Virtual Screening Identifies Chebulagic Acid as an Inhibitor of the M2(S31N) Viral Ion Channel and Influenza A Virus
Abstract
1. Introduction
2. Results
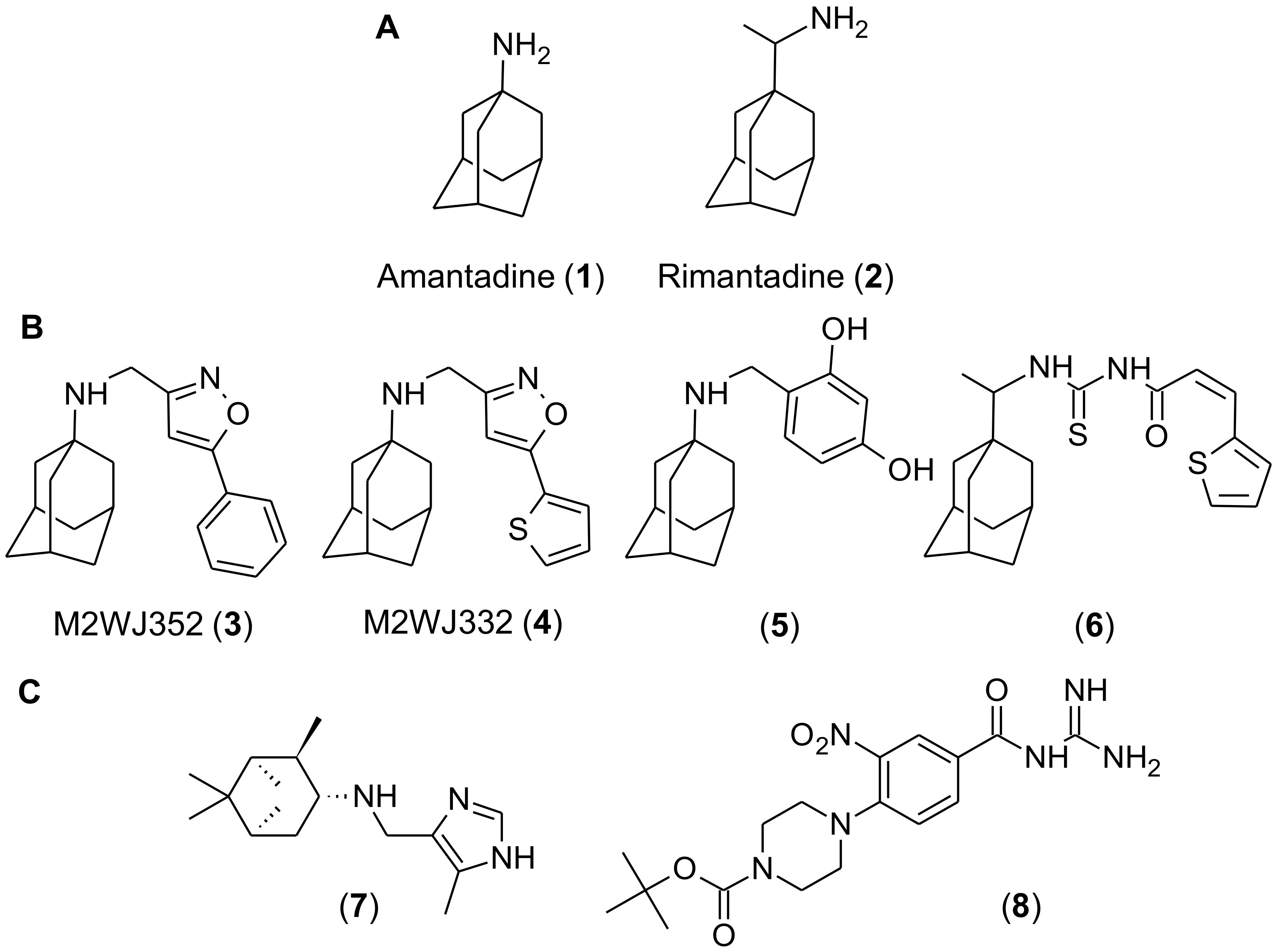
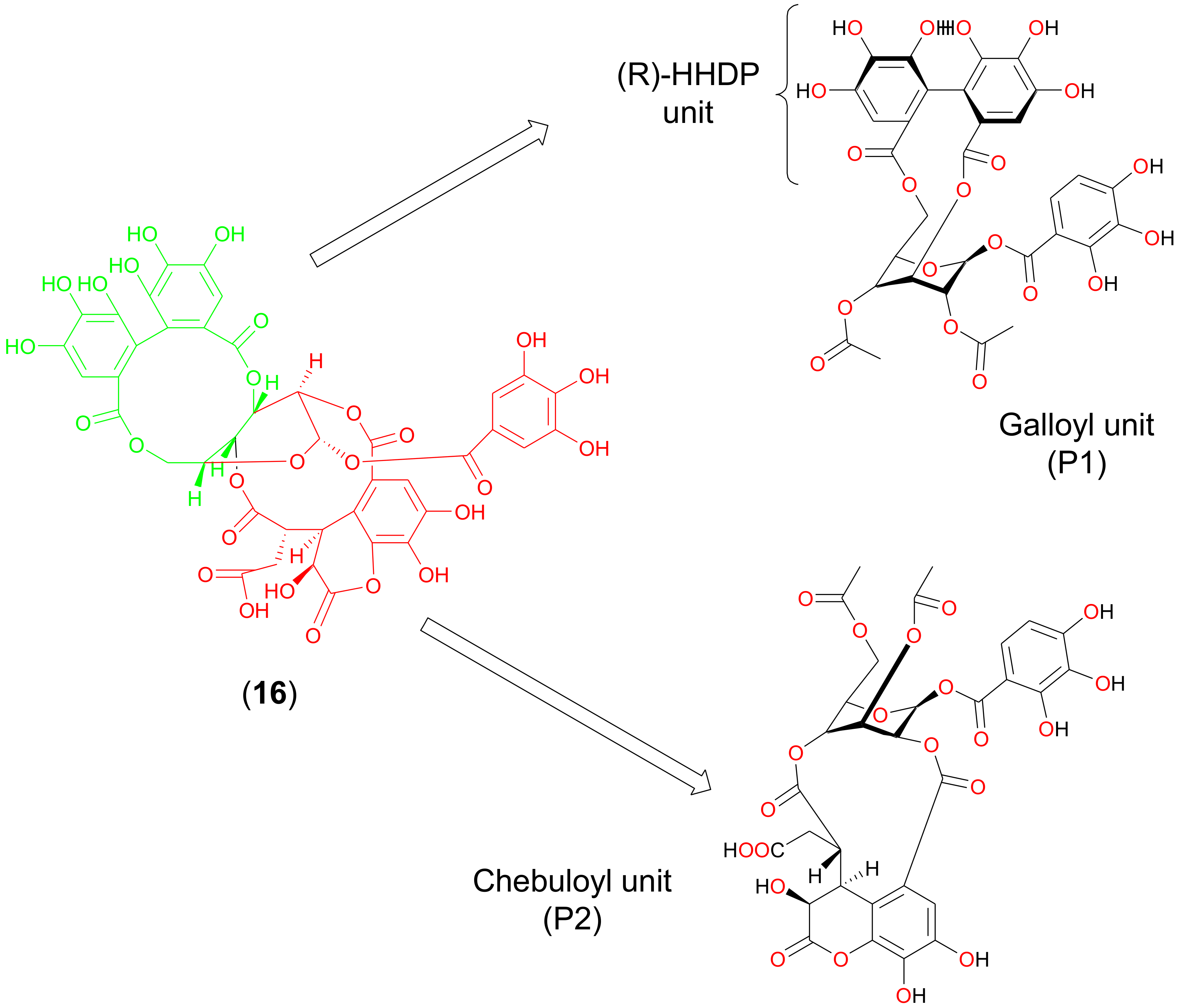
2.1. Discovery of Chebulagic Acid as a Candidate M2(S31N)Inhibitor by Virtual Screening
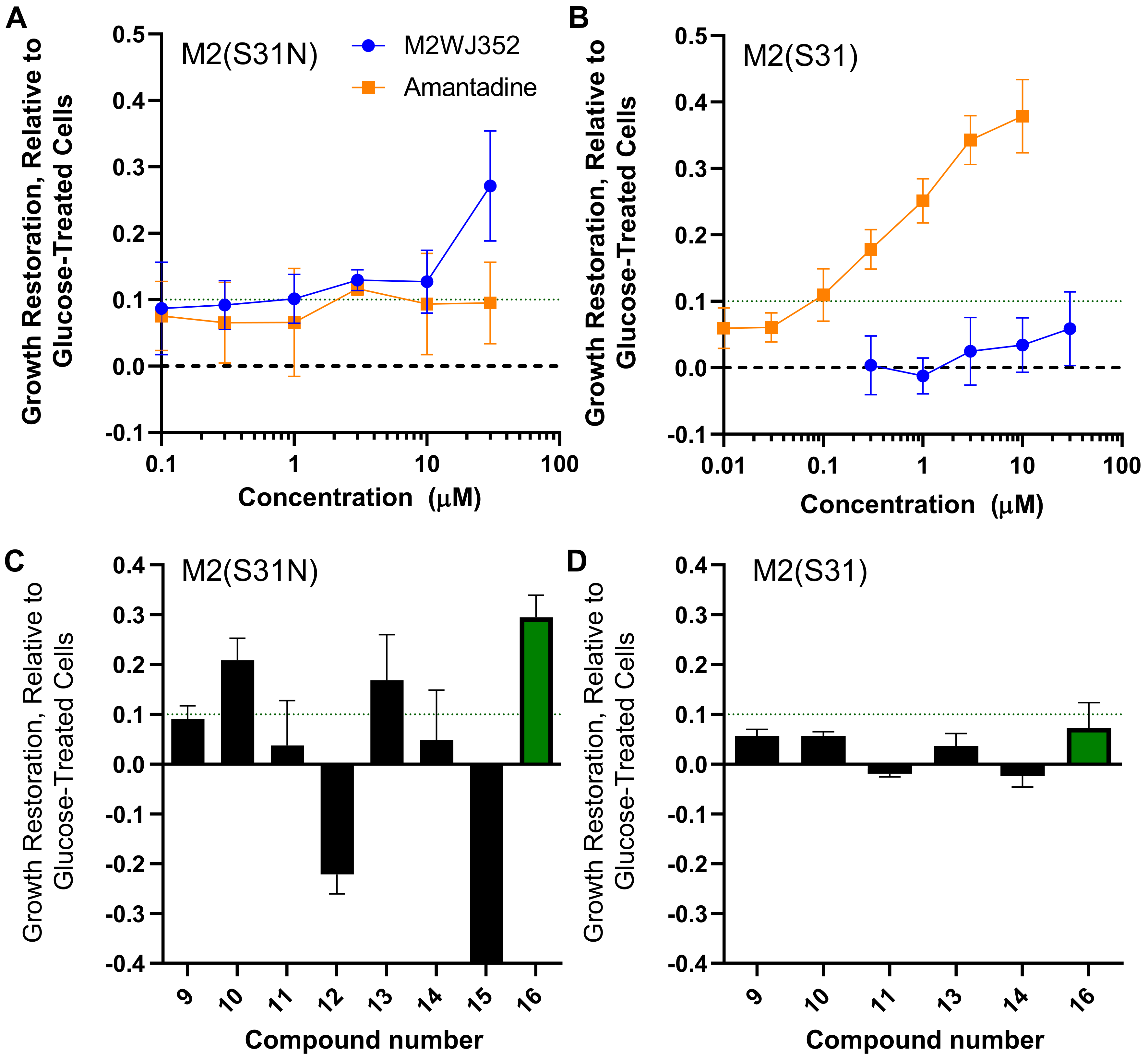
2.2. Chebulagic Acid Inhibits M2(S31N)Activity In Vitro
2.3. Molecular Simulation of Chebulagic Acid with Both Wild-Type and Mutant Forms of M2 Viroporin
2.4. Chebulagic Acid Inhibits Influenza Virus Replication
3. Discussion
4. Materials and Methods
4.1. Materials and Reagents
4.2. Pharmacophore-Based Virtual Screening Methods
4.3. Yeast Growth-Restoration Assays
4.4. Preparation of Chebulagic Acid for Computational Docking Studies
4.5. Preparation of M2(S31N) for Computational Docking Studies
4.6. Docking Calculations
4.7. Generation of Amantadine-Sensitive and -Resistant Influenza Viruses
4.8. Viral Cytopathic Assays
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hayden, F.G.; De Jong, M.D. Emerging influenza antiviral resistance threats. J. Infect. Dis. 2011, 203, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Nieva, J.L.; Madan, V.; Carrasco, L. Viroporins: Structure and biological functions. Nat. Rev. Genet. 2012, 10, 563–574. [Google Scholar] [CrossRef]
- Nieto-Torres, J.L.; Verdiá-Báguena, C.; Castaño-Rodriguez, C.; Aguilella, V.M.; Enjuanes, L. Relevance of Viroporin Ion Channel Activity on Viral Replication and Pathogenesis. Viruses 2015, 7, 3552–3573. [Google Scholar] [CrossRef] [PubMed]
- Jalily, P.H.; Duncan, M.C.; Fedida, D.; Wang, J.; Tietjen, I. Put a cork in it: Plugging the M2 viral ion channel to sink influenza. Antivir. Res. 2020, 178, 104780. [Google Scholar] [CrossRef] [PubMed]
- Sugrue, R.J.; Bahadur, G.; Zambon, M.C.; Hall-Smith, M.; Douglas, A.R.; Hay, A.J. Specific structural alteration of the influenza haemagglutinin by amantadine. EMBO J. 1990, 9, 3469–3476. [Google Scholar] [CrossRef]
- Alvarado-Facundo, E.; Gao, Y.; Ribas-Aparicio, R.M.; Jiménez-Alberto, A.; Weiss, C.D.; Wang, W. Influenza Virus M2 Protein Ion Channel Activity Helps To Maintain Pandemic 2009 H1N1 Virus Hemagglutinin Fusion Competence during Transport to the Cell Surface. J. Virol. 2014, 89, 1975–1985. [Google Scholar] [CrossRef]
- A Bright, R.; Medina, M.-J.; Xu, X.; Perez-Oronoz, G.; Wallis, T.R.; Davis, X.M.; Povinelli, L.; Cox, N.J.; I Klimov, A. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: A cause for concern. Lancet 2005, 366, 1175–1181. [Google Scholar] [CrossRef]
- Fiore, A.E.; Fry, A.; Shay, D.; Gubareva, L.; Bresee, J.S.; Uyeki, T.M. Centers for Disease Control and Prevention (CDC). Antiviral agents for the treatment and chemoprophylaxis of influenza—Recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR. Recomm. Rep. 2011, 60, 1–24. [Google Scholar] [PubMed]
- Chaput, L.; Martinez-Sanz, J.; Saettel, N.; Mouawad, L. Benchmark of four popular virtual screening programs: Construction of the active/decoy dataset remains a major determinant of measured performance. J. Chemin. 2016, 8, 56. [Google Scholar] [CrossRef]
- Chen, Y.; Kops, C.D.B.; Kirchmair, J. Data Resources for the Computer-Guided Discovery of Bioactive Natural Products. J. Chem. Inf. Model. 2017, 57, 2099–2111. [Google Scholar] [CrossRef]
- Cichero, E.; D’Ursi, P.; Moscatelli, M.; Bruno, O.; Orro, A.; Rotolo, C.; Milanesi, L.; Fossa, P. Homology Modeling, Docking Studies and Molecular Dynamic Simulations Using Graphical Processing Unit Architecture to Probe the Type-11 Phosphodiesterase Catalytic Site: A Computational Approach for the Rational Design of Selective Inhibitors. Chem. Boil. Drug Des. 2013, 82, 718–731. [Google Scholar] [CrossRef] [PubMed]
- Franchini, S.; Battisti, U.M.; Prandi, A.; Tait, A.; Borsari, C.; Cichero, E.; Fossa, P.; Cilia, A.; Prezzavento, O.; Ronsisvalle, S.; et al. Scouting new sigma receptor ligands: Synthesis, pharmacological evaluation and molecular modeling of 1,3-dioxolane-based structures and derivatives. Eur. J. Med. Chem. 2016, 112, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Tietjen, I.; Ntie-Kang, F.; Mwimanzi, P.; Onguéné, P.A.; Scull, M.A.; Idowu, T.; Ogundaini, A.; Meva’A, L.M.; Abegaz, B.M.; Rice, C.M.; et al. Screening of the Pan-African Natural Product Library Identifies Ixoratannin A-2 and Boldine as Novel HIV-1 Inhibitors. PLoS ONE 2015, 10, e0121099. [Google Scholar] [CrossRef] [PubMed]
- Divsalar, D.N.; Simoben, C.V.; Schonhofer, C.; Richard, K.; Sippl, W.; Ntie-Kang, F.; Tietjen, I. Novel Histone Deacetylase Inhibitors and HIV-1 Latency-Reversing Agents Identified by Large-Scale Virtual Screening. Front. Pharmacol. 2020, 11. [Google Scholar] [CrossRef]
- Naithani, R.; Huma, L.; Holland, L.; Shukla, D.; McCormick, D.; Mehta, R.; Moriarty, R. Antiviral Activity of Phytochemicals: A Comprehensive Review. Mini Rev. Med. Chem. 2008, 8, 1106–1133. [Google Scholar] [CrossRef]
- Andrae-Marobela, K.; Ghislain, F.W.; Okatch, H.; Majinda, R.R. Polyphenols: A diverse class of multi-target anti-HIV-1 agents. Curr. Drug Metab. 2013, 14, 392–413. [Google Scholar] [CrossRef]
- Tietjen, I.; Williams, D.E.; Read, S.; Kuang, X.T.; Mwimanzi, P.; Wilhelm, E.; Markle, T.; Kinloch, N.N.; Naphen, C.N.; Tenney, K.; et al. Inhibition of NF-κB-dependent HIV-1 replication by the marine natural product bengamide A. Antivir. Res. 2018, 152, 94–103. [Google Scholar] [CrossRef]
- Richard, K.; Williams, D.E.; De Silva, E.D.; Brockman, M.A.; Brumme, Z.L.; Andersen, R.J.; Tietjen, I. Identification of Novel HIV-1 Latency-Reversing Agents from a Library of Marine Natural Products. Viruses 2018, 10, 348. [Google Scholar] [CrossRef]
- Ntie-Kang, F.; Onguéné, P.A.; Fotso, G.W.; Andrae-Marobela, K.; Bezabih, M.; Ndom, J.C.; Ngadjui, B.T.; Ogundaini, A.; Abegaz, B.M.; Meva’A, L.M. Virtualizing the p-ANAPL Library: A Step towards Drug Discovery from African Medicinal Plants. PLoS ONE 2014, 9, e90655. [Google Scholar] [CrossRef]
- Wang, J.; Wu, Y.; Ma, C.; Fiorin, G.; Wang, J.; Pinto, L.H.; Lamb, R.A.; Klein, M.L.; DeGrado, W.F. Structure and inhibition of the drug-resistant S31N mutant of the M2 ion channel of influenza A virus. Proc. Natl. Acad. Sci. USA 2013, 110, 1315–1320. [Google Scholar] [CrossRef]
- Wang, J.; Ma, C.; Wang, J.; Jo, H.; Canturk, B.; Fiorin, G.; Pinto, L.H.; Lamb, R.A.; Klein, M.L.; DeGrado, W.F. Discovery of Novel Dual Inhibitors of the Wild-Type and the Most Prevalent Drug-Resistant Mutant, S31N, of the M2 Proton Channel from Influenza A Virus. J. Med. Chem. 2013, 56, 2804–2812. [Google Scholar] [CrossRef] [PubMed]
- Astrahan, P.; Flitman-Tene, R.; Bennett, E.R.; Krugliak, M.; Gilon, C.; Arkin, I.T. Quantitative analysis of influenza M2 channel blockers. Biochim. Biophys. Acta (BBA) Biomembr. 2011, 1808, 394–398. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Alhadeff, R.; Assa, D.; Astrahan, P.; Krugliak, M.; Arkin, I.T. Computational and experimental analysis of drug binding to the Influenza M2 channel. Biochim. Biophys. Acta (BBA) Biomembr. 2014, 1838, 1068–1073. [Google Scholar] [CrossRef][Green Version]
- Zhao, X.; Jie, Y.; Rosenberg, M.R.; Wan, J.; Zeng, S.; Cui, W.; Xiao, Y.; Li, Z.; Tu, Z.; Casarotto, M.; et al. Design and synthesis of pinanamine derivatives as anti-influenza A M2 ion channel inhibitors. Antivir. Res. 2012, 96, 91–99. [Google Scholar] [CrossRef]
- Jalily, P.H.; Eldstrom, J.; Miller, S.; Kwan, D.C.; Tai, S.S.-H.; Chou, D.; Niikura, M.; Tietjen, I.; Fedida, D. Mechanisms of Action of Novel Influenza A/M2 Viroporin Inhibitors Derived from Hexamethylene Amiloride. Mol. Pharmacol. 2016, 90, 80–95. [Google Scholar] [CrossRef] [PubMed]
- Halgren, T.A. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J. Comput. Chem. 1996, 17, 490–641. [Google Scholar] [CrossRef]
- Daveu, C.; Bureau, R.; Baglin, I.; Prunier, H.; Lancelot, J.-C.; Rault, S. Definition of a pharmacophore for partial agonists of serotonin 5-HT3 receptors. J. Chem. Inf. Comput. Sci. 1999, 39, 362–369. [Google Scholar] [CrossRef]
- Balgi, A.D.; Roberge, M. Screening for Chemical Inhibitors of Heterologous Proteins Expressed in Yeast Using a Simple Growth-Restoration Assay. Adv. Struct. Saf. Stud. 2009, 486, 125–137. [Google Scholar] [CrossRef]
- Balgi, A.D.; Wang, J.; Cheng, D.Y.H.; Ma, C.; Pfeifer, T.A.; Shimizu, Y.; Anderson, H.J.; Pinto, L.H.; Lamb, R.A.; DeGrado, W.F.; et al. Inhibitors of the Influenza A Virus M2 Proton Channel Discovered Using a High-Throughput Yeast Growth Restoration Assay. PLoS ONE 2013, 8, e55271. [Google Scholar] [CrossRef]
- Neumann, G.; Watanabe, T.; Ito, H.; Goto, H.; Gao, P.; Hughes, M.; Perez, D.R.; Donis, R.; Hoffmann, E.; Hobom, G.; et al. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. USA 1999, 96, 9345–9350. [Google Scholar] [CrossRef]
- Niikura, M.; Bance, N.; Mohan, S.; Pinto, B.M. Replication inhibition activity of carbocycles related to oseltamivir on influenza A virus in vitro. Antivir. Res. 2011, 90, 160–163. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-R.; Kaunda, J.S.; Zhu, H.-T.; Wang, D.; Yang, C.-R.; Zhang, Y.-J. The Genus Terminalia (Combretaceae): An Ethnopharmacological, Phytochemical and Pharmacological Review. Nat. Prod. Bioprospect. 2019, 9, 357–392. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-X.; Yu, S.; Liu, N.; Proksch, P.; Lin, W. Inhibitory effects of polyphenols toward HCV from the mangrove plant Excoecaria agallocha L. Bioorg. Med. Chem. Lett. 2012, 22, 1099–1102. [Google Scholar] [CrossRef] [PubMed]
- Vu, T.T.; Kim, H.; Tran, V.K.; Vu, H.D.; Hoang, T.X.; Han, J.W.; Choi, Y.H.; Jang, K.S.; Choi, G.J.; Kim, J.-C. Antibacterial activity of tannins isolated from Sapiumbaccatum extract and use for control of tomato bacterial wilt. PLoS ONE 2017, 12, e0181499. [Google Scholar] [CrossRef] [PubMed]
- Pompermaier, L.; Marzocco, S.; Adesso, S.; Monizi, M.; Schwaiger, S.; Neinhuis, C.; Stuppner, H.; Lautenschläger, T. Medicinal plants of northern Angola and their anti-inflammatory properties. J. Ethnopharmacol. 2018, 216, 26–36. [Google Scholar] [CrossRef]
- Lee, S.-I.; Hyun, P.-M.; Kim, S.-H.; Kim, K.-S.; Lee, S.-K.; Kim, B.-S.; Maeng, P.J.; Lim, J.-S. Suppression of the onset and progression of collagen-induced arthritis by chebulagic acid screened from a natural product library. Arthritis Rheum. 2005, 52, 345–353. [Google Scholar] [CrossRef]
- Liu, Y.; Bao, L.; Xuan, L.; Song, B.; Lin, L.; Han, H. Chebulagic acid inhibits the LPS-induced expression of TNF-α and IL-1β in endothelial cells by suppressing MAPK activation. Exp. Ther. Med. 2015, 10, 263–268. [Google Scholar] [CrossRef] [PubMed]
- Shanmuganathan, S.; Angayarkanni, N. Chebulagic acid Chebulinic acid and Gallic acid, the active principles of Triphala, inhibit TNFα induced pro-angiogenic and pro-inflammatory activities in retinal capillary endothelial cells by inhibiting p38, ERK and NFkB phosphorylation. Vasc. Pharmacol. 2018, 108, 23–35. [Google Scholar] [CrossRef]
- Athira, A.P.; Helen, A.; Saja, K.; Reddanna, P.; Sudhakaran, P.R. Inhibition of Angiogenesis In Vitro by Chebulagic Acid: A COX-LOX Dual Inhibitor. Int. J. Vasc. Med. 2013, 2013, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Basu, S. The natural compound chebulagic acid inhibits vascular endothelial growth factor A mediated regulation of endothelial cell functions. Sci. Rep. 2015, 5, 9642. [Google Scholar] [CrossRef] [PubMed]
- Kashiwada, Y.; Nonaka, G.-I.; Nishioka, I.; Chang, J.-J.; Lee, K.-H. Antitumor Agents, 129. Tannins and Related Compounds as Selective Cytotoxic Agents. J. Nat. Prod. 1992, 55, 1033–1043. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Gangappa, D.; Gupta, G.; Karnati, R. Chebulagic acid from Terminalia chebula causes G1 arrest, inhibits NFκB and induces apoptosis in retinoblastoma cells. BMC Complement. Altern. Med. 2014, 14, 319. [Google Scholar] [CrossRef]
- Gao, H.; Huang, Y.-N.; Gao, B.; Kawabata, J. Chebulagic Acid Is a Potent α-Glucosidase Inhibitor. Biosci. Biotechnol. Biochem. 2008, 72, 601–603. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-N.; Zhao, D.-D.; Gao, B.; Zhong, K.; Zhu, R.-X.; Zhang, Y.; Xie, W.-J.; Jia, L.-R.; Gao, H. Anti-Hyperglycemic Effect of Chebulagic Acid from the Fruits of Terminalia chebula Retz. Int. J. Mol. Sci. 2012, 13, 6320–6333. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.H.; Vasquez, Y.; Ali, Z.; Khan, I.A.; Khan, S.I. Constituents from Terminalia species increase PPARα and PPARγ levels and stimulate glucose uptake without enhancing adipocyte differentiation. J. Ethnopharmacol. 2013, 149, 490–498. [Google Scholar] [CrossRef]
- Shyni, G.L.; Kavitha, S.; Indu, S.; Das Arya, A.; Anusree, S.S.; Vineetha, V.P.; Vandana, S.; Sundaresan, A.; Raghu, K.G. Chebulagic acid from Terminalia chebula enhances insulin mediated glucose uptake in 3T3-L1 adipocytes via PPARγ signaling pathway. BioFactors 2014, 40, 646–657. [Google Scholar] [CrossRef]
- Takechi, M.; Tanaka, Y.; Takehara, M.; Nonaka, G.-I.; Nishioka, I. Structure and antiherpetic activity among the Tannins. Phytochemistry 1985, 24, 2245–2250. [Google Scholar] [CrossRef]
- Lin, L.-T.; Chen, T.-Y.; Chung, C.-Y.; Noyce, R.S.; Grindley, T.B.; McCormick, C.; Lin, T.-C.; Wang, G.-H.; Lin, C.-C.; Richardson, C.D. Hydrolyzable Tannins (Chebulagic Acid and Punicalagin) Target Viral Glycoprotein-Glycosaminoglycan Interactions To Inhibit Herpes Simplex Virus 1 Entry and Cell-to-Cell Spread. J. Virol. 2011, 85, 4386–4398. [Google Scholar] [CrossRef]
- Lin, L.-T.; Chen, T.-Y.; Lin, S.-C.; Chung, C.-Y.; Lin, T.-C.; Wang, G.-H.; Anderson, R.; Lin, C.-C.; Richardson, C.D. Broad-spectrum antiviral activity of chebulagic acid and punicalagin against viruses that use glycosaminoglycans for entry. BMC Microbiol. 2013, 13, 187. [Google Scholar] [CrossRef]
- Kesharwani, A.; Polachira, S.K.; Nair, R.; Agarwal, A.; Mishra, N.N.; Gupta, S.K. Anti-HSV-2 activity of Terminalia chebula Retz extract and its constituents, chebulagic and chebulinic acids. BMC Complement. Altern. Med. 2017, 17, 110. [Google Scholar] [CrossRef]
- Lin, C.-J.; Liu, C.-H.; Jassey, A.; Lin, C.-C.; Li, Y.-F.; Richardson, C.D.; Lin, L.-T. Small molecules targeting coxsackievirus A16 capsid inactivate viral particles and prevent viral binding. Emerg. Microbes Infect. 2018, 7, 1–11. [Google Scholar] [CrossRef]
- Yang, Y.; Xiu, J.; Liu, J.; Zhang, L.; Li, X.; Xu, Y.; Qin, C.; Zhang, L. Chebulagic Acid, a Hydrolyzable Tannin, Exhibited Antiviral Activity in Vitro and in Vivo against Human Enterovirus 71. Int. J. Mol. Sci. 2013, 14, 9618–9627. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Du, R.; Wang, Y.; Hou, X.; Wang, L.; Zhao, X.; Zhan, P.; Liu, X.; Rong, L.; Cui, Q. Identification of Chebulinic Acid and Chebulagic Acid as Novel Influenza Viral Neuraminidase Inhibitors. Front. Microbiol. 2020, 11, 182. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug. Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Tirado-Rives, J. The OPLS [optimized potentials for liquid simulations] potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. J. Am. Chem. Soc. 1988, 110, 1657–1666. [Google Scholar] [CrossRef] [PubMed]
- Labute, P. The generalized Born/volume integral implicit solvent model: Estimation of the free energy of hydration using London dispersion instead of atomic surface area. J. Comput. Chem. 2008, 29, 1693–1698. [Google Scholar] [CrossRef]
- Lenzi, O.; Colotta, V.; Catarzi, D.; Varano, F.; Squarcialupi, L.; Filacchioni, G.; Varani, K.; Vincenzi, F.; Borea, P.A.; Ben, D.D.; et al. Synthesis, structure–affinity relationships, and molecular modeling studies of novel pyrazolo[3,4-c]quinoline derivatives as adenosine receptor antagonists. Bioorg. Med. Chem. 2011, 19, 3757–3768. [Google Scholar] [CrossRef]
- Song, J.-M.; Lee, K.-H.; Seong, B.L. Antiviral effect of catechins in green tea on influenza virus. Antivir. Res. 2005, 68, 66–74. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the authors. |



| Ligand | M2(S31N), Mutant | Interacting Residues in Mutant (M2 Subunits) | M2(S31), Wild Type | Interacting Residues in Wild Type (M2 Subunits) |
|---|---|---|---|---|
| P1 | −39.75 | His37 (B and D), Gly34 (B and D) | −11.78 | Ala30 (A,B and D) |
| P2 | −30.17 | His37 (A and B) | +47.0 | Ala30 (A)His37 (B, C and D) |
| Inhibitory Concentration Required vs. 50× TCID50 (µM) | PR8M2(S31N) | PR8M2(S31) |
|---|---|---|
| Amantadine (1) | >100 | 1.8 ± 2.3 |
| M2WJ352 (3) | 17.6 ± 19.3 | >100 |
| Agathisflavone (10) | >140 | >140 |
| Thiocillin I (13) | >65 | >65 |
| Chebulagic acid (16) | 17.2 ± 15.2 | 32.4 ± 24.7 |
| EC50(µM) | PR8M2(S31N) | PR8M2(S31) |
|---|---|---|
| Amantadine (1) | >5 | 0.16 ± 0.02 |
| M2WJ352 (3) | 3.2 ± 1.2 | 32.7 ± 16.1 |
| Chebulagic acid (16) | 60.9 ± 22.0 | 50.3 ± 26.4 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duncan, M.C.; Onguéné, P.A.; Kihara, I.; Nebangwa, D.N.; Naidu, M.E.; Williams, D.E.; Balgi, A.D.; Andrae-Marobela, K.; Roberge, M.; Andersen, R.J.; et al. Virtual Screening Identifies Chebulagic Acid as an Inhibitor of the M2(S31N) Viral Ion Channel and Influenza A Virus. Molecules 2020, 25, 2903. https://doi.org/10.3390/molecules25122903
Duncan MC, Onguéné PA, Kihara I, Nebangwa DN, Naidu ME, Williams DE, Balgi AD, Andrae-Marobela K, Roberge M, Andersen RJ, et al. Virtual Screening Identifies Chebulagic Acid as an Inhibitor of the M2(S31N) Viral Ion Channel and Influenza A Virus. Molecules. 2020; 25(12):2903. https://doi.org/10.3390/molecules25122903
Chicago/Turabian StyleDuncan, Maggie C., Pascal Amoa Onguéné, Ibuki Kihara, Derrick N. Nebangwa, Maya E. Naidu, David E. Williams, Aruna D. Balgi, Kerstin Andrae-Marobela, Michel Roberge, Raymond J. Andersen, and et al. 2020. "Virtual Screening Identifies Chebulagic Acid as an Inhibitor of the M2(S31N) Viral Ion Channel and Influenza A Virus" Molecules 25, no. 12: 2903. https://doi.org/10.3390/molecules25122903
APA StyleDuncan, M. C., Onguéné, P. A., Kihara, I., Nebangwa, D. N., Naidu, M. E., Williams, D. E., Balgi, A. D., Andrae-Marobela, K., Roberge, M., Andersen, R. J., Niikura, M., Ntie-Kang, F., & Tietjen, I. (2020). Virtual Screening Identifies Chebulagic Acid as an Inhibitor of the M2(S31N) Viral Ion Channel and Influenza A Virus. Molecules, 25(12), 2903. https://doi.org/10.3390/molecules25122903







