Exploiting the Biosynthetic Potency of Taxol from Fungal Endophytes of Conifers Plants; Genome Mining and Metabolic Manipulation
Abstract
1. Introduction
2. Chronology of Taxol, its Derivatives as Antiproliferative Drug
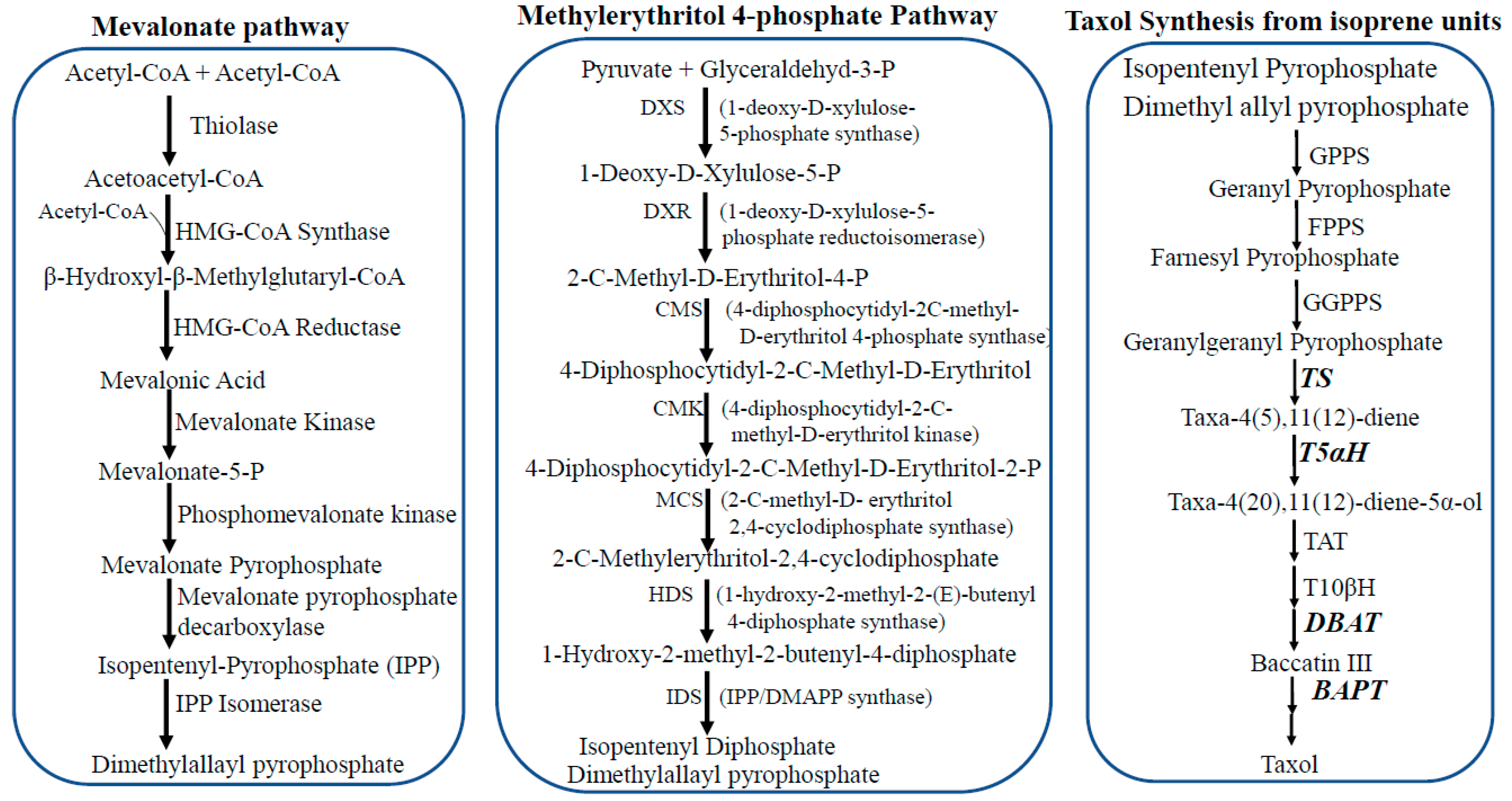
3. Taxol Biosynthesis
4. Mode of Action
5. Sources of Taxol Production
5.1. Natural Source
5.1.1. Family Taxaceae; Taxonomy and Ethnopharmacological Use
5.1.2. Family Podocarpaceae; Taxonomy and Ethnopharmacological Uses
5.2. Taxol-Producing Endophytic Fungi from Taxus and Podocarpus Species
6. Maximizing Taxol Bio-Production Strategies
6.1. Molecular Manipulation of the Microbial Strain
6.2. Bioprocess Optimization Strategy for Taxol Production
7. Co-Cultivation and Mixed Fermentation
8. Genome Mining
8.1. Classical Genome Mining
8.2. Comparative Genome Mining
8.3. Resistance/target Genome Mining
9. Conclusion and Future Directions
Supplementary Materials
Funding
Acknowledgments
Conflicts of Interest
References
- Ali, G.S.; Norman, D.; El-Sayed, A.S. Soluble and Volatile Metabolites of Plant Growth-Promoting Rhizobacteria (PGPRs): Role and Practical Applications in Inhibiting Pathogens and Activating Induced Systemic Resistance (ISR). Adv. Bot. Res. 2015, 75, 241–284. [Google Scholar]
- Walker, K.; Croteau, R. Taxol biosynthesis: Molecular cloning of a benzoyl-CoA:taxane 2alpha-O-benzoyltransferase cDNA from taxus and functional expression in Escherichia coli. Proc. Natl. Acad. Sci. USA 2000, 97, 13591–13596. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, A.S.A.; Ruff, L.E.; Ghany, S.E.A.; Ali, G.S.; Esener, S. Molecular and Spectroscopic Characterization of Aspergillus flavipes and Pseudomonas putida L-Methionine γ-Lyase in Vitro. Appl. Biochem. Biotechnol. 2017, 181, 1513–1532. [Google Scholar] [CrossRef] [PubMed]
- Bode, H.B.; Müller, R. Possibility of bacterial recruitment of plant genes associated with the biosynthesis of secondary metabolites. Plant Physiol. 2003, 132, 1153–1161. [Google Scholar] [CrossRef] [PubMed]
- Han, J.-L.; Li, Z.-Q.; Liu, B.-Y.; Wang, H.; Li, G.-F.; Ye, H.-C. Metabolic engineering of terpenoids in plants. Chin. J. Biotechnol. 2007, 23, 561–569. [Google Scholar]
- Wani, M.C.; Taylor, H.L.; Wall, M.E.; Coggon, P.; McPhail, A.T. Plant Antitumor Agents. VI. The Isolation and Strcture of Taxol, a Novel Antileukemic and Antitumo Agent from Taxus bretvifolia. J. Am. Chem. Soc. 1971, 93, 2325–2327. [Google Scholar] [CrossRef]
- Croteau, R.; Ketchum, R.E.B.; Long, R.M.; Kaspera, R.; Wildung, M.R. Taxol biosynthesis and molecular genetics. Phytochem. Rev. 2006, 5, 75–97. [Google Scholar] [CrossRef]
- Sharma, A.; Mayhew, E.; Straubinger, R.M. Antitumor Effect of Taxol-containing Liposomes in a Taxol-resistant Murine Tumor Model. Cancer Res. 1993, 53, 5877–5881. [Google Scholar]
- Malik, S.; Cusidó, R.M.; Mirjalili, M.H.; Moyano, E.; Palazón, J.; Bonfill, M. Production of the anticancer drug taxol in Taxus baccata suspension cultures: A review. Process Biochem. 2011, 46, 23–34. [Google Scholar] [CrossRef]
- Wang, C.; Wang, Y.; Wang, Y.; Fan, M.; Luo, F.; Qian, Z. Characterization, pharmacokinetics and disposition of novel nanoscale preparations of paclitaxel. Int. J. Pharm. 2011, 414, 251–259. [Google Scholar] [CrossRef]
- El-Sayed, A.S.; Khalaf, S.A.; Abdel-Hamid, G.; El-Batrik, M.I. Screening, Morphological and Molecular characterization of fungi producing cyStathionine γ-lyaSe. Acta Biol. Hung. 2015, 661, 119–132. [Google Scholar] [CrossRef]
- El-Sayed, A.S.A.; Yassin, M.A.; Khalaf, S.A.; El-Batrik, M.; Ali, G.S.; Esener, S. Biochemical and Pharmacokinetic Properties of PEGylated Cystathionine γ-Lyase from Aspergillus carneus KF723837. J. Mol. Microbiol. Biotechnol. 2015, 25, 301–310. [Google Scholar] [CrossRef]
- El-Sayed, A.S.A.; Abdel-Ghany, S.E.; Ali, G.S. Genome editing approaches: Manipulating of lovastatin and taxol synthesis of filamentous fungi by CRISPR/Cas9 system. Appl. Microbiol. Biotechnol. 2017, 101, 3953–3976. [Google Scholar] [CrossRef] [PubMed]
- Holton, R.A.; Kim, H.B.; Somoza, C.; Liang, F.; Biediger, R.J.; Boatman, P.D.; Shindo, M.; Smith, C.C.; Kim, S. First total synthesis of taxol. 2. Completion of the C and D rings. J. Am. Chem. Soc. 1994, 116, 1599–1600. [Google Scholar] [CrossRef]
- Exposito, O.; Bonfill, M.; Moyano, E.; Onrubia, M.; Mirjalili, M.; Cusido, R.; Palazon, J. Biotechnological Production of Taxol and Related Taxoids: Current State and Prospects. Anti-Cancer Agents Med. Chem. 2009, 9, 109–121. [Google Scholar] [CrossRef] [PubMed]
- Nims, E.; Dubois, C.P.; Roberts, S.C.; Walker, E.L. Expression profiling of genes involved in paclitaxel biosynthesis for targeted metabolic engineering. Metab. Eng. 2006, 8, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.J.; Gibson, D.M.; Shuler, M.L. Effect of Subculture and Elicitation on Instability of Taxol Production in Taxus sp. Suspension Cultures. Biotechnol. Progress 2004, 20, 1666–1673. [Google Scholar] [CrossRef] [PubMed]
- Stierle, A.; Strobel, G.; Stierle, D. Taxol and taxane production by Taxomyces andreanae, an endophytic fungus of Pacific yew. Science 1993, 260, 214–216. [Google Scholar] [CrossRef]
- Stierle, A.; Strobel, G.; Stierle, D.; Grothaus, P.; Bignami, G. The Search for a Taxol-Producing Microorganism Among the Endophytic Fungi of the Pacific Yew, Taxus brevifolia. J. Nat. Prod. 1995, 58, 1315–1324. [Google Scholar] [CrossRef]
- Caruso, M.; Colombo, A.L.; Fedeli, L.; Pavesi, A.; Quaroni, S.; Saracchi, M.; Ventrella, G. Isolation of endophytic fungi and Actinomycetes taxane producers. Ann. Microbiol. 2000, 50, 3–13. [Google Scholar]
- El-Sayed, A.S.; Khalaf, S.A.; Aziz, H.A. Characterization of homocysteine γ-lyase from submerged and solid cultures of Aspergillus fumigatus ASH (JX006238). J. Microbiol. Biotechnol. 2013, 23. [Google Scholar] [CrossRef]
- El-Sayed, A.S.; Shindia, A.A. Characterization and immobilization of purified Aspergillus flavipesl-methioninase: Continuous production of methanethiol. J. Appl. Microbiol. 2011, 111, 54–69. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, A.S.A.; Hassan, M.N.; Nada, H.M.S. Purification, immobilization, and biochemical characterization of l-arginine deiminase from thermophilic Aspergillus fumigatus KJ434941: Anticancer activity in vitro. Biotechnol. Prog. 2015, 31. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, A.S.A.; Yassin, M.A.; Ibrahim, H. Coimmobilization of l -methioninase and glutamate dehydrogenase: Novel approach for l -homoalanine synthesis. Biotechnol. Appl. Biochem. 2015, 62. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, A.S.A.; Shad, G. Aspergillus flavipes is a novel e ffi cient biocontrol agent of Phytophthora parasitica. Biol. Control 2020, 140. [Google Scholar] [CrossRef]
- El-Sayed, A.S. Microbial l-methioninase: Production, molecular characterization, and therapeutic applications. Appl. Microbiol. Biotechnol. 2010, 86, 445–467. [Google Scholar] [CrossRef]
- Wang, S.L.; Chen, Y.H.; Luwang, C.; Yen, Y.H.; Kaichern, M. Purification and characterization of a serine protease extracellularly produced by Aspergillus fumigatus in a shrimp and crab shell powder medium. Enzyme Microb. Technol. 2005, 36, 660–665. [Google Scholar] [CrossRef]
- El-Sayed, A.S.A.; Mohamed, N.Z.; Safan, S.; Yassin, M.A.; Shaban, L.; Shindia, A.A.; Shad Ali, G.; Sitohy, M.Z. Restoring the Taxol biosynthetic machinery of Aspergillus terreus by Podocarpus gracilior Pilger microbiome, with retrieving the ribosome biogenesis proteins of WD40 superfamily. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef]
- Heinig, U.; Scholz, S.; Jennewein, S. Getting to the bottom of Taxol biosynthesis by fungi. Fungal Divers. 2013, 60, 161–170. [Google Scholar] [CrossRef]
- El-Naggar, N.E.A.; Moawad, H.; El-Shweihy, N.M.; El-Ewasy, S.M.; Elsehemy, I.A.; Abdelwahed, N.A.M. Process development for scale-up production of a therapeutic L-asparaginase by Streptomyces brollosae NEAE-115 from shake flasks to bioreactor. Sci. Rep. 2019, 9, 1–18. [Google Scholar] [CrossRef]
- Suffness, M.; Wall, E.M. Taxol: Science and Application; CRC Press: Boca Raton, FL, USA, 1995. [Google Scholar]
- El-Sayed, A.S.A.; Fathalla, M.; Yassin, M.A.; Zein, N.; Morsy, S.; Sitohy, M.; Sitohy, B. Conjugation of Aspergillus flavipes taxol with porphyrin increases the anticancer activity of taxol and ameliorates its cytotoxic effects. Molecules 2020, 25, 263. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, A.S.A.; Safan, S.; Mohamed, N.Z.; Shaban, L.; Ali, G.S.; Sitohy, M.Z. Induction of Taxol biosynthesis by Aspergillus terreus, endophyte of Podocarpus gracilior Pilger, upon intimate interaction with the plant endogenous microbes. Process Biochem. 2018, 71, 31–40. [Google Scholar] [CrossRef]
- El-Sayed, A.S.A.; Ali, D.M.I.; Yassin, M.A.; Zayed, R.A.; Ali, G.S. Sterol inhibitor “Fluconazole” enhance the Taxol yield and molecular expression of its encoding genes cluster from Aspergillus flavipes. Process Biochem. 2019, 76, 55–67. [Google Scholar] [CrossRef]
- Crown, J.; O’Leary, M.; Ooi, W. Docetaxel and Paclitaxel in the Treatment of Breast Cancer: A Review of Clinical Experience. Oncologist 2004, 9, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Javeed, A.; Ashraf, M.; Riaz, A.; Ghafoor, A.; Afzal, S.; Mukhtar, M.M. Paclitaxel and immune system. Eur. J. Pharm. Sci. 2009, 38, 283–290. [Google Scholar] [CrossRef]
- De Bono, J.S.; Oudard, S.; Ozguroglu, M.; Hansen, S.; Machiels, J.H.; Shen, L.; Matthews, P.; Sartor, A.O. Cabazitaxel or mitoxantrone with prednisone in patients with metastatic castration-resistant prostate cancer (mCRPC) previously treated with docetaxel: Final results of a multinational phase III trial (TROPIC). J. Clin. Oncol. 2010, 28, 4508. [Google Scholar] [CrossRef]
- Maheshwari, P.; Garg, S.; Kumar, A. Taxoids: Biosynthesis and in vitro production. Biotechnol. Mol. Biol. Rev. 2008, 3, 71–087. [Google Scholar]
- McLaughlin, J.L.; Miller, R.W.; Powell, R.G.; Smith, C.R. 19-hydroxybaccatin iii, 10-deacetylcephalomannine, and 10-deacetyltaxol: New Antitumor Taxanes from Taxus Wallichiana. J. Nat. Prod. 1981, 44, 312–319. [Google Scholar] [CrossRef]
- Kelsey, R.G.; Vance, N.C. Taxol and cephalomannine concentrations in the foliage and bark of shade-grown and sun-exposed taxus brevifolia trees. J. Nat. Prod. 1992, 55, 912–917. [Google Scholar] [CrossRef]
- Singla, A.K.; Garg, A.; Aggarwal, D. Paclitaxel and its formulations. Int. J. Pharm. 2002, 235, 179–192. [Google Scholar] [CrossRef]
- Rook, E.J.; Van Ree, J.M.; Van Den Brink, W.; Hillebrand, M.J.X.; Huitema, A.D.R.; Hendriks, V.M.; Beijnen, J.H. Pharmacokinetics and pharmacodynamics of high doses of pharmaceutically prepared heroin, by intravenous or by inhalation route in opioid-dependent patients. Basic Clin. Pharmacol. Toxicol. 2006, 98, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Han, S.W.; Choi, H.J.; Kim, K. Nanoparticle-Directed Crystallization of Calcium Carbonate. Adv. Mater. 2001, 13, 1617–1620. [Google Scholar] [CrossRef]
- Cavalli, R.; Gasco, M.R.; Chetoni, P.; Burgalassi, S.; Saettone, M.F. Solid lipid nanoparticles (SLN) as ocular delivery system for tobramycin. Int. J. Pharm. 2002, 238, 241–245. [Google Scholar] [CrossRef]
- Khandavilli, S.; Panchagnula, R. Nanoemulsions as versatile formulations for paclitaxel delivery: Peroral and dermal delivery studies in rats. J. Investig. Dermatol. 2007, 127, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Torne, S.J.; Ansari, K.A.; Vavia, P.R.; Trotta, F.; Cavalli, R. Enhanced oral paclitaxel bioavailability after administration of paclitaxel-loaded nanosponges. Drug Deliv. 2010, 17, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Yoncheva, K.; Calleja, P.; Agüeros, M.; Petrov, P.; Miladinova, I.; Tsvetanov, C.; Irache, J.M. Stabilized micelles as delivery vehicles for paclitaxel. Int. J. Pharm. 2012, 436, 258–264. [Google Scholar] [CrossRef]
- Dahmani, F.Z.; Yang, H.; Zhou, J.; Yao, J.; Zhang, T.; Zhang, Q. Enhanced oral bioavailability of paclitaxel in pluronic/LHR mixed polymeric micelles: Preparation, in vitro and in vivo evaluation. Eur. J. Pharm. Sci. 2012, 47, 179–189. [Google Scholar] [CrossRef]
- Nornoo, A.O.; Zheng, H.A.; Lopes, L.B.; Johnson-Restrepo, B.; Kannan, K.; Reed, R. Oral microemulsions of paclitaxel: In situ and pharmacokinetic studies. Eur. J. Pharm. Biopharm. 2009, 71, 310–317. [Google Scholar] [CrossRef]
- Rohmer, M.; Knani, M.; Simonin, P.; Sutter, B.; Sahm, H. Isoprenoid biosynthesis in bacteria: A novel pathway for the early steps leading to isopentenyl diphosphate. Biochem. J. 1993, 295, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Flesch, G.; Rohmer, M. Prokaryotic hopanoids: The biosynthesis of the bacteriohopane skeleton: Formation of isoprenic units from two distinct acetate pools and a novel type of carbon/carbon linkage between a triterpene and d-ribose. Eur. J. Biochem. 1988, 175, 405–411. [Google Scholar] [CrossRef]
- Bach, T.J. Hydroxymethylglutaryl-CoA reductase, a key enzyme in phytosterol synthesis? Lipids 1986, 21, 82–88. [Google Scholar] [CrossRef] [PubMed]
- McGarvey, D.J.; Croteau, R. Terpenoid metabolism. Plant Cell 1995, 7, 1015–1026. [Google Scholar] [PubMed]
- El-Sayed, A.S.; Shindia, A.A.; Diab, A.A.; Rady, A.M. Purification and immobilization of l-arginase from thermotolerant Penicillium chrysogenum KJ185377.1; with unique kinetic properties as thermostable anticancer enzyme. Arch. Pharmacal Res. 2014, 1–10. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, A.S.; Shindia, A.A.; Zaher, Y. L-Amino acid oxidase from filamentous fungi: Screening and optimization. Ann. Microbiol. 2012, 62, 773–784. [Google Scholar] [CrossRef]
- El-Sayed, A.S.; Shindia, A.A.; Zaher, Y.A. Purification and characterization of L-amino acid oxidase from the solid-state grown cultures of Aspergillus oryzae ASH. Microbiology 2013, 82, 762–771. [Google Scholar] [CrossRef]
- El-Sayed, A.S.; Shouman, S.A.; Nassrat, H.M. Pharmacokinetics, immunogenicity and anticancer efficiency of Aspergillus flavipes l-methioninase. Enzyme Microb. Technol. 2012, 51, 200–210. [Google Scholar] [CrossRef]
- El-Sayed, A.S.A. Purification and characterization of a new L-methioninase from solid cultures of Aspergillus flavipes. J. Microbiol. 2011, 49, 130–140. [Google Scholar] [CrossRef]
- El-Sayed, A.S.A.; Abdel-Azeim, S.; Ibrahim, H.M.; Yassin, M.A.; Abdel-Ghany, S.E.; Esener, S.; Ali, G.S. Biochemical stability and molecular dynamic characterization of Aspergillus fumigatus cystathionine γ-lyase in response to various reaction effectors. Enzyme Microb. Technol. 2015, 81, 31–46. [Google Scholar] [CrossRef]
- El-Sayed, A.S.A.; Akbar, A.; Iqrar, I.; Ali, R.; Norman, D.; Brennan, M.; Ali, G.S. A glucanolytic Pseudomonas sp. associated with Smilax bona-nox L. displays strong activity against Phytophthora parasitica. Microbiol. Res. 2018, 207, 140–152. [Google Scholar] [CrossRef]
- El-Sayed, A.S.A.; Hassan, A.E.A.; Shindia, A.A.; Mohamed, S.G.; Sitohy, M.Z. Aspergillus flavipes methionine γ-lyase-dextran conjugates with enhanced structural, proteolytic stability and anticancer efficiency. J. Mol. Catal. B Enzym. 2016, 133, S15–S24. [Google Scholar] [CrossRef]
- El-Sayed, A.S.A.; Hassan, A.E.; Yassin, M.A.; Hassan, A.M.F. Characterization of glutathione-homocystine transhydrogenase as a novel isoform of glutathione s-transferase from Aspergillus flavipes. Pharm. Chem. J. 2015, 49, 373–383. [Google Scholar] [CrossRef]
- El-sayed, A.S.A.; George, N.M.; Yassin, M.A.; Alaidaroos, B.A.; Bolbol, A.A.; Mohamed, M.S.; Rady, A.M.; Aziz, S.W.; Zayed, R.A.; Sitohy, M.Z. Purification and Characterization of Ornithine Decarboxylase From Aspergillus terreus; Kinetics of Inhibition by Various Inhibitors. Molecules 2019, 24, 2756. [Google Scholar] [CrossRef] [PubMed]
- El-sayed, A.S.A.; El Sayed, M.T.; Nada, H.S.; Hassan, A.E.; Yousef, E.K. Production and Characterization of Taxol as Anticancer Agent from Aspergillus terreus. J. Pure Appl. Microbiol. 2019, 13, 2055–2063. [Google Scholar] [CrossRef]
- Jennewein, S.; Rithner, C.D.; Williams, R.M.; Croteau, R.B. Taxol biosynthesis: Taxane 13 -hydroxylase is a cytochrome P450-dependent monooxygenase. Proc. Natl. Acad. Sci. USA 2001, 98, 13595–13600. [Google Scholar] [CrossRef]
- Chau, M.; Jennewein, S.; Walker, K.; Croteau, R. Taxol biosynthesis: Molecular cloning and characterization of a cytochrome P450 taxoid 7 beta-hydroxylase. Chem Biol. 2004, 11, 663–672. [Google Scholar] [CrossRef]
- Jennewein, S.; Croteau, R. Taxol: Biosynthesis, molecular genetics, and biotechnological applications. Appl. Microbiol. Biotechnol. 2001, 57, 13–19. [Google Scholar]
- Kusari, S.; Hertweck, C.; Spiteller, M. Chemical Ecology of Endophytic Fungi: Origins of Secondary Metabolites. Chem. Biol. 2012, 19, 792–798. [Google Scholar] [CrossRef]
- Cragg, G.M.; Schepartz, S.A.; Suffness, M.; Grever, M.R. The taxol supply crisis. New NCI policies for handling the large-scale production of novel natural product anticancer and anti-HIV agents. J. Nat. Prod. 1993, 56, 1657–1668. [Google Scholar] [CrossRef]
- Goldspiel, B.R. Clinical overview of the taxanes. Pharmacotherapy 1997, 17, 110S–125S. [Google Scholar]
- Amos, L.A.; Klug, A. Arrangement of subunits in flagellar microtubules. J. Cell Sci. 1974, 14, 523–549. [Google Scholar]
- Orr, G.A.; Verdier-Pinard, P.; McDaid, H.; Horwitz, S.B. Mechanisms of Taxol resistance related to microtubules. Oncogene 2003, 22, 7280–7295. [Google Scholar] [CrossRef] [PubMed]
- Schiff, P.B.; Fant, J.; Horwitz, S.B. Promotion of microtubule assembly in vitro by taxol. Nature 1979, 277, 665–667. [Google Scholar] [CrossRef] [PubMed]
- Andreu, J.M.; Bordas, J.; Diaz, J.F.; de Ancos, J.G.; Gil, R.; Medrano, F.J.; Nogales, E.; Pantos, E.; Towns-Andrews, E. Low resolution structure of microtubules in solution. Synchrotron X-ray scattering and electron microscopy of taxol-induced microtubules assembled from purified tubulin in comparison with glycerol and MAP-induced microtubules. J. Mol. Biol. 1992, 226, 169–184. [Google Scholar] [CrossRef]
- Kingston, D.G.; Jagtap, P.G.; Yuan, H.; Samala, L. The chemistry of taxol and related taxoids. Fortschr. Chem. Org. Naturst. 2002, 84, 53–225. [Google Scholar]
- Thomas, P.A.; Polwart, A. Taxus baccata L. J. Ecol. 2003, 91, 489–524. [Google Scholar] [CrossRef]
- Staniek, A.; Woerdenbag, H.; Kayser, O. Taxomyces andreanae: A Presumed Paclitaxel Producer Demystified? Planta Medica 2009, 75, 1561–1566. [Google Scholar] [CrossRef]
- El-Sayed, A.S.A.; Shindia, A.A.; AbouZaid, A.A.; Yassin, A.M.; Shad Ali, G.; Sitohy, M.Z. Biochemical characterization of peptidylarginine deiminase-like orthologs from thermotolerant Emericella dentata and Aspergillus nidulans. Enzyme Microb. Technol. 2019, 124, 41–53. [Google Scholar] [CrossRef]
- El-Sayed, A.S.A.; Shindia, A.A.; Zeid, A.A.A.; Yassin, A.M.; Sitohy, M.Z.; Sitohy, B. Aspergillus nidulans thermostable arginine deiminase-Dextran conjugates with enhanced molecular stability, proteolytic resistance, pharmacokinetic properties and anticancer activity. Enzyme Microb. Technol. 2019, 131. [Google Scholar] [CrossRef]
- Abdillahi, H.S.; Stafford, G.I.; Finnie, J.F.; Van Staden, J. Ethnobotany, phytochemistry and pharmacology of Podocarpus sensu latissimo (s.l.). S. Afr. J. Bot. 2010, 76, 1–24. [Google Scholar] [CrossRef]
- Stahlhut, R.; Park, G.; Petersen, R.; Ma, W.; Hylands, P. The occurrence of the anti-cancer diterpene taxol in Podocarpus gracilior Pilger (Podocarpaceae). Biochem. Syst. Ecol. 1999, 27, 613–622. [Google Scholar] [CrossRef]
- Cope, E.A. Taxaceae: The Genera and Cultivated Species. Bot. Rev. 1998, 64, 291–322. [Google Scholar] [CrossRef]
- Raubeson, L.A.; Jansen, R.K. Chloroplast DNA evidence on the ancient evolutionary split in vascular land plants. Science 1992, 255, 1697–1699. [Google Scholar] [CrossRef] [PubMed]
- Chaw, S.M.; Long, H.; Wang, B.S.; Zharkikh, A.; Lie, W.H. The phylogenetic position of Taxaceae based on 18S rRNA sequences. J. Mol. Evol. 1993, 37, 624–630. [Google Scholar] [CrossRef] [PubMed]
- Stewart, W.N. Paleobotany and the Evolution of Plants; Cambridge University Press: Cambridge, UK, 1983. [Google Scholar]
- Keener, C.S.; Gifford, E.M.; Foster, A.S. Morphology and Evolution of Vascular Plants. Syst. Bot. 1990, 15. [Google Scholar] [CrossRef]
- Delong, J.M.; Prange, R.K. Taxus spp.: Botany, Horticulture, and Source of Anti-Cancer Compounds. In Horticultural Reviews; Wiley Blackwell: Hoboken, NJ, USA, 2010; Volume 32, pp. 299–327. [Google Scholar]
- Turner, N.J.; Hebda, R.J. Contemporary use of bark for medicine by two salishan native elders of Southeast Vancouver Island, Canada. J. Ethnopharmacol. 1990, 29, 59–72. [Google Scholar] [CrossRef]
- Kanda, Y.; Nakamura, H.; Umemiya, S.; Puthukanoori, R.K.; Appala, V.R.; Gaddamanugu, J.K.; Paraselli, B.R.; Baran, P.S. Two-Phase Synthesis of Taxol. J. Am. Chem. Soc. 2020, 142, 10526–10533. [Google Scholar] [CrossRef]
- Farjon, A. A Handbook of the World’s Conifers: Revised and Updated Edition; Brill: Leiden, The Netherlands, 2017. [Google Scholar]
- Brummitt, N.A.; Bachman, S.P.; Griffiths-Lee, J.; Lutz, M.; Moat, J.F.; Farjon, A.; Donaldson, J.S.; Hilton-Taylor, C.; Meagher, T.R.; Albuquerque, S.; et al. Green Plants in the Red: A Baseline Global Assessment for the IUCN Sampled Red List Index for Plants. PLoS ONE 2015, 10. [Google Scholar] [CrossRef]
- Abdillahi, H.S.; Stafford, G.I.; Finnie, J.F.; Van Staden, J. Antimicrobial activity of South African Podocarpus species. J. Ethnopharmacol. 2008, 119, 191–194. [Google Scholar] [CrossRef]
- Kuo, Y.J.; Hwang, S.Y.; Wu, M.D.; Liao, C.C.; Liang, Y.H.; Kuo, Y.H.; Ho, H.O. Cytotoxic constituents from Podocarpus fasciculus. Chem. Pharm. Bull. 2008, 56, 585–588. [Google Scholar] [CrossRef]
- Vent, W.; Duke, J.A.; Ayensu, E.S. Medicinal Plants of China. 2 Vols. 705 S., 1300 Strichzeichnungen. Reference Publ., Inc. Algonac. Michigan, 1985. ISBN 0-917266-20-4. Preis: Geb. m. Schutzumschlag $94,95. Feddes Repert. 2008, 98, 398. [Google Scholar] [CrossRef]
- Bok, J.W.; Hoffmeister, D.; Maggio-Hall, L.A.; Murillo, R.; Glasner, J.D.; Keller, N.P. Genomic mining for Aspergillus natural products. Chem. Biol. 2006, 13, 31–37. [Google Scholar] [CrossRef]
- Kupchan, S.M.; Baxter, R.L.; Ziegler, M.F.; Smith, P.M.; Bryan, R.F. Podolide, a new antileukemic norditerpene dilactone from Podocarpus gracilior. Experientia 1975, 31, 137–138. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, A.S.A.; Moustafa, A.H.; Hussein, H.A.; El-Sheikh, A.A.; El-Shafey, S.N.; Fathy, N.A.M.; Enan, G.A. Potential insecticidal activity of Sarocladium strictum, an endophyte of Cynanchum acutum, against Spodoptera littoralis, a polyphagous insect pest. Biocatal. Agric. Biotechnol. 2020, 24. [Google Scholar] [CrossRef]
- Kusari, S.; Pandey, S.P.; Spiteller, M. Untapped mutualistic paradigms linking host plant and endophytic fungal production of similar bioactive secondary metabolites. Phytochemistry 2013, 91, 81–87. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, A.S.A.; Yassin, M.A.; Ali, G.S. Transcriptional and Proteomic Profiling of Aspergillus flavipes in Response to Sulfur Starvation. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, A.S.A. L-methioninase production by Aspergillus flavipes under solid-state fermentation. J. Basic Microbiol. 2009, 49, 331–341. [Google Scholar] [CrossRef]
- El-Sayed, A.S.A. L-glutaminase production by Trichoderma koningii under solid-state fermentation. Indian J. Microbiol. 2009, 49, 243–250. [Google Scholar] [CrossRef]
- El-Sayed, A.S.A.; Ibrahim, H.; Sitohy, M.Z. Co-immobilization of PEGylated Aspergillus flavipes l-methioninase with glutamate dehydrogenase: A novel catalytically stable anticancer consortium. Enzyme Microb. Technol. 2014, 54, 59–69. [Google Scholar] [CrossRef]
- Gamborg, O.; Miller, R.; Ojima, K. Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 1968, 50, 151–158. [Google Scholar] [CrossRef]
- Khan, A.A.; El-Sayed, A.; Akbar, A.; Mangravita-Novo, A.; Bibi, S.; Afzal, Z.; Norman, D.J.; Shad Ali, G. A highly efficient ligation-independent cloning system for CRISPR/Cas9 based genome editing in plants. Plant Methods 2017, 13. [Google Scholar] [CrossRef]
- Zhang, H.W.; Song, Y.C.; Tan, R.X. Biology and chemistry of endophytes. Nat. Prod. Rep. 2006, 23, 753–771. [Google Scholar] [CrossRef]
- Gure, A.; Wahlstrom, K.; Stenlid, J. Pathogenicity of seed-associated fungi to Podocarpus falcatus in vitro. Forest Pathol. 2005, 35, 23–35. [Google Scholar] [CrossRef]
- Li, J.-Y.; Sidhu, R.S.; Bollon, A.; Strobel, G.A. Stimulation of taxol production in liquid cultures of Pestalotiopsis microspora. Mycol. Res. 1998, 102, 461–464. [Google Scholar] [CrossRef]
- Strobel, G.; Yang, X.; Sears, J.; Kramer, R.; Sidhu4, R.S.; Hess5, W.M. Taxol from Pestalotiopsis microspora, an endophytic fungus of Taxus wallachiana. Sun Mkmbiology 2017, 2339, 43–49. [Google Scholar] [CrossRef]
- Li, J.-Y.; Strobel, G.; Sidhu, R.; Hess, W.M.; Ford, E.J. Endophytic taxol-producing fungi from bald cypress, Taxodium distichum. Microbiology 1996, 142, 2223–2226. [Google Scholar] [CrossRef]
- Zhang, P.; Zhou, P.-P.; Yu, L.-J. An endophytic taxol-producing fungus from Taxus x media, Aspergillus candidus MD3. FEBS Microbiol. Lett. 2009, 293. [Google Scholar] [CrossRef]
- Strobel, G.; Yang, X.; Sears, J.; Kramer, R.; Sidhu, R.S.; Hess, W.M.; Young, B. Endophytic fungus of Taxus wallachiana. Microbiology 1996, 142, 3–8. [Google Scholar] [CrossRef]
- Metz, A.M.; Haddad, A.; Worapong, J.; Long, D.M.; Ford, E.J.; Hess, W.M.; Strobel, G.A. Induction of the sexual stage of Pestalotiopsis microspora, a taxol-producing fungus. Microbiology 2000, 146, 2079–2089. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Sun, L.; Ma, X.; Li, X.; Wang, X.; Ping, W.; Zhou, D. Improved taxol production in Nodulisporium sylviforme derived from inactivated protoplast fusion. Afr. J. Biotechnol. 2011, 10, 4175–4182. [Google Scholar] [CrossRef]
- Zhang, P.; Zhou, P.-P.; Yu, L.-J. An Endophytic Taxol-Producing Fungus from Taxus media, Cladosporium cladosporioides MD2. Curr. Microbiol. 2009, 59, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Kumaran, R.S.; Muthumary, J.; Hur, B.K. Production of taxol from Phyllosticta spinarum, an endophytic fungus of Cupressus sp. Eng. Life Sci. 2008, 8, 438–446. [Google Scholar] [CrossRef]
- Deng, B.W.; Liu, K.H.; Chen, W.Q.; Ding, X.W.; Xie, X.C. Fusarium solani, Tax-3, a new endophytic taxol-producing fungus from Taxus chinensis. World J. Microbiol. Biotechnol. 2009, 25, 139–143. [Google Scholar] [CrossRef]
- Zhao, K.; Ping, W.; Li, Q.; Hao, S.; Zhao, L.; Gao, T.; Zhou, D. Aspergillus niger var. taxi, a new species variant of taxol-producing fungus isolated from Taxus cuspidata in China. J. Appl. Microbiol. 2009, 107, 1202–1207. [Google Scholar] [CrossRef]
- Venkatachalam, R.; Subban, K.; Paul, M.J. Taxol from Botryodiplodia theobromae (BT 115)—AN endophytic fungus of Taxus baccata. J. Biotechnol. 2008, 136, S189–S190. [Google Scholar] [CrossRef]
- Qiu, F.; Chen, Y.-R.; Liu, X.; Chu, C.-Y.; Shen, L.-J.; Xu, J.; Gaur, S.; Forman, H.J.; Zhang, H.; Zheng, S.; et al. Arginine Starvation Impairs Mitochondrial Respiratory Function in ASS1-Deficient Breast Cancer Cells. Sci. Signal. 2014, 7. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, C.; Sun, Y.-T.; Sun, C.-Z.; Zhang, Y.; Wang, X.-H.; Zhao, K. Taxol Produced from Endophytic Fungi Induces Apoptosis in Human Breast, Cervical and Ovarian Cancer Cells. Asian Pac. J. Cancer Prev. 2015, 16, 125–131. [Google Scholar] [CrossRef]
- Zhou, X.; Zhu, H.; Liu, L.; Lin, J.; Tang, K. A review: Recent advances and future prospects of taxol-producing endophytic fungi. Appl. Microbiol. Biotechnol. 2010, 86, 1707–1717. [Google Scholar] [CrossRef]
- Chakravarthi, B.V.S.K.; Das, P.; Surendranath, K.; Karande, A.A.; Jayabaskaran, C. Production of paclitaxel by Fusarium solani isolated from Taxus celebica. J. Biosci. 2008, 33, 259–267. [Google Scholar] [CrossRef]
- Guo, B.; Wang, Y.; Sun, X.; Tang, K. Bioactive natural products from endophytes: A review. Appl. Biochem. Microbiol. 2008, 44, 136–142. [Google Scholar] [CrossRef]
- Sun, D.; Ran, X.; Wang, J. Isolation and identification of a taxol-producing endophytic fungus from Podocarpus. Acta Microbiol. Sin. 2008, 48, 589–595. [Google Scholar]
- Yang, Y.; Zhao, H.; Barrero, R.A.; Zhang, B.; Sun, G.; Wilson, I.W.; Xie, F.; Walker, K.D.; Parks, J.W.; Bruce, R.; et al. Genome sequencing and analysis of the paclitaxel-producing endophytic fungus Penicillium aurantiogriseum NRRL 62431. BMC Genom. 2014, 15. [Google Scholar] [CrossRef] [PubMed]
- Bi, J.; Ji, Y.; Pan, J.; Yu, Y.; Chen, H.; Zhu, X. A new taxol-producing fungus (Pestalotiopsis malicola) and evidence for taxol as a transient product in the culture. Afr. J. Biotechnol. 2011, 10, 6647–6654. [Google Scholar] [CrossRef]
- Kusari, S.; Singh, S.; Jayabaskaran, C. Rethinking production of Taxol® (paclitaxel) using endophyte biotechnology. Trends Biotechnol. 2014, 32, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Soliman, S.S.M.; Raizada, M.N. Interactions between Co-Habitating fungi Elicit Synthesis of Taxol from an Endophytic Fungus in Host Taxus Plants. Front. Microbiol. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Flores-Bustamante, Z.R.; Rivera-Orduña, F.N.; Martínez-Cárdenas, A.; Flores-Cotera, L.B. Microbial paclitaxel: Advances and perspectives. J. Antibiot. 2010, 63, 460–467. [Google Scholar] [CrossRef]
- Capasso, F. Medicinal plants: An approach to the study of naturally occurring drugs. J. Ethnopharmacol. 1985, 13, 111–114. [Google Scholar] [CrossRef]
- Shiba, Y.; Paradise, E.M.; Kirby, J.; Ro, D.K.; Keasling, J.D. Engineering of the pyruvate dehydrogenase bypass in Saccharomyces cerevisiae for high-level production of isoprenoids. Metab. Eng. 2007, 9, 160–168. [Google Scholar] [CrossRef]
- Kang, A.; George, K.W.; Wang, G.; Baidoo, E.; Keasling, J.D.; Lee, T.S. Isopentenyl diphosphate (IPP)-bypass mevalonate pathways for isopentenol production. Metab. Eng. 2016, 34, 25–35. [Google Scholar] [CrossRef]
- Roberts, S.C. Production and engineering of terpenoids in plant cell culture. Nat. Chem. Biol. 2007, 3, 387–395. [Google Scholar] [CrossRef]
- Soliman, S.S.M.; Mosa, K.A.; EI-keblawy, A.A.; Husseiny, M.I. Exogenous and endogenous increase in fungal GGPP increased fungal taxol production. Appl. Microbiol. Biotechnol. 2017, 101, 7523–7533. [Google Scholar] [CrossRef]
- Li, Y.C.; Tao, W.Y.; Cheng, L. Paclitaxel production using co-culture of Taxus suspension cells and paclitaxel-producing endophytic fungi in a co-bioreactor. Appl. Microbiol. Biotechnol. 2009, 83, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Brakhage, A.A.; Schuemann, J.; Bergmann, S.; Scherlach, K.; Schroeckh, V.; Hertweck, C. Activation of fungal silent gene clusters: A new avenue to drug discovery. Prog. Drug Res. 2008, 66, 1–12. [Google Scholar]
- Liu, C.; Yu, F.; Liu, Q.; Bian, X.; Hu, S.; Yang, H.; Yin, Y.; Li, Y.; Shen, Y.; Xia, L.; et al. Yield improvement of epothilones in Burkholderia strain DSM7029 via transporter engineering. FEMS Microbiol. Lett. 2018, 365. [Google Scholar] [CrossRef] [PubMed]
- Yamin, W.; Xuanwei, Z.; Lu, L.; Jie, L.; Zinan, W.; Geng, Y.; Lingchuan, H.; Juan, L.; Xiaofen, S.; Kexuan, T. An efficient transformation system of taxol-producing endophytic fungus EFY-21 (Ozonium sp.). Afr. J. Biotechnol. 2010, 9, 1726–1733. [Google Scholar] [CrossRef]
- Liu, L.; Wei, Y.; Zhou, X.; Lin, J.; Sun, X.; Tang, K. Agrobacterium tumefaciens-mediated genetic transformation of the Taxol-producing endophytic fungus Ozonium sp. EFY21. Genet. Mol. Res. 2013, 12, 2913–2922. [Google Scholar] [CrossRef]
- Abdel-Shafi, S.; Al-Mohammadi, A.R.; Hamdi, S.; Moustafa, A.H.; Enan, G. Biological characterization and inhibition of streptococcus pyogenes ZUH1 causing chronic cystitis by crocus sativus methanol extract, bee honey alone or in combination with antibiotics: An in vitro study. Molecules 2019, 24, 2903. [Google Scholar] [CrossRef]
- de Crécy-Lagard, V.; Hanson, A.D. Finding novel metabolic genes through plant-prokaryote phylogenomics. Trends Microbiol. 2007, 15, 563–570. [Google Scholar] [CrossRef]
- Strobel, G.; Daisy, B. Bioprospecting for Microbial Endophytes and Their Natural Products. Microbiol. Mol. Biol. Rev. 2003, 67, 491–502. [Google Scholar] [CrossRef]
- Gond, S.K.; Kharwar, R.N.; White, J.F. Will fungi be the new source of the blockbuster drug taxol? Fungal Biol. Rev. 2014, 28, 77–84. [Google Scholar] [CrossRef]
- Vasundhara, M.; Kumar, A.; Reddy, M.S. Molecular approaches to screen bioactive compounds from endophytic fungi. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Z.-Q.; Yang, Y.-Y.; Zhao, N.; Wang, Y. Diversity of endophytic fungi and screening of fungal paclitaxel producer from Anglojap yew, Taxus x media. BMC Microbiology 2013, 13. [Google Scholar] [CrossRef]
- Khalaf, S.A.; El-Sayed, A.S.A. l-Methioninase Production by Filamentous Fungi: I-Screening and Optimization Under Submerged Conditions. Curr. Microbiol. 2009, 58, 219–226. [Google Scholar] [CrossRef]
- Osbourn, A. Secondary metabolic gene clusters: Evolutionary toolkits for chemical innovation. Trends Genet. 2010, 26, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Brakhage, A.A. Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 2013, 11, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.J.; Wang, C.C.C. Recent advances in genome mining of secondary metabolites in Aspergillus terreus. Front. Microbiol. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Cain, J.W.; Miller, K.I.; Kalaitzis, J.A.; Chau, R.; Neilan, B.A. Genome mining of a fungal endophyte of Taxus yunnanensis (Chinese yew) leads to the discovery of a novel azaphilone polyketide, lijiquinone. Microb. Biotechnol. 2020. [Google Scholar] [CrossRef]
- Wisecaver, J.H.; Rokas, A. Fungal metabolic gene clusters-caravans traveling across genomes and environments. Front. Microbiol. 2015, 6, 1–11. [Google Scholar] [CrossRef]
- Chen, A.J.; Frisvad, J.C.; Sun, B.D.; Varga, J.; Kocsubé, S.; Dijksterhuis, J.; Kim, D.H.; Hong, S.B.; Houbraken, J.; Samson, R.A. Aspergillus section Nidulantes (formerly Emericella): Polyphasic taxonomy, chemistry and biology. Stud. Mycol. 2016, 84, 1–118. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, T.T.; Xu, Q.L.; Xiong, Y.; Zhang, L.; Han, H.; Xu, K.; Guo, W.J.; Xu, Q.; Tan, R.X.; et al. Genome Mining and Comparative Biosynthesis of Meroterpenoids from Two Phylogenetically Distinct Fungi. Angew. Chem.-Int. Ed. 2018, 57, 8184–8188. [Google Scholar] [CrossRef]
- Ziemert, N.; Alanjary, M.; Weber, T. The evolution of genome mining in microbes-a review. Nat. Prod. Rep. 2016, 33, 988–1005. [Google Scholar] [CrossRef] [PubMed]
- Ali, G.S.; El-Sayed, A.S.A.; Patel, J.S.; Green, K.B.; Ali, M.; Brennan, M.; Norman, D. Ex vivo application of secreted metabolites produced by soil-inhabiting Bacillus spp. efficiently controls foliar diseases caused by Alternaria spp. Appl. Environ. Microbiol. 2016, 82, 478–490. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2014, 12, 59–60. [Google Scholar] [CrossRef]
- de Jong, A.; van Heel, A.J.; Kok, J.; Kuipers, O.P. BAGEL2: Mining for bacteriocins in genomic data. Nucleic Acids Res. 2010, 38. [Google Scholar] [CrossRef]
- Weber, T.; Rausch, C.; Lopez, P.; Hoof, I.; Gaykova, V.; Huson, D.H.; Wohlleben, W. CLUSEAN: A computer-based framework for the automated analysis of bacterial secondary metabolite biosynthetic gene clusters. J. Biotechnol. 2009, 140, 13–17. [Google Scholar] [CrossRef]
- Tong, Y.; Charusanti, P.; Zhang, L.; Weber, T.; Lee, S.Y. CRISPR-Cas9 Based Engineering of Actinomycetal Genomes. ACS Synth. Biol. 2015, 4, 1020–1029. [Google Scholar] [CrossRef]
- Medema, M.H.; Blin, K.; Cimermancic, P.; De Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. AntiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39. [Google Scholar] [CrossRef]
- Medema, M.H.; Kottmann, R.; Yilmaz, P.; Cummings, M.; Biggins, J.B.; Blin, K.; De Bruijn, I.; Chooi, Y.H.; Claesen, J.; Coates, R.C.; et al. Minimum Information about a Biosynthetic Gene cluster. Nat. Chem. Biol. 2015, 11, 625–631. [Google Scholar] [CrossRef]
- Hornung, A.; Bertazzo, M.; Dziarnowski, A.; Schneider, K.; Welzel, K.; Wohlert, S.E.; Holzenkämpfer, M.; Nicholson, G.J.; Bechthold, A.; Süssmuth, R.D.; et al. A genomic screening approach to the structure-guided identification of drug candidates from natural sources. ChemBioChem 2007, 8, 757–766. [Google Scholar] [CrossRef]
- Dcosta, V.M.; King, C.E.; Kalan, L.; Morar, M.; Sung, W.W.L.; Schwarz, C.; Froese, D.; Zazula, G.; Calmels, F.; Debruyne, R.; et al. Antibiotic resistance is ancient. Nature 2011, 477, 457–461. [Google Scholar] [CrossRef]
- Cox, G.; Wright, G.D. Intrinsic antibiotic resistance: Mechanisms, origins, challenges and solutions. Int. J. Med. Microbiol. 2013, 303, 287–292. [Google Scholar] [CrossRef]
- Peterson, R.M.; Huang, T.; Rudolf, J.D.; Smanski, M.J.; Shen, B. Mechanisms of self-resistance in the Platensimycin- and platencin-producing streptomyces platensis MA7327 and MA7339 strains. Chem. Biol. 2014, 21, 389–397. [Google Scholar] [CrossRef] [PubMed]

| Family | Fungus | Host | Taxol Yield µg/L Culture | Method of Assay | Reference |
|---|---|---|---|---|---|
| Taxaceae | Taxomyces andreanae | Taxus brevifolia | 0.05 | CIEIA, HPLC | [111] |
| Alternaria alternata | Taxus hicksii | 512 | HPLC | [34] | |
| Pestalothiopsis microspora | Taxus walichiana | 2.9 | CIEIA | [112] | |
| Nodulisporium sylviforme | Taxus cuspidata | 450 | HPLC | [113] | |
| Cladosporium cladosporioides | Taxus media | 800 | TLC, HPLC | [110] | |
| Aspergillus candidus | Taxus media | 112 | TLC, HPLC | [114] | |
| Phomopsis sp. | Taxus cuspidata | 418 | HPLC, TLC | [115] | |
| Fusarium solani | Taxus chinensis | 164 | HPLC | [116] | |
| Mucor rouxianus | Taxus chinensis | 30 | HPLC | [29] | |
| Aspergillus niger | Taxus cuspidata | 273 | HPLC | [117] | |
| Botryodiplodia theobromae | Taxus baccata | 280 | HPLC, MS | [118] | |
| Taxomyces sp. | Taxus yunnanensis | 100 | HPLC, TLC | [119] | |
| Alternaria alternata | T. hicksii | 90 | HPLC, TLC | [34] | |
| Pestalotiopsis microspora | Taxodium distichum | 87 | HPLC, TLC | [109] | |
| Pithomyces sp. | Taxus sumatrana | 84 | HPLC, TLC | [111] | |
| Pestalotiopsis microspora | T. wallichiana | 89 | HPLC, TLC | [111] | |
| Alternaria sp. | T. cuspidata | 19 | HPLC, TLC | [120] | |
| P. microspora | T. baccata | 120 | HPLC, TLC | ||
| Fusarium lateritium | T. baccata | 113 | HPLC, TLC | ||
| Pestalotia bicilia | T. baccata | 125 | HPLC, TLC | ||
| Monochaetia sp. | T. baccata | 190 | HPLC, TLC | ||
| Kitasatospora sp. | T. baccata | 120 | HPLC, TLC | [20] | |
| Penicillium spp. | Taxus species | 111 | HPLC, TLC | [20] | |
| Pestalothiopsis microspora | T. wallichiana | 136 | HPLC, TLC | [112] | |
| Tubercularia sp. | T. mairei | 180 | HPLC, TLC | [120] | |
| Taxomyces sp. | T. yunnanensis | 180 | HPLC, TLC | [10] | |
| Alternaria alternate | T. chinensis | 129 | HPLC, TLC | [34] | |
| Ozonium sp. | T. chinensis | 89 | HPLC, TLC | [34] | |
| Fusarium mairei | T. chinensis | 78 | HPLC, TLC | [34] | |
| Fusarium solani | T. celebica | 75 | HPLC, TLC | [34] | |
| Botryodiplodia theobromae | T. baccata | 45 | HPLC, TLC | [34] | |
| Botrytis sp | T. cuspidata | 65 | HPLC, TLC | [117] | |
| Fusarium arthrosporioides | T. cuspidata | 78 | HPLC, TLC | [109] | |
| Gliocladium sp. | T. baccata | 90 | HPLC, TLC | [34] | |
| Fusarium solani | T. chinensis | 98 | HPLC, TLC | [116] | |
| Mucor rouxianus sp. | T. chinensis | 94 | HPLC, TLC | [116] | |
| Aspergillus niger var taxi | T. cuspidata | 91 | HPLC, TLC | [121] | |
| Phomopsis sp. | T. cuspidata | 82 | HPLC, TLC | [121] | |
| C. cladosporioides | T. media | 72 | HPLC, TLC | [110] | |
| Aspergillus candidus | T. media | 73 | HPLC, TLC | [110] | |
| Phomopsis sp. | T. cuspidata | 70 | HPLC, TLC | [110] | |
| Pithomyces s | T. sumatrana | 20 | HPLC, TLC | [122] | |
| Didymostilbe sp. | T. chinensis | 26 | HPLC, TLC | [120] | |
| Ozonium sp., | T. chinensis | 29 | HPLC, TLC | [121] | |
| Alternaria alternata, | T. chinensis | 30 | HPLC, TLC | [123] | |
| Botrytis sp., | T. chinensis | 36 | HPLC, TLC | ||
| Ectostroma sp., | T. chinensis | 90 | HPLC, TLC | ||
| Podocarpaceae | Aspergillus terreus 1 | Podocarpus gracilior | 20 | HPLC, TLC | [103] |
| A. terreus 2 | Podocarpus gracilior | 14 | HPLC, TLC | ||
| A. terreus 3 | Podocarpus gracilior | 18 | HPLC, TLC | ||
| A. flavus 1 | Podocarpus gracilior | 4.5 | HPLC, TLC | ||
| A. flavus 2 | Podocarpus gracilior | 1.8 | HPLC, TLC | ||
| Penicillium egyptiacum | Podocarpus gracilior | 3.6 | HPLC, TLC | ||
| Aspergillus terreus 1 | Podocarpus gracilior | 20 | HPLC, TLC | ||
| A. terreus 2 | Podocarpus gracilior | 14 | HPLC, TLC | ||
| Aspergillus fumigatus | Podocarpus sp. | 590 | HPLC | [124] | |
| Other plants | Phyllosticta dioscorea | Hibiscus rosa-sinensis | 298 | HPLC, TLC | [115] |
| Phoma betae | Ginkgo biloba | 795 | HPLC | [115] | |
| Phomopsis sp | Ginkgo biloba | 372 | HPLC, MS | ||
| Phomopsis sp. | Larix leptolepis | 334 | HPLC, NMR | ||
| Penicillium aurantiogriseum | Corylus avellana | 70 | LCMS, NMR | [125] | |
| Bartalinia robillardoides | Aegle mamelos | 188 | HPLC, MS | [125] | |
| Phomopsis sp. | Wollemia nobili s | 170 | HPLC, TLC | [77] | |
| Lasiodiplodia theobromae | Morinda citrifolia | 120 | HPLC, TLC | [34] | |
| Phyllostica melochiae | Melochia corchorifolia | 478 | HPLC, TLC | [115] | |
| Phyllosticta spinarum | Cupressus sp. | 235 | HPLC, TLC | ||
| Phyllosticta citricarpa | Citrus media | 265 | HPLC, TLC | ||
| Fusarium proliferatum | Tillandsia usneoides | 165 | HPLC | [34] | |
| Pestalotiopsis sp.107 | Tillandsia usneoides | 89 | HPLC | ||
| Phomopsis sp. 116 | Tillandsia usneoides | 22 | HPLC | ||
| Pestalotiopsis sp., 118 | Tillandsia usneoides | 8.9 | HPLC | ||
| Pestalotiopsis humus 133 | Tillandsia usneoides | 6.1 | HPLC | ||
| Pestalotiopsis humus 154 | Tillandsia usneoides | 5.7 | HPLC | ||
| Pestalotiopsis sp.155 | Tillandsia usneoides | 4.3 | HPLC | ||
| Pestalotiopsis sp.163 | Tillandsia usneoides | 4.0 | HPLC | ||
| Rhizosphere | Aspergillus flavipes | Rhizosphere | 850 | HPLC, TLC | [34] |
| Aspergillus flavus | Rhizosphere | 2.8 | HPLC, TLC | ||
| Aspergillus oryzae | Rhizosphere | 3.2 | HPLC, TLC | ||
| Alternaria sp. | Rhizosphere | 4.2 | HPLC, TLC | ||
| Penicillium chrysogenum | Rhizosphere | 85 | HPLC, TLC | ||
| Pestalotiopsis malicola | Rhizosphere | 186 | HPLC, LCMS | [126] |
| Improvement Approach | Wild-Type Strain | Method | Taxol Increasing (Folds) | Reference |
|---|---|---|---|---|
| Mutagenesis and molecular manipulation | Nodulisporium sylviforme | UV, EMS, 60Co, NTG | 2.5 | [121] |
| Fusarium maire | UV + DES | 8.6 | [34] | |
| Nodulisporium sylviforme | Genome shuffling | 0.5 | [113] | |
| Ozonium sp. | PEG-transformation | 5 | [34] | |
| Ozonium sp. | ATMT | 6 | [121] | |
| Ozonium sp. | ATMT | N.A | [120] | |
| Cladosporium cladosporioides | ATMT | N.A | [117] | |
| Cultural nutritional optimization | Fusarium mairei | pH, temperature, carbon, nitrogen source, fermentation period (Single factor) | 10.2 | [121] |
| F. maire | Nitrogen source (Plackett Burman design) | 1.3 | [121] | |
| Nodulisporium sylviforme | pH, temperature, fermentation period (Single factor) | 1.15 | [113] | |
| Pestalotiopsis microspora | Monobasic sodium phosphate (Single factor) | 2.2 | [107] | |
| Aspergillus terreus | ||||
| Elicitation/Inhibition Strategy | Nodulisporium sylviforme | Serine, SA, silver nitrate, ammonium acetate | 1.1 | [113] |
| Periconia sp. | Serinol, p-hydroxy benzoic acid, β-resorcyclic acid, gallic acid, Benzoic acid | 8 | [107] | |
| Periconia sp. | Benzoate | 8 | [121] | |
| Fusarium maire | Sodium acetate | 11 | [121] | |
| Epicoccum nigrum | Serine | 29 | [121] | |
| Pestalotiopsis microspora | Fluconazole | 50 | [107] | |
| Aspergillus flavipes | Fluconazole | 50 | [34] | |
| Co-cultivation/mixed fermentation | Paraconiothyrium sp. | Alternaria sp. | 2.7 | [134] |
| Phomopsis sp. | 3.8 | |||
| Alternaria sp. and Phomopsis sp. | 7.8 | |||
| Fusarium sp. | Taxus suspension cells | 38 | [135] | |
| Aspergillus terreus | surface sterilized leaves of P. gracilior | 2.5 | [102] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
El-Sayed, A.S.A.; El-Sayed, M.T.; Rady, A.M.; Zein, N.; Enan, G.; Shindia, A.; El-Hefnawy, S.; Sitohy, M.; Sitohy, B. Exploiting the Biosynthetic Potency of Taxol from Fungal Endophytes of Conifers Plants; Genome Mining and Metabolic Manipulation. Molecules 2020, 25, 3000. https://doi.org/10.3390/molecules25133000
El-Sayed ASA, El-Sayed MT, Rady AM, Zein N, Enan G, Shindia A, El-Hefnawy S, Sitohy M, Sitohy B. Exploiting the Biosynthetic Potency of Taxol from Fungal Endophytes of Conifers Plants; Genome Mining and Metabolic Manipulation. Molecules. 2020; 25(13):3000. https://doi.org/10.3390/molecules25133000
Chicago/Turabian StyleEl-Sayed, Ashraf S.A., Manal T. El-Sayed, Amgad M. Rady, Nabila Zein, Gamal Enan, Ahmed Shindia, Sara El-Hefnawy, Mahmoud Sitohy, and Basel Sitohy. 2020. "Exploiting the Biosynthetic Potency of Taxol from Fungal Endophytes of Conifers Plants; Genome Mining and Metabolic Manipulation" Molecules 25, no. 13: 3000. https://doi.org/10.3390/molecules25133000
APA StyleEl-Sayed, A. S. A., El-Sayed, M. T., Rady, A. M., Zein, N., Enan, G., Shindia, A., El-Hefnawy, S., Sitohy, M., & Sitohy, B. (2020). Exploiting the Biosynthetic Potency of Taxol from Fungal Endophytes of Conifers Plants; Genome Mining and Metabolic Manipulation. Molecules, 25(13), 3000. https://doi.org/10.3390/molecules25133000





