and endothelial progenitor cells. Cancer Sci. 2009, 100, 1963–1970. [Google Scholar] [CrossRef] [Green Version]
- Thambyrajah, R.; Fadlullah, M.Z.; Proffitt, M.; Patel, R.; Cowley, S.M.; Kouskoff, V.; Lacaud, G. HDAC1 and HDAC2 Modulate TGF-β Signaling during Endothelial-to-Hematopoietic Transition. Stem Cell Rep. 2018, 10, 1369–1383. [Google Scholar] [CrossRef] [Green Version]
- Park, D.; Park, H.; Kim, Y.; Kim, H.; Jeoung, D. HDAC3 acts as a negative regulator of angiogenesis. BMB Rep. 2014, 47, 227–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zampetaki, A.; Zeng, L.; Margariti, A.; Xiao, Q.; Li, H.; Zhang, Z.; Pepe, A.E.; Wang, G.; Habi, O.; Defalco, E.; et al. Histone Deacetylase 3 Is Critical in Endothelial Survival and Atherosclerosis Development in Response to Disturbed Flow. Circulation 2010, 121, 132–142. [Google Scholar] [CrossRef] [Green Version]
- Urbich, C.; Rössig, L.; Kaluza, D.; Potente, M.; Boeckel, J.N.; Knau, A.; Diehl, F.; Geng, J.G.; Hofmann, W.K.; Zeiher, A.M.; et al. HDAC5 is a repressor of angiogenesis and determines the angiogenic gene expression pattern of endothelial cells. Blood 2009, 113, 5669–5679. [Google Scholar] [CrossRef] [Green Version]
- Margariti, A.; Zampetaki, A.; Xiao, Q.; Zhou, B.; Karamariti, E.; Martin, D.; Yin, X.; Mayr, M.; Li, H.; Zhang, Z.; et al. Histone Deacetylase 7 Controls Endothelial Cell Growth Through Modulation of β-Catenin. Circ. Res. 2010, 106, 1202–1211. [Google Scholar] [CrossRef] [Green Version]
- Oya, Y.; Mondal, A.; Rawangkan, A.; Umsumarng, S.; Iida, K.; Watanabe, T.; Kanno, M.; Suzuki, K.; Li, Z.; Kagechika, H.; et al. Down-regulation of histone deacetylase 4, −5 and −6 as a mechanism of synergistic enhancement of apoptosis in human lung cancer cells treated with the combination of a synthetic retinoid, Am80 and green tea catechin. J. Nutr. Biochem. 2017, 42, 7–16. [Google Scholar] [CrossRef]
- Yoon, H.G. EGCG suppresses prostate cancer cell growth modulating acetylation of androgen receptor by anti-histone acetyltransferase activity. Int. J. Mol. Med. 2012, 30, 69–74. [Google Scholar] [CrossRef] [Green Version]
- Deb, G.; Shankar, E.; Thakur, V.S.; Ponsky, L.E.; Bodner, D.R.; Fu, P.; Gupta, S. Green tea-induced epigenetic reactivation of tissue inhibitor of matrix metalloproteinase-3 suppresses prostate cancer progression through histone-modifying enzymes. Mol. Carcinog. 2019, 58, 1194–1207. [Google Scholar] [CrossRef]
- Bochynńska, A.; Lüscher-Firzlaff, J.; Lüscher, B. Modes of Interaction of KMT2 Histone H3 Lysine 4 Methyltransferase/COMPASS Complexes with Chromatin. Cells 2018, 7, 17. [Google Scholar]
- Gu, F.; Lin, Y.; Wang, Z.; Wu, X.; Ye, Z.; Wang, Y.; Lan, H. Biological roles of LSD1 beyond its demethylase activity. Cell Mol. Life Sci. 2020. [Google Scholar] [CrossRef] [PubMed]
- Karakaidos, P.; Verigos, J.; Magklara, A. LSD1/KDM1A, a Gate-Keeper of Cancer Stemness and a Promising Therapeutic Target. Cancers 2019, 11, 1821. [Google Scholar] [CrossRef] [Green Version]
- Wojtala, M.; Dąbek, A.; Rybaczek, D.; Śliwińska, A.; Świderska, E.; Słapek, K.; El-Osta, A.; Balcerczyk, A. Silencing Lysine-Specific Histone Demethylase 1 (LSD1) Causes Increased HP1-Positive Chromatin, Stimulation of DNA Repair Processes, and Dysregulation of Proliferation by Chk1 Phosphorylation in Human Endothelial Cells. Cells 2019, 8, 1212. [Google Scholar] [CrossRef] [Green Version]
- Quivy, J.-P.; Gérard, A.; Cook, A.J.L.; Roche, D.; Almouzni, G. The HP1–p150/CAF-1 interaction is required for pericentric heterochromatin replication and S-phase progression in mouse cells. Nat. Struct. Mol. Biol. 2008, 15, 972–979. [Google Scholar] [CrossRef]
- Polioudaki, H.; Kourmouli, N.; Drosou, V.; Bakou, A.; Theodoropoulos, P.A.; Singh, P.B.; Giannakouros, T.; Georgatos, S.D. Histones H3/H4 form a tight complex with the inner nuclear membrane protein LBR and heterochromatin protein 1. EMBO Rep. 2001, 2, 920–925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balcerczyk, A.; Rybaczek, D.; Wojtala, M.; Pirola, L.; Okabe, J.; El-Osta, A. Pharmacological inhibition of arginine and lysine methyltransferases induces nuclear abnormalities and suppresses angiogenesis in human endothelial cells. Biochem. Pharmacol. 2016, 121, 18–32. [Google Scholar] [CrossRef] [PubMed]
- Wojtala, M.; Macierzyńska-Piotrowska, E.; Rybaczek, D.; Pirola, L.; Balcerczyk, A. Pharmacological and transcriptional inhibition of the G9a histone methyltransferase suppresses proliferation and modulates redox homeostasis in human microvascular endothelial cells. Pharmacol. Res. 2018, 128, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Okabe, J.; Fernandez, A.Z.; Ziemann, M.; Keating, S.T.; Balcerczyk, A.; El-Osta, A. Endothelial Transcriptome in Response to Pharmacological Methyltransferase Inhibition. ChemMedChem 2014, 9, 1755–1762. [Google Scholar] [CrossRef] [PubMed]
- Chriett, S.; Dąbek, A.; Wojtala, M.; Vidal, H.; Balcerczyk, A.; Pirola, L. Prominent action of butyrate over β-hydroxybutyrate as histone deacetylase inhibitor, transcriptional modulator and anti-inflammatory molecule. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Sample Availability: Samples of the compound are commercially available as indicated in Materials and Methods section, not provided by the authors. |









| Gene | Primer Sequence |
|---|---|
| HPRT1 | F–TCCATTCCTATGACTGTAGATTT R–AACTTTTATGTCCCCCGTTGATT |
| HDAC1 | F–ACTGGTGGTCTGTGTTCTGTGA R–GATGCCAGTCTTACTCATAGCTAC |
| HDAC3 | F–TGGTGAATGGACACCAACTC R–TAGCATGCTTCGATGTGGCA |
| HDAC5 | F–ATGCCAACCTCCTCAACGACC R–TCTGTTCCTCGCAGACCTCCA |
| HDAC7 | F–GCCTGTACTGAGCTGGGCAAA R–TTTTGGCTGCAGAGAGGTGCA |
| CREBP | F–ATGCCAACCTCCTCAACGACC R–TCTGTTCCTCGCAGACCTCCA |
| p300 | F–CCAGACCAGCATGACAGATTTC R–GCTTCCTCTTGGAGCAGATCAG |
| G9a | R–TGGGGCATTGATTGCATCTGG F–TCTCAACTGAAGCTCGCACT |
| KMT2A | F–ACTGGTGGTCTGTGTTCTGTGA R–GATGCCAGTCTTACTCATAGCTAC |
| SET7 | F–ATGCCAACCTCCTCAACGACC R–TCTGTTCCTCGCAGACCTCCA |
| LSD1 | F–ATGCCAACCTCCTCAACGACC R–TCTGTTCCTCGCAGACCTCCA |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ciesielski, O.; Biesiekierska, M.; Balcerczyk, A. Epigallocatechin-3-gallate (EGCG) Alters Histone Acetylation and Methylation and Impacts Chromatin Architecture Profile in Human Endothelial Cells. Molecules 2020, 25, 2326. https://doi.org/10.3390/molecules25102326
Ciesielski O, Biesiekierska M, Balcerczyk A. Epigallocatechin-3-gallate (EGCG) Alters Histone Acetylation and Methylation and Impacts Chromatin Architecture Profile in Human Endothelial Cells. Molecules. 2020; 25(10):2326. https://doi.org/10.3390/molecules25102326
Chicago/Turabian StyleCiesielski, Oskar, Marta Biesiekierska, and Aneta Balcerczyk. 2020. "Epigallocatechin-3-gallate (EGCG) Alters Histone Acetylation and Methylation and Impacts Chromatin Architecture Profile in Human Endothelial Cells" Molecules 25, no. 10: 2326. https://doi.org/10.3390/molecules25102326
APA StyleCiesielski, O., Biesiekierska, M., & Balcerczyk, A. (2020). Epigallocatechin-3-gallate (EGCG) Alters Histone Acetylation and Methylation and Impacts Chromatin Architecture Profile in Human Endothelial Cells. Molecules, 25(10), 2326. https://doi.org/10.3390/molecules25102326







