Chemometric and Transcriptomic Profiling, Microtubule Disruption and Cell Death Induction by Secalonic Acid in Tumor Cells
Abstract
:1. Introduction
2. Results
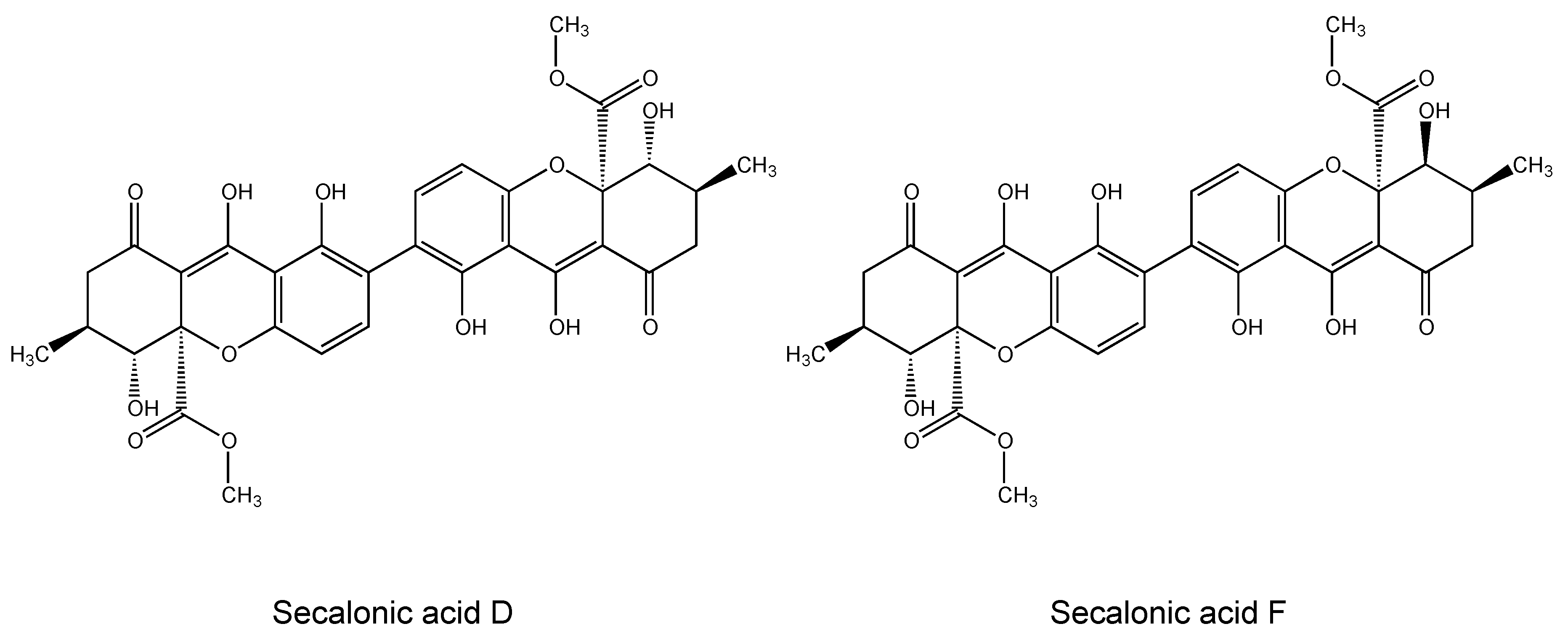
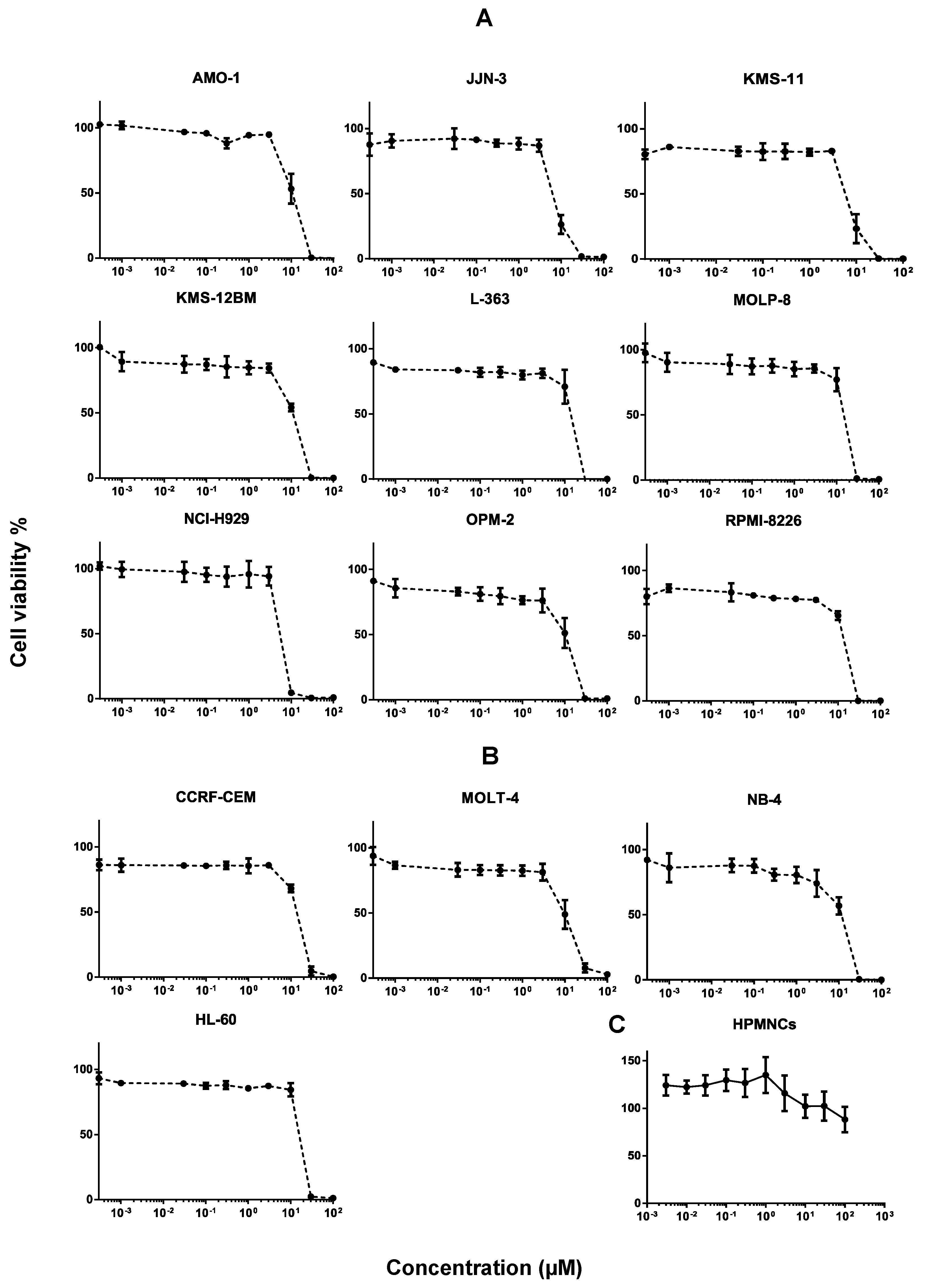
2.1. Cytotoxicity of Secalonic Acid F toward Leukemia and Multiple Myeloma Cells
2.2. Toxicity of Secalonic Acid F in Healthy Cells
2.3. Cross-Resistance of Secalonic Acid to Established Anticancer Drugs
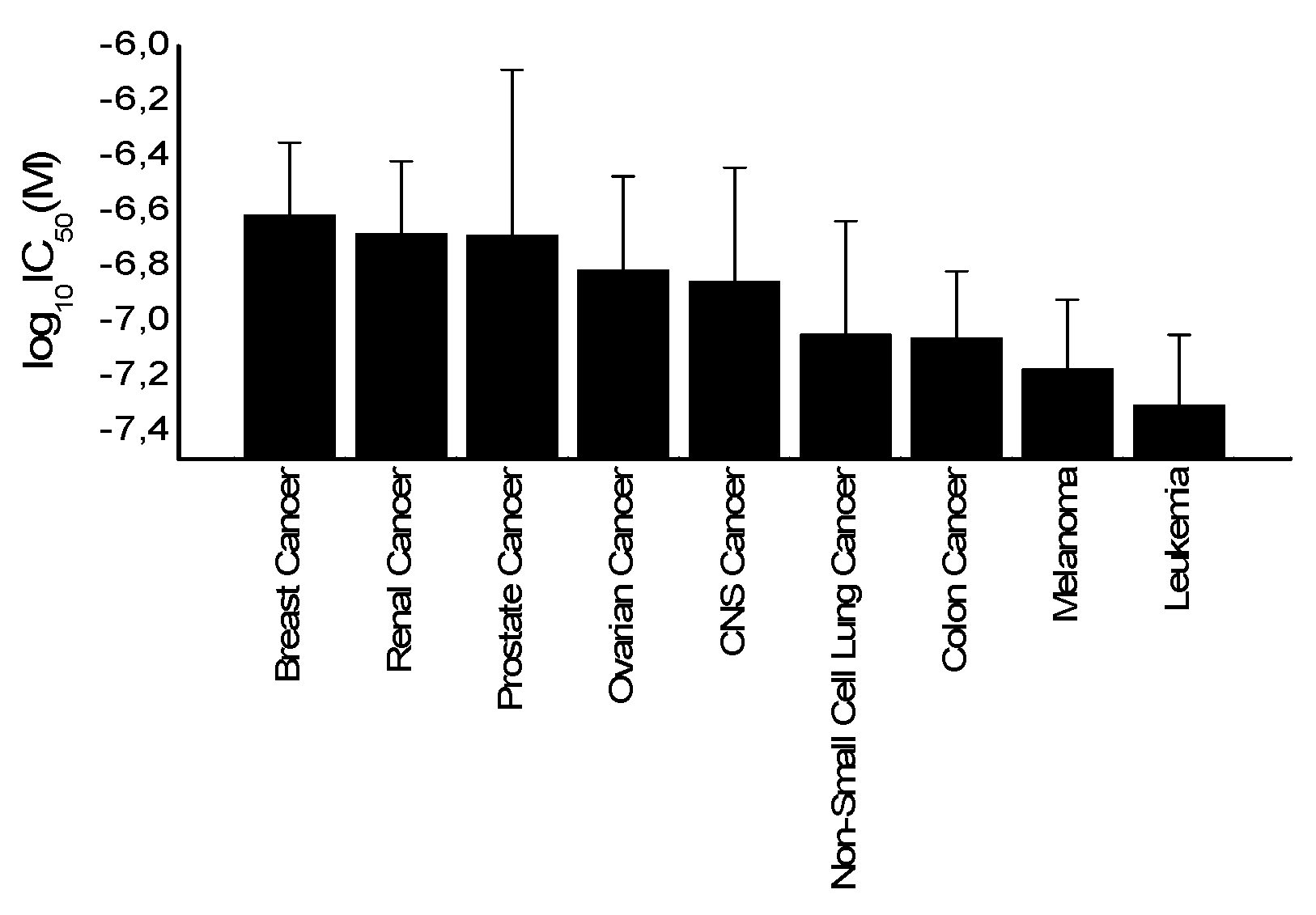
2.4. Tumor-Type Dependent Response toward Secalonic Acid
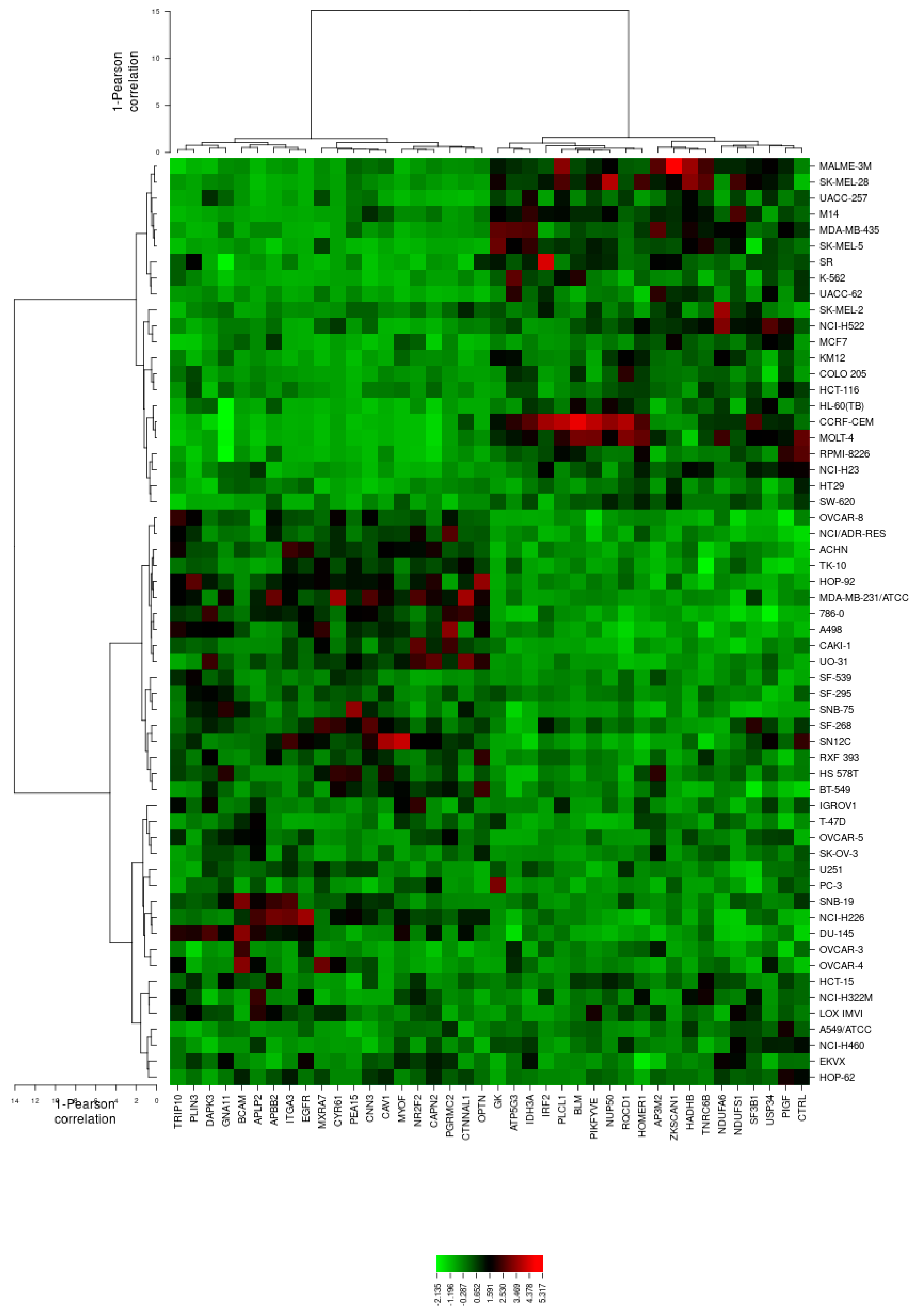
2.5. Microarray-Based Expression Profiling to Predict Sensitivity and Resistance to Secalonic Acid
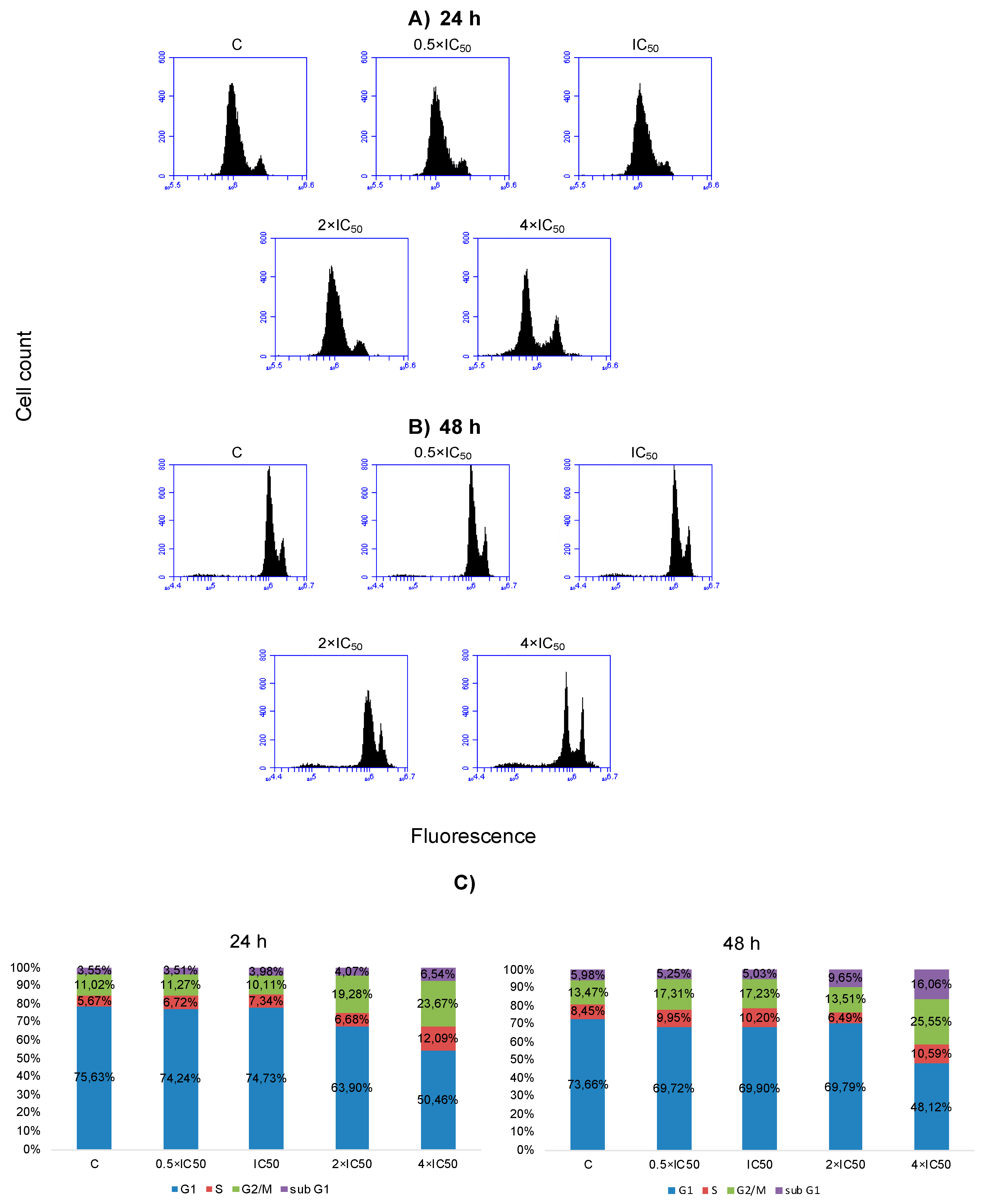
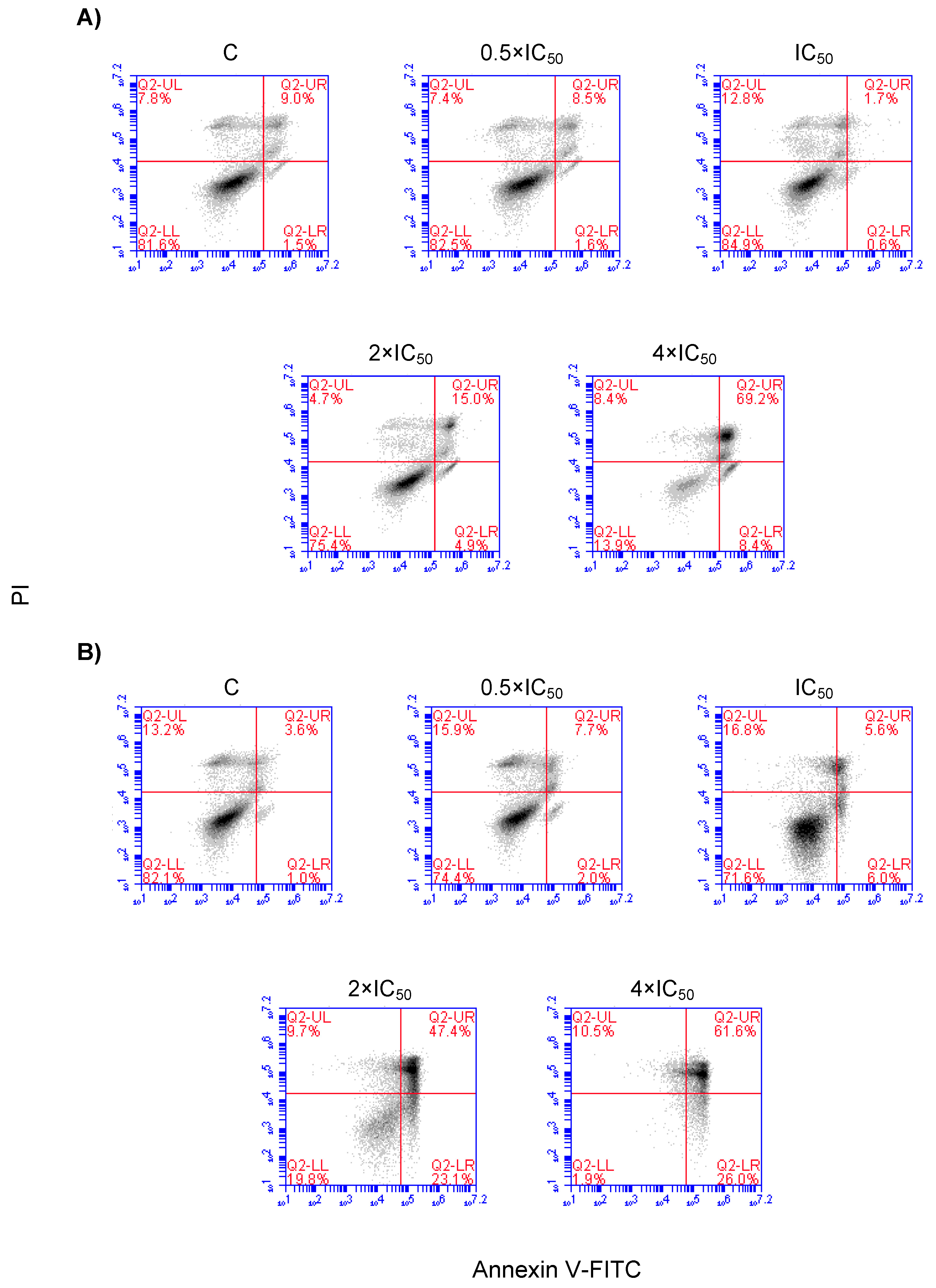
2.6. Cell Cycle Distribution and Apoptosis
2.7. Influence of Secalonic Acid F (SAF) on Microtubules
3. Discussion
3.1. Cytotoxicity of Secalonic Acid F toward Leukemia and Multiple Myeloma Cells
3.2. Toxicity of Secalonic Acid F in Healthy Cells
3.3. COMPARE and Cluster Analyses of Microarray Data
3.4. Cross-Resistance Pattern of the National Cancer Institute (NCI) Cell Line Panel between Secalonic Acid and Standard Drugs
3.5. Cell Cycle Distribution and Apoptosis/Necrosis
4. Materials and Methods
4.1. Cell Lines
4.2. Cytotoxicity Assay
4.3. Toxicity of Secalonic Acid F in Healthy Cells
4.4. Analysis of Cell Cycle Distribution by Flow Cytometry
4.5. Detection of Apoptosis/Necrosis by Annexin V/PI Staining
4.6. COMPARE and Hierarchical Cluster Analyses of Microarray Data
4.7. Imaging of Structure and Dynamics of the Microtubule Cytoskeleton by Fluorescence Microscopy
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- World Health Organization. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 25 May 2020).
- Abe, J.-I.; Martin, J.F.; Yeh, E.T.H. The future of onco-cardiology: We are not just “side effect hunters”. Circ. Res. 2016, 119, 896–899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tewari, D.; Rawat, P.; Singh, P.K. Adverse drug reactions of anticancer drugs derived from natural sources. Food Chem. Toxicol. 2019, 123, 522–535. [Google Scholar] [CrossRef]
- Henning, R.J.; Harbison, R.D. Cardio-oncology: Cardiovascular complications of cancer therapy. Future Cardiol. 2017, 13, 379–396. [Google Scholar] [CrossRef]
- Rajkumar, S.V.; Dimopoulos, M.A.; Palumbo, A.; Blade, J.; Merlini, G.; Mateos, M.V.; Kumar, S.; Hillengass, J.; Kastritis, E.; Richardson, P.; et al. International myeloma working group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 2014, 15, e538–e548. [Google Scholar] [CrossRef]
- Vincent, R.S. Multiple myeloma: 2016 update on diagnosis, risk-stratification, and management. Am. J. Hematol. 2016, 91, 719–734. [Google Scholar] [CrossRef] [Green Version]
- Palumbo, A.; Anderson, K. Multiple myeloma. N. Engl. J. Med. 2011, 364, 1046–1060. [Google Scholar] [CrossRef] [Green Version]
- Brenner, H.; Gondos, A.; Pulte, D. Recent major improvement in long-term survival of younger patients with multiple myeloma. Blood 2008, 111, 2521–2526. [Google Scholar] [CrossRef]
- Lida, S. Mechanisms of action and resistance for multiple myeloma novel drug treatments. Int. J. Hematol. 2016, 104, 271–272. [Google Scholar] [CrossRef] [Green Version]
- Nass, J.; Efferth, T. Drug targets and resistance mechanisms in multiple myeloma. Cancer Drug Resist. 2018, 1, 87–117. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.K.; Dispenzieri, A.; Lacy, M.Q.; Gertz, M.A.; Buadi, F.K.; Pandey, S.; Kapoor, P.; Dingli, D.; Hayman, S.R.; Leung, N.; et al. Continued improvement in survival in multiple myeloma: Changes in early mortality and outcomes in older patients. Leukemia 2014, 28, 1122–1128. [Google Scholar] [CrossRef] [Green Version]
- Juliusson, G.; Hough, R. Leukemia. Prog. Tumor. Res. 2016, 43, 87–100. [Google Scholar] [CrossRef] [PubMed]
- National Cancer Institute. Available online: https://www.cancer.gov/types/leukemia (accessed on 27 June 2020).
- Terwilliger, T.; Abdul-Hay, M. Acute lymphoblastic leukemia: A comprehensive review and 2017 update. Blood Cancer J. 2017, 7, e577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jabbour, E.; O’Brien, S.; Konopleva, M.; Kantarjian, H. New insights into the pathophysiology and therapy of adult acute lymphoblastic leukemia. Cancer 2015, 121, 2517–2528. [Google Scholar] [CrossRef] [PubMed]
- Döhner, H.; Estey, E.H.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Burnett, A.K.; Dombret, H.; Fenaux, P.; Grimwade, D.; Larson, R.A.; et al. Diagnosis and management of acute myeloid leukemia in adults: Recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood 2010, 115, 453–474. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamazaki, M.; Maebayashi, Y.; Miyaki, K. The isolation of secalonic acid A from Aspergillus ochraceus cultured on rice. Chem. Pharm. Bull. 1971, 19, 199–201. [Google Scholar] [CrossRef] [Green Version]
- Andersen, R.; Buechi, G.; Kobbe, B.; Demain, A.L. Secalonic acids D and F are toxic metabolites of Aspergillus aculeatus. J. Org. Chem. 1977, 42, 352–353. [Google Scholar] [CrossRef]
- Lazarovits, G.; Steele, R.W.; Higgins, V.J.; Stoessl, A. Tricyclazole as an inhibitor of polyketide metabolism in the onion pink root rot pathogen, Pyrenochaeta terrestris. Pestic. Biochem. Phys. 1989, 34, 277–287. [Google Scholar] [CrossRef]
- Luisa, B.G. Handbook of Toxic Fungal Metabolites; Elsevier: New York, NY, USA, 2012. [Google Scholar]
- Steyn, P.S. The isolation, structure and absolute configuration of secalonic acid D, the toxic metabolite of Penicillium oxalicum. Tetrahedron 1970, 26, 51–57. [Google Scholar] [CrossRef]
- Betina, V. Mycotoxins: Production, Isolation, Separation, and Purification, 8th ed.; Elsevier: New York, NY, USA, 1984. [Google Scholar]
- Zhang, W.; Krohn, K.; Ullah, Z.; Flörke, U.; Pescitelli, G.; Di Bari, L.; Antus, S.; Kurtán, T.; Rheinheimer, J.; Draeger, S.; et al. New mono- and dimeric members of the secalonic acid family: Blennolides A–G isolated from the fungus Blennoria sp. Chemistry 2008, 14, 4913–4923. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; She, Z.; Shao, C.; Wen, L.; Liu, F.; Zheng, Z.; Lin, Y. 1H and 13C NMR signal assignments of paecilin A and B, two new chromone derivatives from mangrove endophytic fungus Paecilomyces sp. (tree 1–7). Magn. Reson. Chem. 2007, 45, 777–780. [Google Scholar] [CrossRef]
- Guru, S.K.; Pathania, A.S.; Kumar, S.; Ramesh, D.; Kumar, M.; Rana, S.; Kumar, A.; Malik, F.; Sharma, P.R.; Chandan, B.K.; et al. Secalonic acid-d represses hif1α/vegf-mediated angiogenesis by regulating the Akt/mTOR/p70S6K signaling cascade. Cancer Res. 2015, 75, 2886–2896. [Google Scholar] [CrossRef] [Green Version]
- Harada, M.; Yano, S.; Watanabe, H.; Yamazaki, M.; Miyaki, K. Phlogistic activity of secalonic acid A. Chem. Pharm. Bull 1974, 22, 1600–1606. [Google Scholar] [CrossRef] [Green Version]
- Iwaguchi, T.; Kitagawa, H.; Hirose, K.; Ishida, T.; Yamamoto, T. 5-di-(2′-tetrahydropyranyl)secalonic acid D as a new antibiotic derivative with anticancer activity. GANN 1980, 71, 900–906. [Google Scholar]
- Kurobane, I.; Iwahashi, S.; Fukuda, A. Cytostatic activity of naturally isolated isomers of secalonic acids and their chemically rearranged dimers. Drugs Exp. Clin. Res. 1987, 13, 339–344. [Google Scholar]
- Shimizu, M.; Nakamura, M.; Kataoka, T.; Iwaguchi, T. Mechanism of the antitumor activity of 5,5′-bis(2′-tetrahydropyranyl) secalonic acid D against Meth-A. Cancer Chemother. Pharmacol. 1983, 11, 144–146. [Google Scholar] [CrossRef]
- Cai, X.L.; Gao, J.P.; Li, Q.; Wen, L.; She, Z.G.; Lin, Y.C. Cytotoxicity of the secondary metabolites of marine mangrove fungus Paecilomyces sp. tree 1-7 on human hepatoma cell line HepG2. Zhong Yao Cai 2008, 31, 864–865. [Google Scholar] [PubMed]
- Hong, R. Secalonic acid D as a novel DNA topoisomerase I inhibitor from marine lichen-derived fungus Gliocladium sp. T31. Pharm. Biol. 2011, 49, 796–799. [Google Scholar] [CrossRef]
- Ren, H.; Tian, L.; Gu, Q.; Zhu, W. Secalonic acid D.; A cytotoxic constituent from marine lichen-derived fungus Gliocladium sp. T31. Arch. Pharm. Res. 2006, 29, 59–63. [Google Scholar] [CrossRef]
- Tang, R.; Kimishima, A.; Setiawan, A.; Arai, M. Secalonic acid D as a selective cytotoxic substance on the cancer cells adapted to nutrient starvation. J. Nat. Med. 2020, 74, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.P.; Tao, L.Y.; Wang, F.; Zhang, J.Y.; Liang, Y.J.; Fu, L.W. Secalonic acid D reduced the percentage of side populations by down-regulating the expression of ABCG2. Biochem. Pharmacol. 2013, 85, 1619–1625. [Google Scholar] [CrossRef] [PubMed]
- Liao, G.; Zhou, J.; Wang, H.; Mao, Z.; Xiao, W.; Wang, H.; She, Z.; Zhu, Y. The cell toxicity effect of secalonic acid D on GH3 cells and the related mechanisms. Oncol. Rep. 2010, 23, 387–395. [Google Scholar] [PubMed]
- Zhang, J.Y.; Tao, L.Y.; Liang, Y.J.; Yan, Y.Y.; Dai, C.L.; Xia, X.K.; She, Z.G.; Lin, Y.C.; Fu, L.W. Secalonic acid D induced leukemia cell apoptosis and cell cycle arrest of G(1) with involvement of GSK-3beta/beta-catenin/c-Myc pathway. Cell Cycle 2009, 8, 2444–2450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Huang, L.; Tao, L.; Zhang, J.; Wang, F.; Zhang, X.; Fu, L. Secalonic acid D induces cell apoptosis in both sensitive and ABCG2-overexpressing multidrug resistant cancer cells through upregulating c-Jun expression. Acta Pharm. Sin. B. 2019, 9, 516–525. [Google Scholar] [CrossRef]
- Cherigo, L.; Lopez, D.; Martinez-Luis, S. Marine natural products as breast cancer resistance protein inhibitors. Mar. Drugs 2015, 13, 2010–2029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aberhart, D.; Chen, Y.; De Mayo, P.; Stothers, J. Mould metabolites—IV: The isolation and constitution of some ergot pigments. Tetrahedron 1965, 21, 1417–1432. [Google Scholar] [CrossRef]
- Zeng, R.; Luo, S.; Shi, M.; Shi, Y.; Zeng, Q.; Tan, H. Allelopathy of Aspergillus japonicus on crops. Agron. J. 2001, 93, 60–64. [Google Scholar] [CrossRef]
- Zeng, R.S.; Luo, S.M.; Shi, Y.H.; Shi, M.B.; Tu, C.Y. Physiological and biochemical mechanism of allelopathy of secalonic acid F on higher plants. Agron. J. 2001, 93, 72–79. [Google Scholar] [CrossRef] [Green Version]
- Gao, X.; Sun, H.L.; Liu, D.S.; Zhang, J.R.; Zhang, J.; Yan, M.M.; Pan, X.H. Secalonic acid- F inhibited cell growth more effectively than 5-fluorouracil on hepatocellular carcinoma in vitro and in vivo. Neoplasma 2017, 64, 344–350. [Google Scholar] [CrossRef]
- Xie, L.; Li, M.; Liu, D.; Wang, X.; Wang, P.; Dai, H.; Yang, W.; Liu, W.; Hu, X.; Zhao, M. Secalonic acid-F, a novel mycotoxin, represses the progression of hepatocellular carcinoma via MARCH1 regulation of the PI3K/AKT/β-catenin signaling pathway. Molecules 2019, 24, 393. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Yi, Z.; Wang, Y.; Zhang, Q.; Zhong, T.; Qiu, Y.; Wu, Z.; Tang, X. Differential proteomic analysis of HL60 cells treated with secalonic acid F reveals caspase 3-induced cleavage of Rho GDP dissociation inhibitor 2. Oncol. Rep. 2012, 28, 2016–2022. [Google Scholar] [CrossRef] [PubMed]
- Reddy, C.S.; Hayes, A.W.; Williams, W.L.; Ciegler, A. Toxicity of secalonic acid D. J. Toxicol. Environ. Health 1979, 5, 1159–1169. [Google Scholar] [CrossRef] [PubMed]
- Ross, D.T.; Scherf, U.; Eisen, M.B.; Perou, C.M.; Rees, C.; Spellman, P.; Iyer, V.; Jeffrey, S.S.; Van de Rijn, M.; Waltham, M.; et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat. Genet. 2000, 24, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Scherf, U.; Ross, D.T.; Waltham, M.; Smith, L.H.; Lee, J.K.; Tanabe, L.; Kohn, K.W.; Reinhold, W.C.; Myers, T.G.; Andrews, D.T.; et al. A gene expression database for the molecular pharmacology of cancer. Nat. Genet. 2000, 24, 236–244. [Google Scholar] [CrossRef]
- Weinstein, J.N. Fishing expeditions. Science 1998, 282, 627. [Google Scholar] [CrossRef]
- Dumontet, C.; Jordan, M.A. Microtubule-binding agents: A dynamic field of cancer therapeutics. Nat. Rev. Drug Discov. 2010, 9, 790–803. [Google Scholar] [CrossRef] [Green Version]
- Jordan, M.A.; Wilson, L. Microtubules as a target for anticancer drugs. Nat. Rev. Cancer 2004, 4, 253–265. [Google Scholar] [CrossRef]
- Oshiro, C.; Marsh, S.; McLeod, H.; Carrillo, M.W.; Klein, T.; Altman, R. Taxane pathway. Pharmacogenet. Genom. 2009, 19, 979–983. [Google Scholar] [CrossRef]
- Kaverina, I.; Straube, A. Regulation of cell migration by dynamic microtubules. Semin. Cell Dev. Biol. 2011, 22, 968–974. [Google Scholar] [CrossRef] [Green Version]
- Alley, M.C.; Scudiero, D.A.; Monks, A.; Hursey, M.L.; Czerwinski, M.J.; Fine, D.L.; Abbott, B.J.; Mayo, J.G.; Shoemaker, R.H.; Boyd, M.R. Feasibility of drug screening with panels of human tumor cell lines using a microculture tetrazolium assay. Cancer Res. 1988, 48, 589–601. [Google Scholar]
- Rubinstein, L.V.; Shoemaker, R.H.; Paull, K.D.; Simon, R.M.; Tosini, S.; Skehan, P.; Scudiero, D.A.; Monks, A.; Boyd, M.R. Comparison of in vitro anticancer-drug-screening data generated with a tetrazolium assay versus a protein assay against a diverse panel of human tumor cell lines. J. Natl. Cancer Inst. 1990, 82, 1113–1118. [Google Scholar] [CrossRef] [PubMed]
- Kuete, V.; Mbaveng, A.T.; Nono, E.C.; Simo, C.C.; Zeino, M.; Nkengfack, A.E.; Efferth, T. Cytotoxicity of seven naturally occurring phenolic compounds towards multi-factorial drug-resistant cancer cells. Phytomedicine 2016, 23, 856–863. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Wilson, I.; Orton, T.; Pognan, F. Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur. J. Biochem. 2000, 267, 5421–5426. [Google Scholar] [CrossRef] [PubMed]
- Saeed, M.E.M.; Mahmoud, N.; Sugimoto, Y.; Efferth, T.; Abdel-Aziz, H. Molecular determinants of sensitivity or resistance of cancer cells toward sanguinarine. Front. Pharmacol. 2018, 9, 136. [Google Scholar] [CrossRef]
- Paull, K.D.; Shoemaker, R.H.; Hodes, L.; Monks, A.; Scudiero, D.A.; Rubinstein, L.; Plowman, J.; Boyd, M.R. Display and analysis of patterns of differential activity of drugs against human tumor cell lines: Development of mean graph and COMPARE algorithm. J. Natl. Cancer Inst. 1989, 81, 1088–1092. [Google Scholar] [CrossRef]
Sample Availability: The sample of the compound secalonic acid F is available from the authors. |


| R Value | Gene Symbol | Genbank Accession Number | Pattern ID | Gene Name | Gene Function |
|---|---|---|---|---|---|
| Standard COMPARE (Resistance Genes) | |||||
| 0.503 | PIKFYVE | AB023198 | GC35064 | Phosphoinositide kinase, FYVE finger containing | Required for endocytic-vacuolar pathway and nuclear migration |
| 0.502 | NDUFS1 | X61100 | GC28806 | NADH dehydrogenase (ubiquinone) Fe-S protein (NADH-coenzyme Q reductase) | Core subunit of the mitochondrial membrane respiratory chain |
| 0.499 | PIGF | D13435 | GC32669 | Phosphatidylinositol glycan anchor biosynthesis, class F | Involved in GPI-anchor biosynthesis |
| 0.499 | NUP50 | N42007 | GC30995 | Nucleoporin | Component of the nuclear pore complex that has a direct role in nuclear protein import |
| 0.481 | HADHB | D16481 | GC30165 | Hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), b | Encodes a protein, which catalyzes the last three steps of mitochondrial beta-oxidation of long chain fatty acids |
| 0.474 | RQCD1 | AA160724 | GC36841 | RCD1 required for cell differentiation1 homolog (S. pombe) | Involved in various cellular processes including bulk mRNA degradation, miRNA-mediated repression, translational repression and general transcription regulation |
| 0.474 | PLCL1 | D42108 | GC33166 | Phospholipase C-like 1 | Involved in an inositol phospholipid-based intracellular signaling cascade. Regulates the turnover of receptors and contributes to the maintenance of GABA-mediated synaptic inhibition |
| 0.468 | GK | L13943 | GC28538 | Glycerol kinase | Key enzyme in the regulation of glycerol uptake and metabolism |
| 0.454 | AP3M2 | D38293 | GC27638 | Adaptor-related protein complex 3, µ2 subunit | Involved in the budding of vesicles from the Golgi membrane and trafficking to lysosomes |
| 0.453 | IRF2 | X15949 | GC33202 | Interferon regulatory factor 2 | Involved in cell cycle regulation and antagonizes IRF1 transcriptional activation |
| 0.45 | CTRL | X71877 | GC30632 | Chymotrypsin-like | Encodes a serine-type endopeptidase with chymotrypsin- and elastase-2-like activities. The gene encoding this zymogen is expressed specifically in the pancreas and likely functions as a digestive enzyme |
| 0.449 | ATP5G3 | U09813 | GC37818 | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) | Produces ATP from ADP in the presence of a proton gradient across the membrane |
| 0.448 | SF3B1 | AF054248 | GC29865 | Splicing factor 3b, subunit 1 | Required for A complex assembly and involved in the assembly of the E complex |
| 0.446 | TNRC6B | AB029016 | GC27890 | Trinucleotide repeat containing 6B | Plays a role in RNA-mediated gene silencing and is required for miRNA-dependent translational repression and siRNA-dependent endonucleolytic cleavage of complementary mRNAs by argonaute family proteins |
| 0.445 | HOMER1 | Y17829 | GC31446 | Homer homolog 1 (Drosophila) | Involved in the regulation the trafficking and surface expression of GRM5 and the structural changes that occur at synapses during long-lasting neuronal plasticity and development |
| 0.445 | USP34 | AB018272 | GC31656 | Ubiquitin specific peptidase 34 | Regulates Wnt signaling pathway and involved in the processing of poly-ubiquitin precursors as well as that of ubiquitinated proteins |
| 0.444 | BLM | U39817 | GC33552 | Bloom syndrome, RecQ helicase-like | Participates in DNA replication and repair |
| 0.443 | IDH3A | U07681 | GC39215 | Isocitrate dehydrogenase 3 (NAD+) α | Catalyze the oxidative decarboxylation of isocitrate to 2-oxoglutarate |
| 0.441 | ZKSCAN1 | U09848 | GC32155 | Zinc finger with KRAB and SCAN domains 1 | Regulates the expression of GABA type-A receptors in the brain |
| 0.436 | NDUFA6 | AI223047 | GC29399 | NADH dehydrogenase (ubiquinone) 1 α subcomplex 6 | Accessory subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (Complex I) |
| Reverse COMPARE (Sensitive Genes) | |||||
| −0.567 | APBB2 | AL080130 | GC30961 | Amyloid beta (A4) precursor protein-binding, family B, member 2 | Interacting with the cytoplasmic domains of amyloid beta (A4) precursor protein and amyloid beta (A4) precursor-like protein 2 involved in signal transduction |
| −0.548 | ITGA3 | M59911 | GC32775 | Integrin, α3 (antigen CD49C, α3 subunit of VLA-3 receptor) | Participates in the adhesion, formation of invadopodia and matrix degradation processes, promoting cell invasion |
| −0.532 | TRIP10 | AJ000414 | GC39136 | Thyroid hormone receptor interactor 10 | Required for translocation of GLUT4 to the plasma membrane and coordinating membrane tubulation with reorganization of the actin cytoskeleton |
| −0.514 | DAPK3 | AB007144 | GC36901 | Death-associated protein kinase 3 | Regulates apoptosis, autophagy, transcription, translation and actin cytoskeleton reorganization |
| −0.506 | GNA11 | N36926 | GC31915 | Guanine nucleotide binding protein (G protein), α11 (Gq class) RNA | Functioning as modulators or transducers in various transmembrane signaling systems |
| −0.505 | NR2F2 | M64497 | GC29817 | Nuclear receptor subfamily 2, group F, member 2 | Regulates the apolipoprotein A-I gene transcription |
| −0.501 | PGRMC2 | AJ002030 | GC29236 | Progesterone receptor membrane component 2 | Receptor for steroids. Up-regulated during the secretory phase of the menstrual cycle. |
| −0.489 | CTNNAL1 | U97067 | GC38343 | Catenin (cadherin-associated protein), α-like 1 | Modulates the Rho pathway signaling by providing a scaffold for the Lbc Rho guanine nucleotide exchange factor (ARHGEF1) |
| −0.488 | OPTN | AF070533 | GC32186 | Optineurin | Plays an important role in the maintenance of the Golgi complex, in membrane trafficking, in exocytosis, through its interaction with myosin VI and Rab8. |
| −0.488 | CNN3 | S80562 | GC31388 | Calponin 3, acidic | Regulates and modulates smooth muscle contraction |
| −0.486 | PEA15 | X86809 | GC35241 | Phosphoprotein enriched in astrocytes 15 | Blocks Ras-mediated inhibition of integrin activation and modulates the ERK MAP kinase cascade. Inhibits RPS6KA3 (Ribosomal Protein S6 Kinase A3), CASP8 activities and apoptosis and regulates glucose transport |
| −0.486 | CYR61 | Y11307 | GC29186 | Cysteine-rich, angiogenic inducer, 61 | Promotes cell proliferation, chemotaxis, angiogenesis and cell adhesion |
| −0.484 | EGFR | X00588 | GC33544 | Epidermal growth factor receptor | Activates several signaling cascades such as RAS-RAF-MEK-ERK, PI3 kinase-AKT to convert extracellular cues into appropriate cellular responses |
| −0.484 | MXRA7 | AL046940 | GC31711 | Matrix-remodeling associated 7 | Acts on tissue remodeling and may be associated with diseases like endometriosis of ovary |
| −0.479 | PLIN3 | AF057140 | GC30596 | Perilipin 3 | Required for the transport of mannose 6-phosphate receptors |
| −0.479 | MYOF | AL096713 | GC37683 | Myoferlin | Plays a role in the plasmalemma repair mechanism of endothelial cells Involved in endocytic recycling and VEGF signal transduction |
| −0.476 | CAPN2 | M23254 | GC27400 | Calpain 2, (m/II) large subunit | Involved in cytoskeletal remodeling and signal transduction |
| −0.472 | BCAM | X83425 | GC30519 | Basal cell adhesion molecule (Lutheran blood group) | Mediate intracellular signaling and play a role in epithelial cell cancer and in vaso-occlusion of red blood cells in sickle cell disease |
| −0.471 | APLP2 | S60099 | GC36942 | Amyloid β (A4) precursor-like protein 2 | Play a role in the regulation of hemostasis |
| −0.467 | CAV1 | AF070648 | GC39139 | Caveolin 1, caveolae protein | Regulates G-protein activity |
| Secalonic Acid | Sensitive | Resistant |
|---|---|---|
| Partition (log10IC50) | <−7.03 M | ≥−7.03 M |
| Cluster 1 | 8 | 1 |
| Cluster 2 | 10 | 3 |
| Cluster 3 | 3 | 15 |
| Cluster 4 | 7 | 11 |
| χ2 test | p = 4.29 × 10−4 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Özenver, N.; Dawood, M.; Fleischer, E.; Klinger, A.; Efferth, T. Chemometric and Transcriptomic Profiling, Microtubule Disruption and Cell Death Induction by Secalonic Acid in Tumor Cells. Molecules 2020, 25, 3224. https://doi.org/10.3390/molecules25143224
Özenver N, Dawood M, Fleischer E, Klinger A, Efferth T. Chemometric and Transcriptomic Profiling, Microtubule Disruption and Cell Death Induction by Secalonic Acid in Tumor Cells. Molecules. 2020; 25(14):3224. https://doi.org/10.3390/molecules25143224
Chicago/Turabian StyleÖzenver, Nadire, Mona Dawood, Edmond Fleischer, Anette Klinger, and Thomas Efferth. 2020. "Chemometric and Transcriptomic Profiling, Microtubule Disruption and Cell Death Induction by Secalonic Acid in Tumor Cells" Molecules 25, no. 14: 3224. https://doi.org/10.3390/molecules25143224






