Identification and Characterization of a Novel N- and O-Glycosyltransferase from Saccharopolyspora erythraea
Abstract
:1. Introduction
2. Results
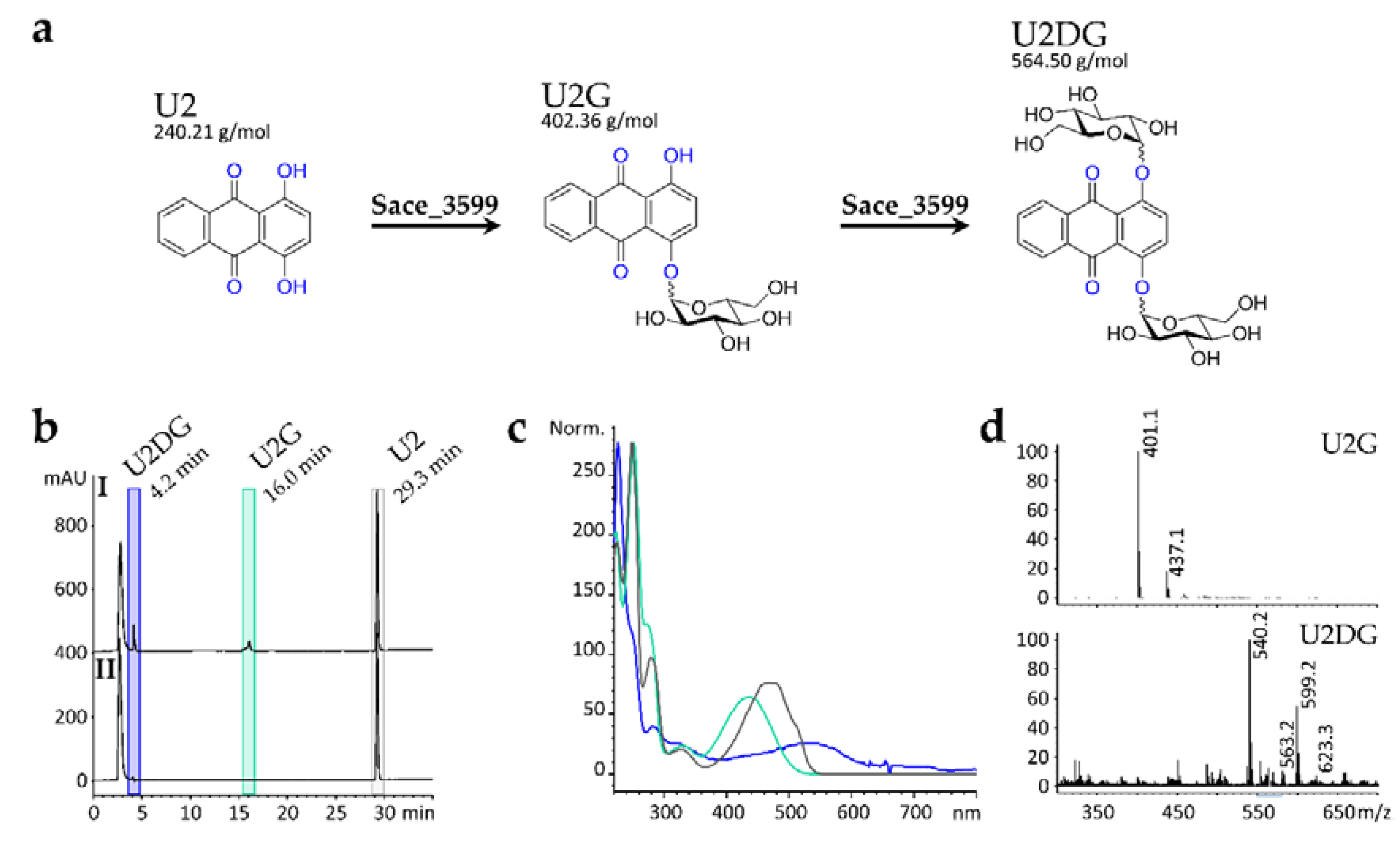
2.1. In Vivo Biotransformation of Anthraquinone Derivatives
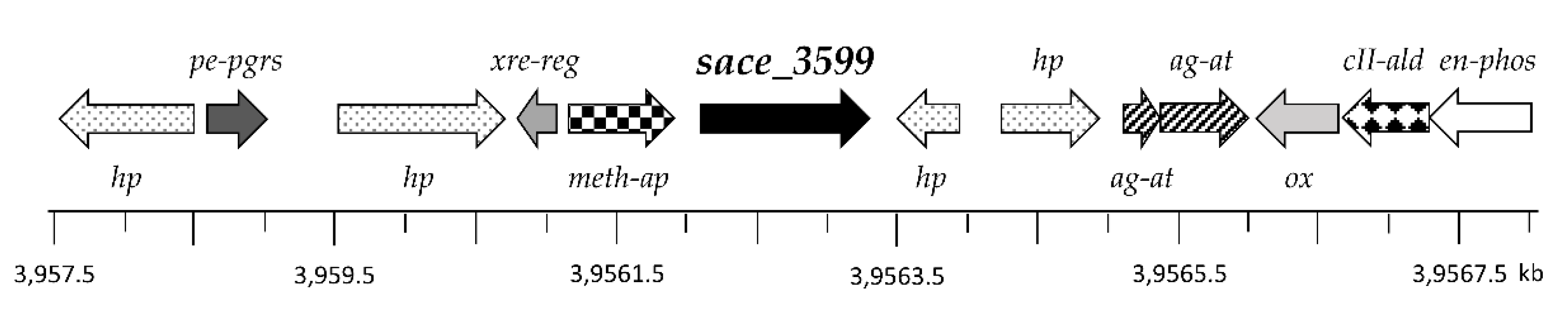
2.2. Genes Encoding for Glycosyltransferases in the Genome of S. erythraea
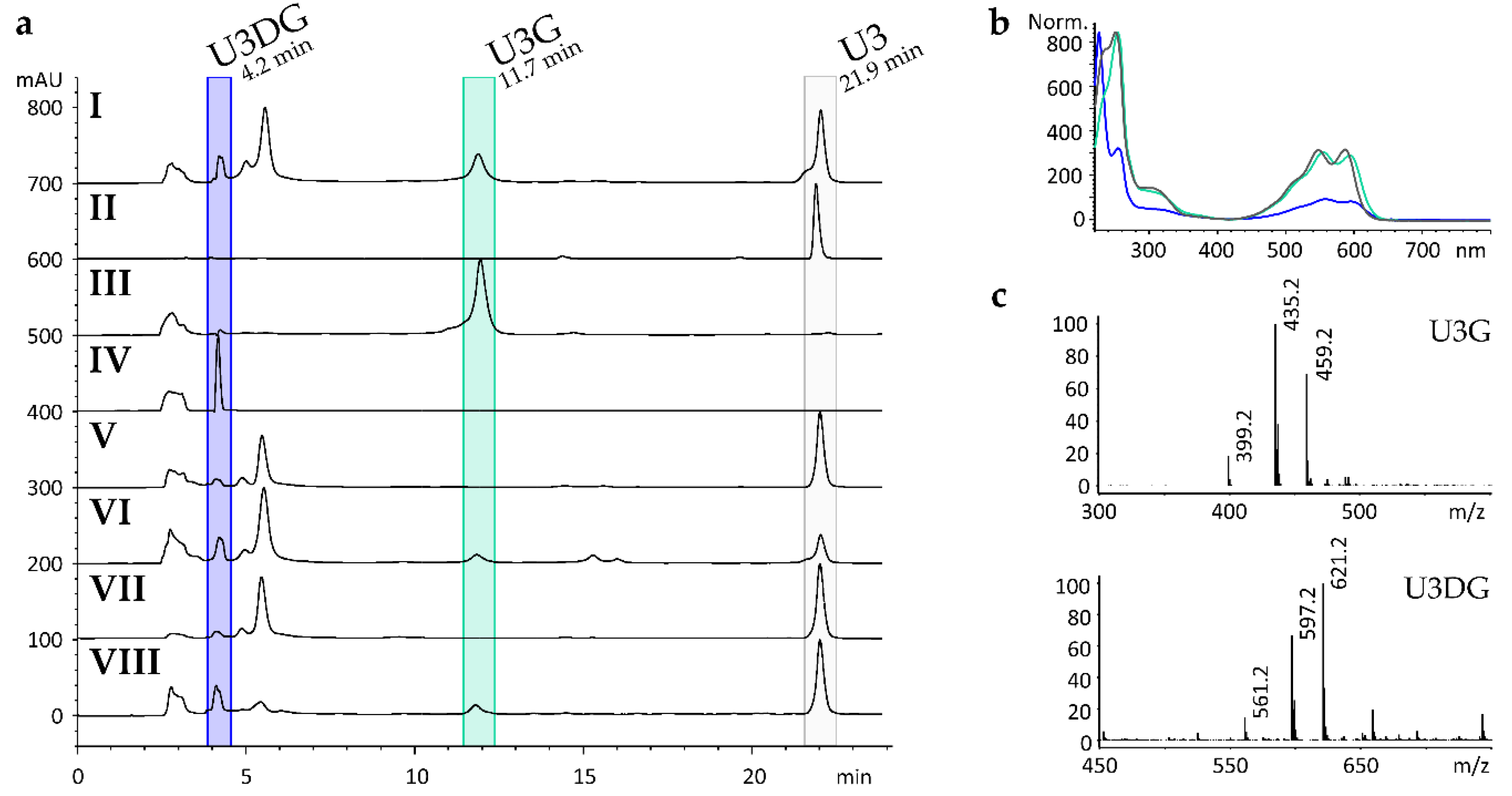
2.3. Heterologous Expression of Sace_3599 in S. albus Gluc
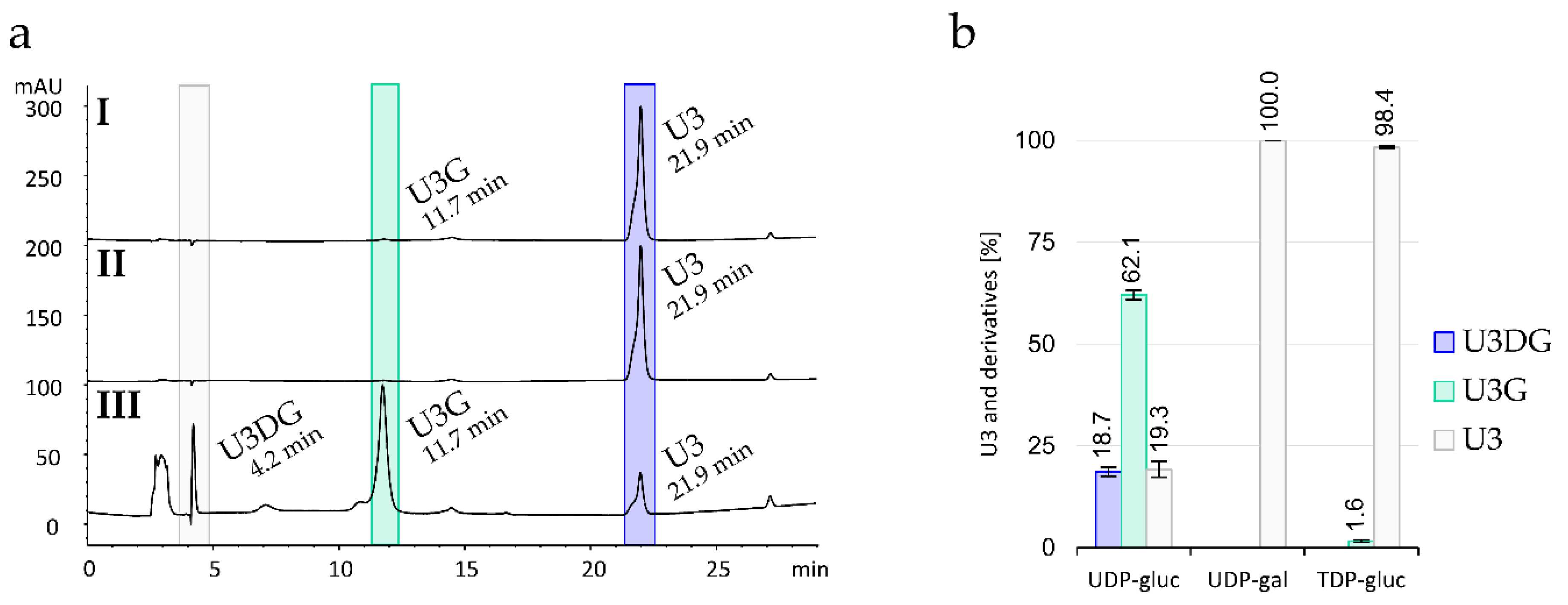
2.4. Gene Deletion and Complementation Experiments
2.5. Structure Elucidation of U3G and U3DG
2.6. Production, Purification and In Vitro Activity Assay of Sace_3599
2.7. Genome Sequence Analysis
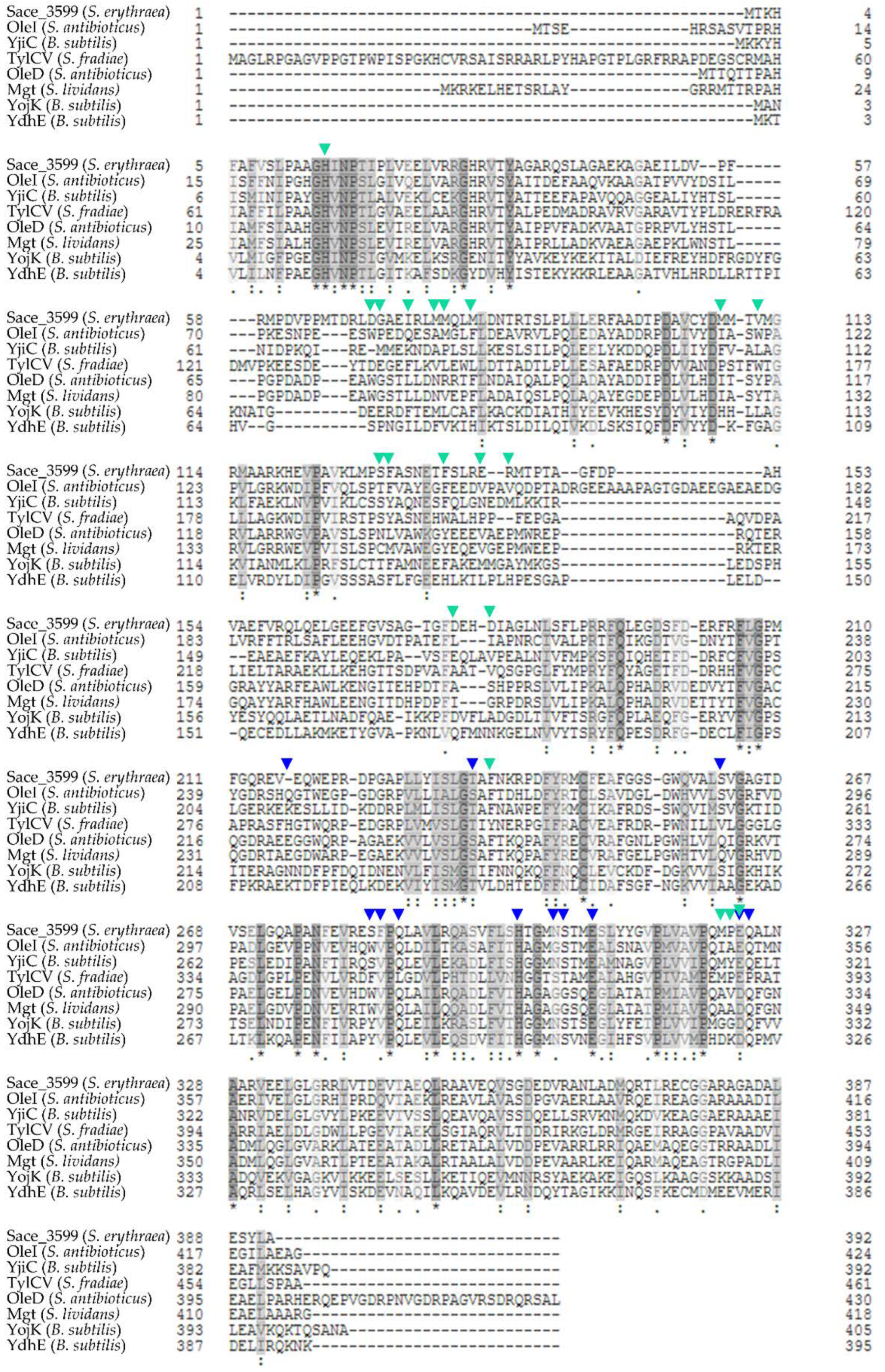
2.8. Homologous Proteins to Sace_3599
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Plasmid Construction
4.3. Whole-Cell Feeding Experiments
4.4. Heterologous Test System
4.5. Inactivation of Sace_3599 in the Genome of S. erythraea
4.6. Complementation of S. erythraea Δsace_3599 with an Intact Copy of Sace_3599
4.7. Purification of Biotransformation Products
4.8. Sample Analysis by HPLC-MS
4.9. Structure Elucidation Using NMR Spectroscopy
4.10. Separation of Mono-Glucosylated Products Using Capillary Electrophoresis
4.11. Expression of Sace_3599 in E. coli and Purification of the Protein
4.12. In Vitro Glycosylation Activity Assay
4.12.1. Variation of Time and Temperature
4.12.2. Variation of Sugar Donor
4.12.3. Variation of Buffer
4.12.4. Variation of Metal Ions as Putative Cofactors
4.12.5. Variation of Substrate
4.12.6. Calculation of Conversion of Substrate to Glucosylated Product
4.13. In Vitro Glycohydrolase Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Khan, H.; Saeedi, M.; Nabavi, S.M.; Mubarak, M.S.; Bishayee, A. Glycosides from Medicinal Plants as Potential Anticancer Agents: Emerging Trends towards Future Drugs. Curr. Med. Chem. 2019, 26, 2389–2406. [Google Scholar] [CrossRef] [PubMed]
- Ushasree, M.V.; Lee, E.Y. Flavonoids, terpenoids, and polyketide antibiotics: Role of glycosylation and biocatalytic tactics in engineering glycosylation [published online ahead of print, 2020 Apr 30]. Biotechnol. Adv. 2020, 41, 107550. [Google Scholar] [CrossRef]
- Kren, V.; Rezanka, T. Sweet antibiotics—The role of glycosidic residues in antibiotic and antitumor activity and their randomization. FEMS Microbiol. Rev. 2008, 32, 858–889. [Google Scholar] [CrossRef] [Green Version]
- Kren, V.; Martínková, L. Glycosides in medicine: “The role of glycosidic residue in biological activity”. Curr. Med. Chem. 2001, 8, 1303–1328. [Google Scholar] [CrossRef] [PubMed]
- Illidge, T.; Klein, C.; Sehn, L.H.; Davies, A.; Salles, G.; Cartron, G. Obinutuzumab in hematologic malignancies: Lessons learned to date. Cancer Treat Rev. 2015, 41, 784–792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weymouth-Wilson, A.C. The role of carbohydrates in biologically active natural products. Nat. Prod. Rep. 1997, 14, 99–110. [Google Scholar] [CrossRef]
- Elshahawi, S.I.; Shaaban, K.A.; Kharel, M.K.; Thorson, T. A comprehensive review of glycosylated bacterial natural products. Chem. Soc. Rev. 2015, 44, 7591–7697. [Google Scholar] [CrossRef] [Green Version]
- Huang, G.; Lv, M.; Hu, J.; Huang, K.; Xu, H. Glycosylation and Activities of Natural Products. Med. Chem. 2016, 16, 1013–1016. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [Green Version]
- Coutinho, P.M.; Deleury, E.; Davies, G.J.; Henrissat, B. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 2003, 328, 307–317. [Google Scholar] [CrossRef]
- Lairson, L.L.; Henrissat, B.; Davies, G.J.; Withers, S.G. Glycosyltransferases: Structures, functions, and mechanisms. Annu. Rev. Biochem. 2008, 77, 521–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourne, Y.; Henrissat, B. Glycoside hydrolases and glycosyltransferases: Families and functional modules. Curr. Opin. Struct. Biol. 2001, 11, 593–600. [Google Scholar] [CrossRef]
- Henrissat, B.; Sulzenbacher, G.; Bourne, Y. Glycosyltransferases, glycoside hydrolases: Surprise, surprise! Curr. Opin. Struct. Biol. 2008, 18, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, F.; Aebi, M. Mechanisms and principles of N-linked protein glycosylation. Curr. Opin. Struct. Biol. 2011, 21, 576–582. [Google Scholar] [CrossRef] [PubMed]
- Eida, A.A.; Abugrain, M.E.; Brumsted, C.J.; Mahmud, T. Glycosylation of acyl carrier protein-bound polyketides during pactamycin biosynthesis. Nat. Chem. Biol. 2019, 15, 795–802. [Google Scholar] [CrossRef]
- Kong, Y.; Li, J.; Hu, X.; Wang, Y.; Meng, Q.; Gu, G.; Wang, P.G.; Chen, M. N-Glycosyltransferase from Aggregatibacter aphrophilus synthesizes glycopeptides with relaxed nucleotide-activated sugar donor selectivity. Carbohydr. Res. 2018, 462, 7–12. [Google Scholar] [CrossRef]
- Zhao, P.; Bai, L.; Ma, J.; Zeng, Y.; Li, L.; Zhang, Y.; Lu, C.; Dai, H.; Wu, Z.; Wu, X.; et al. Amide N-glycosylation by Asm25, an N-glycosyltransferase of ansamitocins. Chem. Biol. 2008, 15, 863–874. [Google Scholar] [CrossRef]
- Guo, Z.; Li, J.; Qin, H.; Wang, M.; Lv, X.; Li, X.; Chen, Y. Biosynthesis of the carbamoylated D-gulosamine moiety of streptothricins: Involvement of a guanidino-N-glycosyltransferase and an N-acetyl-D-gulosamine deacetylase. Angew. Chem. Int. Ed. Engl. 2015, 54, 5175–5178. [Google Scholar] [CrossRef]
- Zhu, L.; Cao, X.; Chen, W.; Zhang, G.; Sun, D.; Wang, P.G. Syntheses and biological activities of daunorubicin analogs with uncommon sugars. Bioorg. Med. Chem. 2005, 13, 6381–6387. [Google Scholar] [CrossRef]
- Battisti, R.F.; Zhong, Y.; Fang, L.; Gibbs, S.; Shen, J.; Nadas, J.; Zhang, G.; Sun, D. Modifying the sugar moieties of daunorubicin overcomes P-gp-mediated multidrug resistance. Mol. Pharm. 2007, 4, 140–153. [Google Scholar] [CrossRef] [Green Version]
- Han, A.R.; Park, J.W.; Lee, M.K.; Ban, Y.H.; Yoo, Y.J.; Kim, E.J.; Kim, E.; Kim, B.-G.; Sohng, J.K.; Yoon, Y.J. Development of a Streptomyces venezuelae-based combinatorial biosynthetic system for the production of glycosylated derivatives of doxorubicin and its biosynthetic intermediates. Appl. Environ. Microbiol. 2011, 77, 4912–4923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boons, G.J. Strategies in Oligosaccharide Synthesis. Tetrahedron 1996, 52, 1095–1121. [Google Scholar] [CrossRef]
- Suzuki, K. Lessons from total synthesis of hybrid natural products. Chem. Rec. 2010, 10, 291–307. [Google Scholar] [CrossRef] [PubMed]
- Gaisser, S.; Böhm, G.A.; Cortés, J.; Leadlay, P.F. Analysis of seven genes from the eryAI-eryK region of the erythromycin biosynthetic gene cluster in Saccharopolyspora erythraea. Mol. Gen. Genet. 1997, 256, 239–251. [Google Scholar] [CrossRef]
- Fernández, E.; Weißbach, U.; Reillo, C.S.; Braña, A.F.; Méndez, C.; Rohr, J.; Salas, J.A. Identification of two genes from Streptomyces argillaceus encoding glycosyltransferases involved in transfer of a disaccharide during biosynthesis of the antitumor drug mithramycin. J. Bacteriol. 1998, 180, 4929–4937. [Google Scholar] [CrossRef] [Green Version]
- Olano, C.; Rodriguez, A.M.; Michel, J.M.; Méndez, C.; Raynal, M.C.; Salas, J.A. Analysis of a Streptomyces antibioticus chromosomal region involved in oleandomycin biosynthesis, which encodes two glycosyltransferases responsible for glycosylation of the macrolactone ring. Mol. Gen. Genet. 1998, 259, 299–308. [Google Scholar] [CrossRef]
- Salah-Bey, K.; Doumith, M.; Michel, J.M.; Haydock, S.; Cortes, J.; Leadlay, P.F.; Raynal, M.C. Targeted gene inactivation for the elucidation of deoxysugar biosynthesis in the erythromycin producer Saccharopolyspora erythraea. Mol. Gen. Genet. 1998, 257, 542–553. [Google Scholar] [CrossRef]
- Westrich, L.; Domann, S.; Faust, B.; Bedford, D.; Hopwood, D.A.; Bechthold, A. Cloning and characterization of a gene cluster from Streptomyces cyanogenus S136 probably involved in landomycin biosynthesis. FEMS Microbiol. Lett. 1999, 170, 381–387. [Google Scholar] [CrossRef]
- Blanco, G.; Fernández, E.; Fernández, M.; Braña, A.F.; Weissbach, U.; Künzel, E.; Rohr, J.; Méndez, C.; Salas, J.A. Characterization of two glycosyltransferases involved in early glycosylation steps during biosynthesis of the antitumor polyketide mithramycin by Streptomyces argillaceus. Mol. Gen. Genet. 2000, 262, 991–1000. [Google Scholar] [CrossRef]
- Trefzer, A.; Hoffmeister, D.; Künzel, E.; Stockert, S.; Weitnauer, G.; Westrich, L.; Rix, U.; Fuchser, J.; Bindseil, K.U.; Bechthold, A.; et al. Function of glycosyltransferase genes involved in urdamycin A biosynthesis. Chem. Biol. 2000, 7, 133–142. [Google Scholar] [CrossRef] [Green Version]
- Wohlert, S.E.; Blanco, G.; Lombó, F.; Fernández, E.; Brana, A.F.; Reich, S.; Udvarnoki, G.; Méndez, C.; Decker, H.; Salas, J.A.; et al. Novel Hybrid Tetracenomycins through Combinatorial Biosynthesis a Glycosyltransferase Encoded by the elm Genes in Cosmid 16F4 and Which Shows a Broad Sugar Substrate Specificity. J. Am. Chem. Soc. 1998, 120, 10596–10601. [Google Scholar] [CrossRef]
- Hoffmeister, D.; Wilkinson, B.; Foster, G.; Sidebottom, P.J.; Ichinose, K.; Bechthold, A. Engineered urdamycin glycosyltransferases are broadened and altered in substrate specificity. Chem. Biol. 2002, 9, 287–295. [Google Scholar] [CrossRef] [Green Version]
- Dürr, C.; Hoffmeister, D.; Wohlert, S.E.; Ichinose, K.; Weber, M.; Von Mulert, U.; Thorson, J.S.; Bechthold, A. The glycosyltransferase UrdGT2 catalyzes both C- and O-glycosidic sugar transfers. Angew. Chem. Int. Ed. Engl. 2004, 43, 2962–2965. [Google Scholar] [CrossRef] [PubMed]
- Salas, A.P.; Zhu, L.; Sánchez, C.; Braña, A.F.; Rohr, J.; Méndez, C.; Salas, J.A. Deciphering the late steps in the biosynthesis of the anti-tumour indolocarbazole staurosporine: Sugar donor substrate flexibility of the StaG glycosyltransferase. Mol. Microbiol. 2005, 58, 17–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Staunton, J.; Weissman, K.J. Polyketide biosynthesis: A millennium review. Nat. Prod. Rep. 2001, 18, 380–416. [Google Scholar] [CrossRef] [PubMed]
- Oliynyk, M.; Samborskyy, M.; Lester, J.B.; Mironenko, T.; Scott, N.; Dickens, S.; Haydock, S.F.; Leadlay, P.F. Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nat. Biotechnol. 2007, 25, 447–453. [Google Scholar] [CrossRef]
- Schmitt-John, T.; Engels, J.W. Promoter constructions for efficient secretion expression in Streptomyces lividans. Appl. Microbiol. Biotechnol. 1992, 36, 493–498. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Rivolta, C.; Soldo, B.; Lazarevic, V.; Joris, B.; Mauël, C.; Karamata, D. A 35.7 kb DNA fragment from the Bacillus subtilis chromosome containing a putative 12.3 kb operon involved in hexuronate catabolism and a perfectly symmetrical hypothetical catabolite-responsive element. Microbiology 1998, 144, 877–884. [Google Scholar] [CrossRef] [Green Version]
- Kunst, F.; Ogasawara, N.; Moszer, I.; Albertini, A.M.; Alloni, G.O.; Azevedo, V.; Bertero, M.G.; Bessières, P.; Bolotin, A.; Borriss, R.; et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 1997, 390, 249–256. [Google Scholar] [CrossRef] [Green Version]
- Quirós, L.M.; Carbajo, R.J.; Braña, A.F.; Salas, J.A. Glycosylation of macrolide antibiotics. Purification and kinetic studies of a macrolide glycosyltransferase from Streptomyces antibioticus. J. Biol. Chem. 2000, 275, 11713–11720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolam, D.N.; Roberts, S.; Proctor, M.R.; Turkenburg, J.P.; Dodson, E.J.; Martinez-Fleites, C.; Yang, M.; Davis, B.G.; Davies, G.J.; Gilbert, H.J. The crystal structure of two macrolide glycosyltransferases provides a blueprint for host cell antibiotic immunity [published correction appears in Proc Natl Acad Sci U S A. 2007 Jun 5;104(23):9911]. Proc. Natl. Acad. Sci. USA 2007, 104, 5336–5341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quirós, L.M.; Carbajo, R.J.; Salas, J.A. Inversion of the anomeric configuration of the transferred sugar during inactivation of the macrolide antibiotic oleandomycin catalyzed by a macrolide glycosyltransferase. FEBS Lett. 2000, 476, 186–189. [Google Scholar] [CrossRef] [Green Version]
- Bate, N.; Butler, A.R.; Smith, I.P.; Cundliffe, E. The mycarose-biosynthetic genes of Streptomyces fradiae, producer of tylosin. Microbiology 2000, 146, 139–146. [Google Scholar] [CrossRef] [Green Version]
- Jenkins, G.; Cundliffe, E. Cloning and characterization of two genes from Streptomyces lividans that confer inducible resistance to lincomycin and macrolide antibiotics. Gene 1991, 108, 55–62. [Google Scholar] [CrossRef]
- Barbe, V.; Cruveiller, S.; Kunst, F.; Lenoble, P.; Meurice, G.; Sekowska, A.; Vallenet, D.; Wang, T.; Moszer, I.; Danchin, A.; et al. From a consortium sequence to a unified sequence: The Bacillus subtilis 168 reference genome a decade later. Microbiology 2009, 155, 1758–1775. [Google Scholar] [CrossRef] [Green Version]
- Sadaie, Y.; Yata, K.; Fujita, M.; Sagai, H.; Itaya, M.; Kasahara, Y.; Ogasawara, N. Nucleotide sequence and analysis of the phoB-rrnE-groESL region of the Bacillus subtilis chromosome. Microbiology 1997, 143, 1861–1866. [Google Scholar] [CrossRef] [Green Version]
- Berghot, M.A.; Hanna, M.A.; Girges, M.M. Synthesis and biological activity of some heterocyclic systems containing anthraquinone. Pharmazie 1992, 47, 340–343. [Google Scholar]
- Huang, H.S.; Chiu, H.F.; Lu, W.C.; Yuan, C.L. Synthesis and antitumor activity of 1,8-diamino-anthraquinone derivatives. Chem. Pharm. Bull. (Tokyo) 2005, 53, 1136–1139. [Google Scholar] [CrossRef] [Green Version]
- Almutairi, M.S.; Hegazy, G.H.; Haiba, M.E.; Ali, H.I.; Khalifa, N.M.; Soliman, A.-M. Synthesis, docking and biological activities of novel hybrids celecoxib and anthraquinone analogs as potent cytotoxic agents. Int. J. Mol. Sci. 2014, 15, 22580–22603. [Google Scholar] [CrossRef] [Green Version]
- Gachon, C.M.; Langlois-Meurinne, M.; Saindrenan, P. Plant secondary metabolism glycosyltransferases: The emerging functional analysis. Trends Plant Sci. 2005, 10, 542–549. [Google Scholar] [CrossRef] [PubMed]
- Cundliffe, E. Glycosylation of macrolide antibiotics in extracts of Streptomyces lividans. Antimicrob. Agents Chemother. 1992, 36, 348–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, K.; Chen, R.; Chen, D.; Li, J.; Wang, R.; Yang, L.; Dai, J. Enzymatic N-Glycosylation of Diverse Arylamine Aglycones by a Promiscuous Glycosyltransferase from Carthamus tinctorius. Adv. Synth. Catal. 2017, 359, 603–608. [Google Scholar] [CrossRef]
- Levanova, N.; Steinemann, M.; Böhmer, K.E.; Schneider, S.; Belyi, Y.; Schlosser, A.; Aktories, K.; Jank, T. Characterization of the glucosyltransferase activity of Legionella pneumophila effector SetA. Naunyn. Schmiedebergs Arch. Pharmacol. 2019, 392, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Endres, R.O.; Lucas, D.O. Lymphocyte surface glycosyltransferases. In Regulatory Mechanisms in Lymphocyte Activation; Lucas, D.O., Ed.; Academic Press: Cambridge, MA, USA, 1977; pp. 423–425. [Google Scholar] [CrossRef]
- Isbell, H.S.; Frush, H.L. Mutarotation, Hydrolysis, and Rearrangement Reactions of Glycosylamines. J. Org. Chem. 1958, 23, 1309–1319. [Google Scholar] [CrossRef]
- Labeda, D.P. Transfer of the Type Strain of Streptomyces erythraeus (Waksman 1923) Waksman and Henrici 1948 to the Genus Saccharopolyspora Lacey and Goodfellow 1975 as Saccharopolyspora erythraea sp. nov., and Designation of a Neotype Strain for Streptomyces erythraeus. Int. J. Syst. Bacteriol. 1987, 37, 19–22. [Google Scholar] [CrossRef] [Green Version]
- Chater, K.F.; Wildem, L.C. Streptomyces albus G mutants defective in the SalGI restriction-modification system. J. Gen. Microbiol. 1980, 116, 323–334. [Google Scholar] [CrossRef] [Green Version]
- Hobbs, G.; Frazer, C.M.; Gardner, D.J.; Cullum, J.A.; Oliver, S.G. Dispersed growth of Streptomyces in liquid culture. Appl. Microbiol. Biotechnol. 2004, 31, 272–277. [Google Scholar] [CrossRef]
- Abbott, J.D.; Graham, J.M. Colicine typing of Shigella sonnei. Mon. Bull. Minist. Health Public Health Lab. Serv. 1961, 20, 51–58. [Google Scholar]
- Herrmann, S.; Siegl, T.; Luzhetska, M.; Petzke, L.; Jilg, C.; Welle, E.; Erb, A.; Leadlay, P.F.; Bechthold, A.; Luzhetskyy, A. Site-specific recombination strategies for engineering actinomycete genomes. Appl. Environ. Microbiol. 2012, 78, 1804–1812. [Google Scholar] [CrossRef] [Green Version]
- Bertani, G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 1951, 62, 293–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lennox, E.S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology 1955, 1, 190–206. [Google Scholar] [CrossRef]
- MacNeil, D.J.; Gewain, K.M.; Ruby, C.L.; Dezeny, G.; Gibbons, P.H.; MacNeil, T. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 1992, 111, 61–68. [Google Scholar] [CrossRef]
- Paget, M.S.; Chamberlin, L.; Atrih, A.; Foster, S.J.; Buttner, M.J. Evidence that the extracytoplasmic function sigma factor sigmaE is required for normal cell wall structure in Streptomyces coelicolor A3(2). J. Bacteriol. 1999, 181, 204–211. [Google Scholar] [CrossRef] [Green Version]
- Strobel, T.; Schmidt, Y.; Linnenbrink, A.; Luzhetskyy, A.; Luzhetska, M.; Taguchi, T.; Brötz, E.; Paululat, T.; Stasevych, M.; Novikov, V.; et al. Tracking down biotransformation to the genetic level: Identification of a highly flexible glycosyltransferase from Saccharothrix espanaensis. Appl. Environ. Microbiol. 2013, 79, 5224–5232. [Google Scholar] [CrossRef] [Green Version]
- Erb, A.; Luzhetskyy, A.; Hardter, U.; Bechthold, A. Cloning and sequencing of the biosynthetic gene cluster for saquayamycin Z and galtamycin B and the elucidation of the assembly of their saccharide chains. Chembiochem 2009, 10, 1392–1401. [Google Scholar] [CrossRef]
- Bierman, M.; Logan, R.; O’Brien, K.; Seno, E.T.; Rao, R.N.; Schoner, B.E. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 1992, 116, 43–49. [Google Scholar] [CrossRef]
- Heitzler, T. Sekundärstoffe aus Aktinomyceten: Untersuchungen zur Biosynthese und Regulation Sowie Verifizierung eines Clustom-Sequencing-Verfahrens. Ph.D. Thesis, Albert-Ludwigs-University Freiburg, Freiburg, Germany, 2015. [Google Scholar]
- Siegl, T.; Tokovenko, B.; Myronovskyi, M.; Luzhetskyy, A. Design, construction and characterisation of a synthetic promoter library for fine-tuned gene expression in actinomycetes. Metab. Eng. 2013, 19, 98–106. [Google Scholar] [CrossRef]
- Dürr, C.; Schnell, H.J.; Luzhetskyy, A.; Murillo, R.; Weber, M.; Welzel, K.; Vente, A.; Bechthold, A. Biosynthesis of the terpene phenalinolactone in Streptomyces sp. Tü6071: Analysis of the gene cluster and generation of derivatives. Chem. Biol. 2006, 13, 365–377. [Google Scholar] [CrossRef] [Green Version]
Sample Availability: Samples of the compounds are available from the authors. |



| Strain or Plasmid | Description 1 | Source or Reference (s) |
|---|---|---|
| Strains | ||
| S. albus J1074 | WT strain, heterologous host | [58] |
| S. albus Gluc | WT expressing the biosynthetic gene for dTDP-d-glucose (oleS) | This study |
| S. albus Gluc x pUWL-A-sace_3599 | S. albus Gluc expressing the gene sace_3599 | This study |
| S. erythraea NRRL2338 | WT strain, biotransformation host | [57] |
| S. erythraea Δsace_3599 | WT with deletion of the gene sace_3599 | This study |
| S. erythraea Δsace_3599 x pTOS(z)-sace_3599 | Deletion mutant of sace_3599 complemented with sace_3599 | This study |
| E. coli Turbo | General cloning host | NEB, Frankfurt am Main, Germany |
| E. coli ET12567 x pUZ8002 | Strain for intergeneric conjugation | [64,65] |
| E. coli Rosetta™ 2 | Heterologous expression host containing seven tRNAs for rarely used codons | Novagen, Darmstadt, Germany |
| Plasmids | ||
| pTOS-Rham | pTOS derivative with oleS, oleE, oleL, and oleU under ermE* promoter | [66] |
| pTOS-Gluc | pTOS derivative with oleS under ermE* promoter | This study |
| pUWL-A [pUWL-oriT-aac(3)IV] | replicative vector for actinomycetes; oriT, bla, aac(3)IV, ermE (pUWL201) | [67] |
| pUWL-A-sace_1884 | pUWL-A derivative with sace_1884 under ermE promoter | This study |
| pUWL-A-sace_3599 | pUWL-A derivative with sace_3599 under ermE promoter | This study |
| pUWL-A-sace_4470 | pUWL-A derivative with sace_4470 under ermE promoter | This study |
| pUWL-Dre | Replicative vector for actinomycetes; oriT, bla, tsr, and dre gene under ermE promoter (pUWL201) | [61] |
| pKC1132 | Replicative vector in E. coli, non-replicative in actinomycetes; lacZa, aac(3)IV, and oriT | [68] |
| pLERE-spec-oriT | Cloning vector with bla, aadA, and oriT flanked by two loxLE sites and two loxRE sites | [66] |
| pKCΔsace_3599 | Vector for deletion of sace_3599, based on pKC1132 | This study |
| pTOS(z) | integrative vector, containing oriT, int, and attP (VWB), aac(3)IV, lacZ gene, and ermEp1* promoter | [69] |
| pTOS(z)-sace_3599 | pTOS(z) derivative with sace_3599 | This study |
| pBluescript II SK(−) | cloning vector for E. coli, with a MCS (SacI to KpnI) flanked by T3 and T7 RNA polymerase promoters; lacZ gene, lac promoter, pUC origin, bla | Agilent Technologies, Santa Clara, CA, USA |
| pET28a(+) | Expression vector with aphII and N-terminal hexahistidine affinity tag | Novagen, Darmstadt, Germany |
| pET28a-sace_3599-N-his6 | pET28a(+) derivative for expression of sace_3599 | This study |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gutacker, F.; Schmidt-Bohli, Y.-I.; Strobel, T.; Qiu, D.; Jessen, H.; Paululat, T.; Bechthold, A. Identification and Characterization of a Novel N- and O-Glycosyltransferase from Saccharopolyspora erythraea. Molecules 2020, 25, 3400. https://doi.org/10.3390/molecules25153400
Gutacker F, Schmidt-Bohli Y-I, Strobel T, Qiu D, Jessen H, Paululat T, Bechthold A. Identification and Characterization of a Novel N- and O-Glycosyltransferase from Saccharopolyspora erythraea. Molecules. 2020; 25(15):3400. https://doi.org/10.3390/molecules25153400
Chicago/Turabian StyleGutacker, Fabienne, Yvonne-Isolde Schmidt-Bohli, Tina Strobel, Danye Qiu, Henning Jessen, Thomas Paululat, and Andreas Bechthold. 2020. "Identification and Characterization of a Novel N- and O-Glycosyltransferase from Saccharopolyspora erythraea" Molecules 25, no. 15: 3400. https://doi.org/10.3390/molecules25153400






